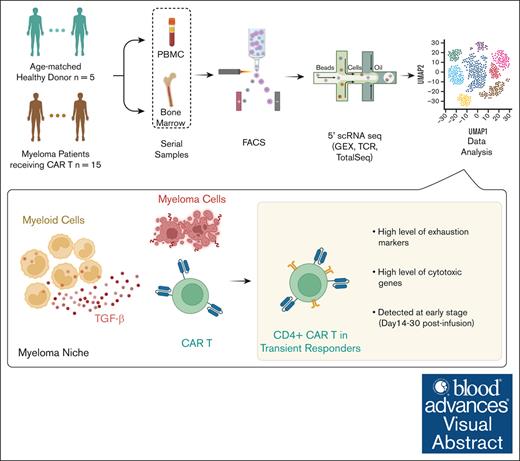
Key Points
Exhausted CD4+ CAR T cells, identified by single-cell assays in the patients’ blood and marrow, is linked to early relapse.
Myeloid cells in the myeloma niche blunt CD4+ CAR T-cell killing via transforming growth factor β.
Visual Abstract
Multiple myeloma is characterized by frequent clinical relapses after conventional therapy. Recently, chimeric antigen receptor (CAR) T cells targeting B-cell maturation antigen (BCMA) has been established as a treatment option for patients with relapsed or refractory disease. However, although >70% of patients initially respond to this treatment, clinical relapse and disease progression occur in most cases. Recent studies showed persistent expression of BCMA at the time of relapse, indicating that immune-intrinsic mechanisms may contribute to this resistance. Although there were no preexisting T-cell features associated with clinical outcomes, we found that patients with a durable response to CAR T-cell treatment had greater persistence of their CAR T cells than patients with transient clinical responses. They also possessed a significantly higher proportion of CD8+ T-effector memory cells. In contrast, patients with short-lived responses to treatment have increased frequencies of cytotoxic CD4+ CAR T cells. These cells expand in vivo early after infusion but express exhaustion markers (hepatitis A virus cellular receptor 2 [HAVCR2] and T-cell immunoglobulin and mucin domain-containing-3 [TIGIT]) and remain polyclonal. Finally, we demonstrate that nonclassical monocytes are enriched in the myeloma niche and may induce CAR T-cell dysfunction through mechanisms that include transforming growth factor β. These findings shed new light on the role of cytotoxic CD4+ T cells in disease progression after CAR T-cell therapy.
Introduction
Multiple myeloma (MM), a plasma cell neoplasm, accounts for 10% of hematologic malignancies in developed countries.1 Despite considerable advances in the last decade, it remains largely incurable, and relapse is considered an inevitable part of the disease course.2,3 B-cell maturation antigen (BCMA) has emerged as a promising therapeutic target in myeloma in the relapse/refractory (R/R) setting.4 BCMA is expressed preferentially by mature B lymphocytes and plasma cells.5 Due to its specificity, BCMA has been an attractive candidate to be targeted by chimeric antigen receptor (CAR) T cells.6-9 These genetically modified T cells express anti-BCMA single-chain variable fragment linked to costimulatory and signaling domains, and upon binding to cell surface BCMA, activation followed by effector function leads to the lysis of myeloma cells.10 Despite the promising results achieved by anti-BCMA CAR T cells in R/R MM in terms of response rates, the absence of durable remissions is common.11 Possible reasons for this phenomenon include CAR T-cell–intrinsic qualities such as expansion capacity and T-cell exhaustion/dysfunction cell state, in addition to CAR T-cell–extrinsic factors such as antigen escape and immune suppression by the hostile bone marrow (BM) microenvironment.12,13 However, previous studies have demonstrated that the immune microenvironment might dictate the response to bispecific T-cell engager antibodies but not to CAR T cells.14 To gain insight into the factors involved in long-term response to anti-BCMA CAR T cells in R/R MM and functional high-risk MM, we performed single-cell sequencing of 690 939 T cells, coupled with T-cell receptor (TCR) clonality assessment, and additional 227 420 non-B non-T immune cells from the myeloma niche, in a longitudinal fashion from 15 patients (78 samples; mean 5 samples per patient), who had received anti-BCMA CAR T-cell therapy; and 5 age-matched control participants. We developed a computational method to identify anti-BCMA CAR T cells in silico and by targeted sequencing and show that, in patients with an early relapse, CD4+ CAR T cells in vivo express cytotoxicity and exhaustion markers and have a polyclonal TCR profile.
Methods
Human patients and participants
A summary of patient characteristics is presented in Table 1. Table 2 provides detailed characteristics by patient.
Patient’s characteristics
| Characteristics . | Total (N = 15) . |
|---|---|
| Age, median (range), y | 66 (34-74) |
| Male, n (%) | 6 (40) |
| ECOG PS 0, n (%) | 9 (60) |
| High-risk cytogenetics∗, n (%) | 8 (53) |
| BM PC ≥50%† | 9 (69) |
| Extramedullary disease, n (%)‡ | 5 (55) |
| Prior antimyeloma regimens, median (range) | 6 (1-12) |
| Penta-refractory§, n (%) | 14 (93) |
| Prior ASCT, n (%) | 14 (93) |
| Bridging therapy, n (%) | 8 (53) |
| Characteristics . | Total (N = 15) . |
|---|---|
| Age, median (range), y | 66 (34-74) |
| Male, n (%) | 6 (40) |
| ECOG PS 0, n (%) | 9 (60) |
| High-risk cytogenetics∗, n (%) | 8 (53) |
| BM PC ≥50%† | 9 (69) |
| Extramedullary disease, n (%)‡ | 5 (55) |
| Prior antimyeloma regimens, median (range) | 6 (1-12) |
| Penta-refractory§, n (%) | 14 (93) |
| Prior ASCT, n (%) | 14 (93) |
| Bridging therapy, n (%) | 8 (53) |
ASCT, autologous stem cell transplantation; ECOG PS, Eastern Cooperative Oncology Group performance status; PC, plasma cells; PET-CT, positron emission tumography-computed tumography.
del17p, t(4:14), t(14:16), 1q abnormalities.
Percentage of 13 patients with adequate BM sample for evaluation.
Percentage of 9 patients with PET-CT scans at baseline.
Exposure or refractoriness to lenalidomide, pomalidomide, bortezomib, carfilzomib, and daratumumab or isatuximab.
Detailed demographics and disease characteristics by patient
| ID . | Age (y) . | Sex . | Prior lines . | ECOG . | MM type . | BMPC (%) . | FISH . | κκLC (mg/L) . | λλLC (mg/L) . | FLC ratio . | M protein (g/dL) . | Bridging . | CAR T-cell product . | CRS grade . | ICANS grade . | Best response by IMWG . | Sustained MRD-neg . | Time to best response (d) . | Time to progression (d) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM1 | 55 | F | 6 | 0 | IgG kappa | na | Hyperploid, 13q- | 139 | 2 | 69.5 | 3.6 | Y | bb2121 | 2 | 0 | sCR MRDneg | N | 231 | 351 |
| MM2 | 66 | F | 12 | 1 | IgG lambda | 50 | Normal | 1 | 318 | 0.03 | 4 | N | bb2121 | 1 | 1 | PR | N | 93 | 232 |
| MM3 | 63 | F | 10 | 0 | IgG kappa | 50 | 1q21 amp | 1228 | 2 | 614 | 1.9 | Y | bb2121 | 1 | 0 | PR | N | 14 | 87 |
| MM4 | 61 | M | 5 | 0 | IgG kappa | 95 | del 17p, 1q21 amp, t(11:14) | 1497 | 2 | 748.5 | 5.2 | N | bb2121 | 2 | 0 | sCR MRDneg | Y | 270 | 335 |
| MM5 | 67 | F | 8 | 0 | IgG kappa | na | t(14:16); 1q21 amo, 13q- | 942 | 2 | 471 | 2.7 | Y | bb21217 | 1 | 1 | PR | N | 28 | 177 |
| MM6 | 71 | F | 1 | 0 | IgG lambda | 5 | Normal | 10.4 | 8 | 0.77 | 0.3 | N | bb2121 | 1 | 0 | sCR MRDneg | Y | 106 | >1000 |
| MM7 | 71 | M | 6 | 1 | IgG kappa | 80 | 1q21 amp, del 17p | 652 | 2 | 326 | 0 | Y | JCARH125 | 1 | 1 | sCR MRDneg | N | 30 | 165 |
| MM8 | 51 | M | 7 | 1 | IgG kappa | 50 | Tetraploidy, t(11:14) | 155 | 2 | 77.5 | 4 | Y | JCARH125 | 2 | 2 | sCR MRDneg | N | 331 | 331 |
| MM9 | 73 | M | 5 | 1 | IgG kappa | 40 | 1q21 amp | 1.5 | 2 | 0.75 | 1.2 | N | JCARH125 | 2 | 0 | PR | N | 21 | 338 |
| MM10 | 69 | F | 5 | 0 | IgG kappa | 10 | 1q21 amp, del 17p | 121 | 2 | 60.5 | 4.3 | Y | JCARH125 | 1 | 1 | VGPR MRDneg | N | 87 | 175 |
| MM11 | 74 | F | 4 | 1 | IgG kappa | 50 | 1q21 amp | 671 | 2.2 | 305 | 1.4 | N | JCARH125 | 0 | 0 | sCR MRDneg | Y | 363 | 1098 |
| MM12 | 52 | F | 7 | 1 | IgG kappa | 5 | 1q21 amp, del 17p | 144 | 2 | 72 | 2 | Y | JCARH125 | 2 | 0 | sCR MRDneg | Y | 221 | >1000 |
| MM13 | 68 | F | 6 | 0 | IgA kappa | 50 | 1q21 amp, del 17p | 717 | 2 | 358.5 | 0 | Y | JCARH125 | 0 | 0 | sCR MRDneg | Y | 27 | 264 |
| MM14 | 34 | M | 6 | 0 | LC kappa | 95 | 1q21 amp, 13q- | 11054 | 2 | 5527 | neg | N | JCARH125 | 2 | 0 | sCR MRDneg | N | 17 | 148 |
| MM15 | 55 | M | 5 | 0 | LC kappa | 70 | t(11:14), del 17q | 1060 | 2 | 530 | neg | N | JCARH125 | 1 | 0 | sCR MRDneg | N | 28 | 125 |
| ID . | Age (y) . | Sex . | Prior lines . | ECOG . | MM type . | BMPC (%) . | FISH . | κκLC (mg/L) . | λλLC (mg/L) . | FLC ratio . | M protein (g/dL) . | Bridging . | CAR T-cell product . | CRS grade . | ICANS grade . | Best response by IMWG . | Sustained MRD-neg . | Time to best response (d) . | Time to progression (d) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM1 | 55 | F | 6 | 0 | IgG kappa | na | Hyperploid, 13q- | 139 | 2 | 69.5 | 3.6 | Y | bb2121 | 2 | 0 | sCR MRDneg | N | 231 | 351 |
| MM2 | 66 | F | 12 | 1 | IgG lambda | 50 | Normal | 1 | 318 | 0.03 | 4 | N | bb2121 | 1 | 1 | PR | N | 93 | 232 |
| MM3 | 63 | F | 10 | 0 | IgG kappa | 50 | 1q21 amp | 1228 | 2 | 614 | 1.9 | Y | bb2121 | 1 | 0 | PR | N | 14 | 87 |
| MM4 | 61 | M | 5 | 0 | IgG kappa | 95 | del 17p, 1q21 amp, t(11:14) | 1497 | 2 | 748.5 | 5.2 | N | bb2121 | 2 | 0 | sCR MRDneg | Y | 270 | 335 |
| MM5 | 67 | F | 8 | 0 | IgG kappa | na | t(14:16); 1q21 amo, 13q- | 942 | 2 | 471 | 2.7 | Y | bb21217 | 1 | 1 | PR | N | 28 | 177 |
| MM6 | 71 | F | 1 | 0 | IgG lambda | 5 | Normal | 10.4 | 8 | 0.77 | 0.3 | N | bb2121 | 1 | 0 | sCR MRDneg | Y | 106 | >1000 |
| MM7 | 71 | M | 6 | 1 | IgG kappa | 80 | 1q21 amp, del 17p | 652 | 2 | 326 | 0 | Y | JCARH125 | 1 | 1 | sCR MRDneg | N | 30 | 165 |
| MM8 | 51 | M | 7 | 1 | IgG kappa | 50 | Tetraploidy, t(11:14) | 155 | 2 | 77.5 | 4 | Y | JCARH125 | 2 | 2 | sCR MRDneg | N | 331 | 331 |
| MM9 | 73 | M | 5 | 1 | IgG kappa | 40 | 1q21 amp | 1.5 | 2 | 0.75 | 1.2 | N | JCARH125 | 2 | 0 | PR | N | 21 | 338 |
| MM10 | 69 | F | 5 | 0 | IgG kappa | 10 | 1q21 amp, del 17p | 121 | 2 | 60.5 | 4.3 | Y | JCARH125 | 1 | 1 | VGPR MRDneg | N | 87 | 175 |
| MM11 | 74 | F | 4 | 1 | IgG kappa | 50 | 1q21 amp | 671 | 2.2 | 305 | 1.4 | N | JCARH125 | 0 | 0 | sCR MRDneg | Y | 363 | 1098 |
| MM12 | 52 | F | 7 | 1 | IgG kappa | 5 | 1q21 amp, del 17p | 144 | 2 | 72 | 2 | Y | JCARH125 | 2 | 0 | sCR MRDneg | Y | 221 | >1000 |
| MM13 | 68 | F | 6 | 0 | IgA kappa | 50 | 1q21 amp, del 17p | 717 | 2 | 358.5 | 0 | Y | JCARH125 | 0 | 0 | sCR MRDneg | Y | 27 | 264 |
| MM14 | 34 | M | 6 | 0 | LC kappa | 95 | 1q21 amp, 13q- | 11054 | 2 | 5527 | neg | N | JCARH125 | 2 | 0 | sCR MRDneg | N | 17 | 148 |
| MM15 | 55 | M | 5 | 0 | LC kappa | 70 | t(11:14), del 17q | 1060 | 2 | 530 | neg | N | JCARH125 | 1 | 0 | sCR MRDneg | N | 28 | 125 |
BMPC, bone marrow plasma cells; CRS, cytokine release syndrome; ECOG, Eastern Cooperative Oncology Group performance status; FISH, fluorescent in situ hybridization; F, female; FLC, serum free light chain; ICANS, immune effector cell-associated neurotoxicity syndrome; IgG, immunoglobulin G; IMWG, International Myeloma Working Group; LC, light chain; M, male; MRD, minimal residual disease; N, no; na, not available; neg, negative; PR, partial response; sCR, stringent complete response; VGPR, very good partial response; Y, yes.
Sustained MRD negativity defined at MRD negativity at D28, D90 and D180.
Processing of human BM and PB samples
BM aspirates were diluted 1:1 in T-cell immunoglobulin and mucin domain-containing-3 (FACS) buffer (phosphate-buffered saline with 0.1% bovine serum albumin and 2 mM EDTA). Peripheral blood (PB) samples were diluted 1:2 in FACS buffer. Peripheral blood mononuclear cell separation was performed by density centrifugation (Ficoll-Pauqe, GE Healthcare) with diluted BM cells (centrifugation for 25 minutes at 500g). Cells were manually aspirated and washed with FACS buffer (5 minutes at 450g). Red blood cells were lysed using RBC lysis buffer (Invitrogen) for 5 minutes at 4°C. BM cells were washed (5 minutes, 300g), resuspended in FACS buffer, and kept on ice.
Cell sorting
Within 1 hour after processing, cells were stained with Total-Seq C antibodies (BioLegend) and flow cytometry antibodies, including biotinylated BCMA protein (Acro Biosystems) followed by streptavidin. Cells were sorted (Aria Fusion, BD) based on forward scatter, side scatter, CD45+ CD19– CD3+ (T cells) and CD45+ CD19– CD3– (non-B non-T cells; supplemental Figure 1).
Single-cell library preparation
Sorted cells were washed, counted, and loaded onto Chromium platform (10x Genomics), with the Single Cell 5ʹ v1 kit, allowing up to 50 000 cells per well. Libraries were sequenced using Illumina NovaSeq 6000.
Single-cell transcriptional analyses
Raw single-cell data were processed by Cell Ranger (version 5, Genome Research Consortium Human Build 38). Doublet detection was performed on the filtered gene expression matrices using DoubletDetection15 with default parameters and then analyzed by Scanpy (Single-Cell Analysis in Python) pipeline,16 as previously described.17 Principle component analysis was performed with top 50 principal components, followed by sample-wise batch correction Combat18 and Harmony,19 Leiden clustering,20 and uniform manifold approximation and projection (UMAP) plotting.21 Annotation of each unbiased population was performed by manual inspection of the top-ranked genes of each cluster. Analysis of cell density was carried out by Scanpy. Analysis of variance with Tukey honestly significant difference test was performed followed by Benjamini-Hochberg adjustment.
For non-B non-T cells, to decrease batch effects, we performed “in silico sorting” for CD14. The expression of CD14 was measured by both frequency distribution density plot and 2-axis plot of CD14 vs CD16. Then a threshold of 0.6 was set to retain only CD14+ cells. Raw expression matrix of CD14+ cells was extracted and processed in the same manner followed by Leiden clustering.
Single-cell TCR analysis
Single-cell RNA sequencing (scRNA-seq) reads were processed by Cell Ranger VDJ. For each cell barcode, the T-cell receptor alpha or T-cell receptor beta clonotype with the most abundant unique molecule identifiers (UMIs) was selected. Clones with either α chain or β chain or paired α:β chains by amino acid sequences were kept for downstream analysis. Expanded clones were defined as >1 cell with the same clonotype. Gini coefficient was calculated using ineq in R.22 Only cells with matched cell barcodes from scRNA-seq reads were used. Comparison of Gini coefficients were carried out by Wilcoxon rank-sum test followed by Benjamini-Hochberg adjustment. When assessing clones longitudinally, only paired pretreatment and posttreatment samples were included.
DGE analysis
Differential gene expression (DGE) analysis was performed using DESeq2 in R.23 For each gene, the raw counts in CD4 CAR T cells (from CD4 T-naïve and CD4 effector memory T cells [Tem] clusters) with low CD8 expression from each sample were aggregated as 1 value to represent this sample. The matrix of gene by sample was used for comparison between durable responders (DRs) and transient responders (TRs). The same was done for CD8 CAR T cells (CD8 GZMB+ Tem, CD8 GZMK+ tissue resident memory T cells, CD8 GZMB+ effector T cells, GNLY+ T, and CD8 T-naïve clusters). Significant genes were selected based on a P value <.05 and fold change >1.5.
DGE analysis was also performed using model-based analysis of single-cell transcriptomics,24 a generalized linear model tailored for scRNA-seq data, whereas the number of genes detected per cell was controlled as a covariate. Multiple test adjustment was performed using p.adjust function in R by controlling false discovery rate (FDR). Significant genes were selected based on an adjusted P value < .05 and fold change >1.5.
Targeted amplification of cDNA libraries for CAR T-cell detection
To annotate CAR T cells, targeted amplification was used on complementary DNA (cDNA) libraries generated from 10x scRNA-seq platform. Lentiviral specific transcripts expressed from bulk RNA sequencing data were identified. Reverse primer (5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTGTTTCTTTCCCCCTGGCCTTAAC3') that is compatible with the 10x Genomics 5′ V1 kit was designed to selectively amplify CAR-specific transcripts from cDNA libraries, while preserving UMI and cell barcodes. A total of 20 ng of cDNA libraries were linearly amplified using reverse primer only, followed by purification using solid-phase reversible immobilization (paramagnetic beads)-select beads at 1.5× (Beckman Coulter), followed by 35 cycles of polymerase chain reaction using target-specific reverse primer and universal forward primer (10x Genomics). We performed sample indexing and attachment of flow cell binding sequences using SI polymerase chain reaction primer and I7 index primers. The next-generation sequencing libraries were pooled and sequenced on MINIseq with 300-cycle kit (Illumina).
CAR T-cell annotation from targeted sequencing data
CAR T cells were identified from targeted amplification sequencing data of the lentiviral GAG gene. The sequence (5'CGAATTTTTTCCCATTTATCTAATTCTCCCCCGCTTAATACCGACGCTCTCGCACCCTAC3') was found on Read 2 (R2), followed by extraction of cell barcodes (1-16nt) and UMI sequence (17-26nt) and calculating the UMI count in each unique cell barcode. The UMI count per cell for GAG– cells was performed in the same manner. Then the UMI per cell distribution was plotted and examined for GAG+ and GAG– cells (supplemental Figure 2). A threshold of UMI per cell of ≥2 was selected to define CAR T cells.
CAR T-cell annotation of fully humanized CAR T cell with modified lentiviral genes
For in silico CAR T-cell identification, we flow-sorted a humanized CAR T-cell sample and created full-length cDNA libraries using SMART-seq.25 We then created a de novo transcriptome assembly using Trinity26 v.2.9.1. From the resulting transcriptome, we identified 1 CAR transcript based on it containing a 3′ viral untranslated region sequence “TGGAAGGGCTAATTCACTCCCAAAGAAGACAAGAT.” We used Kallisto27 v.0.46.0 to align each scRNA-seq read to the de novo transcriptome generated by Trinity. We identified CAR T cells as cells with more than 0 (>0) counts of the associated CAR transcript.
CAR T-cell scores
Activation score was calculated by Scanpy function “scanpy.tl.score_genes” with this gene signature: PDCD1, CTLA4, HAVCR2, LAG3, ENTPD1, TOX, LAYN, CD160, CD244, and TIGIT; whereas cytotoxicity score was calculated in the same way by this gene signature: NKG7, PRF1, GZMB, GZMK, TNF, and IFNG.
Cell interaction analysis
To explore cell-cell communications between nonclassical monocyte population and early stage (day 14 and day 30) CD4+ and CD8+ CAR T cells, CellPhoneDB28 v2 was performed by the function “statistical_analysis” with default parameters. Only significant gene pairs by default threshold from the output “significant_means.csv” were visualized by heat map with Python package Seaborn.29
Enrichment scores for interferon gamma and transforming growth factor β (TGF-β) pathways
Pathway enrichment analysis on differentially expressed genes was performed using gene set enrichment analysis30 4.2.3. The differential expression (DE) genes of CD4+ CAR T cells between DR and TR groups were ranked by “−log(P value) fold change” and analyzed using “G SEAPreranked” tool. Analysis was performed using MSigDB with 1000 permutations. FDR <0.25 was selected as cutoff for statistical significance.
Coculture with restimulation
We generated anti-BCMA CAR T cells as previously described,31 with BCMA-50 single-chain variable fragment sequence,32 CD8 hinge and transmembrane domain, 4-1BB costimulatory domain, CD3 , T2A, and enhanced green fluorescent protein (EGFP). Lentivirus containing the construct was produced, concentrated, and titrated as previously described.31 CD4+ T cells were isolated from apheresis collections of 2 healthy donors using CD4 negative selection kits (STEMCELL Technologies) and were viably frozen in media containing 10% dimethyl sulfoxide. CD4+ T cells were thawed and stimulated for 24 hours using anti-CD3/CD28 Dynabeads (Thermo Fisher) before lentiviral transduction. Seventy-two hours after transduction, CAR T cells were sorted based on EGFP levels. On day 7, CD4+ CAR T cells were cocultured with the target myeloma cell line, MM1.S expressing red fluorescent protein, at an effector-to-target (E:T) ratio of 1:4 with or without 10 ng/mL recombinant human TGF-β (R&D Systems). Fresh media were resupplanted every 3 days. During the coculture, cells were counted, and E:T ratio was assessed by comparing the GFP+ and red fluorescent protein+ populations every 3 days. Fresh target cells were fed when E:T ratio was higher than 1:1. After 3-week coculture, CD4+ CAR T cells were isolated, and the purity of CAR T cells were evaluated by fluorescence microscopy before subsequent assays.
In vitro killing assay
Three technical replicates were performed for each condition and for each donor.
The biobanking study on which patient samples were acquired was approved by the University of California, San Francisco IRB, according to the principles in the Declaration of Helsinki. All patients and healthy donors undergoing hip replacement surgery were provided written informed consent for genomic sequencing and review of electronic medical records (IRB number 10-00545). The patients were enrolled in 4 clinical trials of lentiviral-transduced anti-BCMA CAR T cell containing a 4-1BB costimulatory domain. Patients were enrolled in clinical trials NCT02658929, NCT03274219, NCT03361748, and NCT03430011.
Results
Myeloma BM niche is enriched with GZMB CD8+ T effector cells
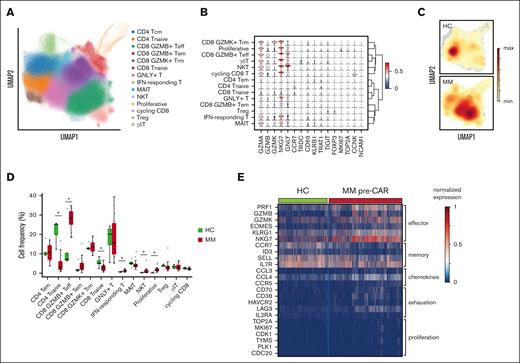
To understand the cell state of T-cell states in patients with myeloma, we collected BM and PB samples at baseline (before lymphodepletion) and longitudinally after infusion (78 samples from 15 patients; median 3 additional time points). Additionally, T cells from 5 participants undergoing hip replacement surgery due to isolated osteoarthritis were collected in the operating room. We performed fresh sorting (within 4 hours from sample collection) of CD45+ CD19– CD3+ lymphocytes (see “Methods”; supplemental Figure 1). Freshly sorted cells were subjected to 10x Genomics microfluidic platform for single-cell gene expression, coupled with TCR and oligo-labeled surface marker antibodies33-35 (CITEseq; see “Methods”). After the removal of doublets and filtering, 690 939 T cells were clustered. Cell state annotation was performed according to canonical markers and revealed 14 different T-cell states36 (Figure 1A). Representative genes of the different cell states include granzymes (cytotoxicity), CCR7 (naïve), FOXP3 (regulatory T cells), and MKI67 and TOP2A (proliferation or cycling; Figure 1B). To compare patients with MM with the participants who underwent hip replacement, we generated density dimensionality reduction map (Figure 1C) and compared the cell fractions of each group within every cluster21 (Figure 1D). Patients with MM were enriched in the GZMB CD8+ T effector cell cluster, whereas participants who underwent hip replacement were enriched in CD4 naïve cluster. Heat map of selected genes shows a preferential expression of exhaustion markers (CD70, CD38, HAVCR2, LAG3, and IL2RA) in T cells derived from patients with MM (Figure 1E).
T-cell states in myeloma and healthy age-matched controls. (A) UMAP of total 690 939 T cells with annotated cell states. (B) Violin plot of normalized expression of selected representative genes for each cell state. (C) Density map of T cells from healthy controls (HCs) and patients with MM. (D) Bar plot showing proportions of each cell state in HC and MM. Significance level was determined using Wilcoxon rank-sum test (P < .001). (E) Heat map of normalized expression of selected differentially expressed genes between HC and MM. IFN, interferon; MAIT, mucosal-associated invariant T cell; NKT, natural killer T cell; Tcm, central memory T cell.
T-cell states in myeloma and healthy age-matched controls. (A) UMAP of total 690 939 T cells with annotated cell states. (B) Violin plot of normalized expression of selected representative genes for each cell state. (C) Density map of T cells from healthy controls (HCs) and patients with MM. (D) Bar plot showing proportions of each cell state in HC and MM. Significance level was determined using Wilcoxon rank-sum test (P < .001). (E) Heat map of normalized expression of selected differentially expressed genes between HC and MM. IFN, interferon; MAIT, mucosal-associated invariant T cell; NKT, natural killer T cell; Tcm, central memory T cell.
CAR T-cell annotation, cell states, and clonality
To understand the specific cell states of CAR T cells, we identified them using a combination of 2 methods (see “Methods”; Figure 2A): In silico generation of a de novo transcriptome from full-length RNA sequencing reads (for CAR T-cell–containing modified lentiviral genes), and a targeted amplification of integrated lentiviral genes (for CAR T-cell–containing unmodified lentiviral genes). A total of 100 157 CAR T cells were identified from all 15 patients across different time points (Figure 2B). Single-cell CAR T-cell annotation correlated with flow cytometry staining for the CAR T cells (supplemental Figure 1). The fraction of CD8+ CAR T cells predominated for up to 30 days after infusion, regardless of the product used (Figure 2C). CD4+ CAR T-naïve cells were enriched in participants who underwent hip replacement (P < .027; P < .00084), whereas CAR T cells and non-CAR T cells from patients with myeloma were enriched for GZMB CD8+ effector T (P = .027; P = .015). There was no statistically significant difference in the distribution of CD8+ effector CAR T cells and CD4+ CAR T cells (naïve or effector memory) for patients dosed with different CAR T-cell products (supplemental Figure 1F). Surprisingly, the Gini index (correlated with TCR clonality) was lower for both CD4+ and CD8+ CAR T cells than non-CAR T cells37,38 (P = .0064 and P < .001, respectively; Figure 2E; supplemental Figure 2). Comparing expanded clones (≥2 cells with the same TCR sequence) in CAR T cells reveals that CD8+ CAR T cells are more clonally expanded than CD4+ CAR T cells (P < .001; Figure 2F).
Annotated CAR T cells from 15 patients with myeloma, their cell states, and TCR clonality. (A) Scheme showing annotation of CAR T cells by de novo transcriptome assembly and targeted amplification. (B) UMAP of 690 939 T cells with projection of annotated CAR T cells (n = 100 157). (C) Fraction of CD4+ and CD8+ CAR T cells over time (mean ± standard error [SE]). (D) Bar plot showing proportions of annotated CAR T-cell states. Significance level was determined using Wilcoxon rank-sum test (P < .001). (E) Gini coefficients (mean ± SE) for non-CAR T cells and CAR T cells, by CD4+ and CD8+, over time. Baseline represents before CAR T-cell infusion; early, day 14 and day 30 after infusion; and late, day ≥90 after infusion. (F) Pie charts of expanded CAR T-cell clonotypes (same TCR clonotype in >1 cell), by CD4+ and CD8+. MAIT, mucosal-associated invariant T cell; NKT, natural killer T cell.
Annotated CAR T cells from 15 patients with myeloma, their cell states, and TCR clonality. (A) Scheme showing annotation of CAR T cells by de novo transcriptome assembly and targeted amplification. (B) UMAP of 690 939 T cells with projection of annotated CAR T cells (n = 100 157). (C) Fraction of CD4+ and CD8+ CAR T cells over time (mean ± standard error [SE]). (D) Bar plot showing proportions of annotated CAR T-cell states. Significance level was determined using Wilcoxon rank-sum test (P < .001). (E) Gini coefficients (mean ± SE) for non-CAR T cells and CAR T cells, by CD4+ and CD8+, over time. Baseline represents before CAR T-cell infusion; early, day 14 and day 30 after infusion; and late, day ≥90 after infusion. (F) Pie charts of expanded CAR T-cell clonotypes (same TCR clonotype in >1 cell), by CD4+ and CD8+. MAIT, mucosal-associated invariant T cell; NKT, natural killer T cell.
Longitudinal analysis of CAR T cells in vivo according to clinical outcome
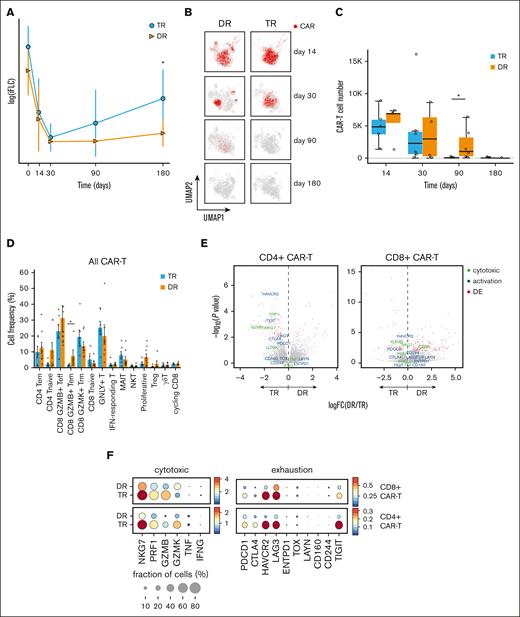
To probe into the differences and similarities between patients with an early relapse compared with those with durable remissions after CAR T-cell infusion, we divided the patients into 2 groups: transient responders (TRs) and durable responders (DRs). Response was assessed by International Myeloma Working Group criteria. To classify patients into DR group, the 6 months mark was used, for which the patient maintained at least a partial response, without rising involved light chains or conversion to minimal residual disease positivity. Patients were generally heavily pretreated, with a median 6 lines of prior antimyeloma therapies (Table 1). Involved serum free light chain levels were increased in patients in the TR group at day 180 (P = .0113; Figure 3A). Overall, CAR T cells were heterogenous, with a representation in all the T-cell clusters (Figure 3B,D). In the TR group, CAR T cells were decreased on days 90 and 180 more than 100-fold, whereas in the DR group, those were decreased by less than onefold on day 90 and more than 100-fold on day 180 (Figure 3C). CAR T cells in the DR group were enriched for CD8+ effector memory cell states (P = .030; Figure 3D). We compared the DGE between DR and TR groups in both CD4+ and CD8+ CAR T-cell compartments23 (see “Methods”). Although in the CD8+ compartment, only HAVCR2 gene (encoding for T-cell immunoglobulin and mucin domain-containing protein 3 [TIM3]) was significantly elevated in the TR group (P = .00011; log2FC = 1.1), in the CD4+ compartment, both HAVCR2 and TIGIT were significantly increased (P < .001; log2FC = 1.8 and P < .001; log2FC = 1.6, respectively), along with cytotoxicity markers NKG7 and PRF1 (P = .0013 and P < .001, respectively) in the TR group (Figures 1F and 3E).
Analysis of transcriptional programs in CAR T cells with correlation to clinical response. (A) Involved serum free light chain (FLC) levels of patients with myeloma over time after CAR T-cell therapy (day 0 = CAR T-cell infusion), by group. Statistical significance is marked by asterisk and determined by Wilcoxon rank-sum test (P = .0113). (B) UMAP of T cells, with projection of annotated CAR T cells from TR and DR groups at different time points (days) after infusion. (C) Bar plot showing the sum of CAR T cells from 15 patients, by group, at different time points (days) after infusion. (D) Bar plots showing proportions of CAR T cells (mean ± SE) at different cell states, by group. Statistical significance is marked by asterisk and determined by Wilcoxon rank-sum test (P = .030) (E) Volcano plots showing differentially expressed genes between patient groups (DR vs TR) in CD4+ CAR T cells and CD8+ CAR T cells. Annotated genes in red have a statistically significant (P < .05) fold change (FC) >1.5. (F) Dot plots showing normalized expression of cytotoxicity and exhaustion genes in CD4+ and CD8+ CAR T cells, by group. Dot size is proportional to fraction of cells. MAIT, mucosa-associated invariant T cell; NKT, natural killer T cell.
Analysis of transcriptional programs in CAR T cells with correlation to clinical response. (A) Involved serum free light chain (FLC) levels of patients with myeloma over time after CAR T-cell therapy (day 0 = CAR T-cell infusion), by group. Statistical significance is marked by asterisk and determined by Wilcoxon rank-sum test (P = .0113). (B) UMAP of T cells, with projection of annotated CAR T cells from TR and DR groups at different time points (days) after infusion. (C) Bar plot showing the sum of CAR T cells from 15 patients, by group, at different time points (days) after infusion. (D) Bar plots showing proportions of CAR T cells (mean ± SE) at different cell states, by group. Statistical significance is marked by asterisk and determined by Wilcoxon rank-sum test (P = .030) (E) Volcano plots showing differentially expressed genes between patient groups (DR vs TR) in CD4+ CAR T cells and CD8+ CAR T cells. Annotated genes in red have a statistically significant (P < .05) fold change (FC) >1.5. (F) Dot plots showing normalized expression of cytotoxicity and exhaustion genes in CD4+ and CD8+ CAR T cells, by group. Dot size is proportional to fraction of cells. MAIT, mucosa-associated invariant T cell; NKT, natural killer T cell.
Exhausted cytotoxic CD4+ CAR T cells are associated with transient, short-lived clinical response
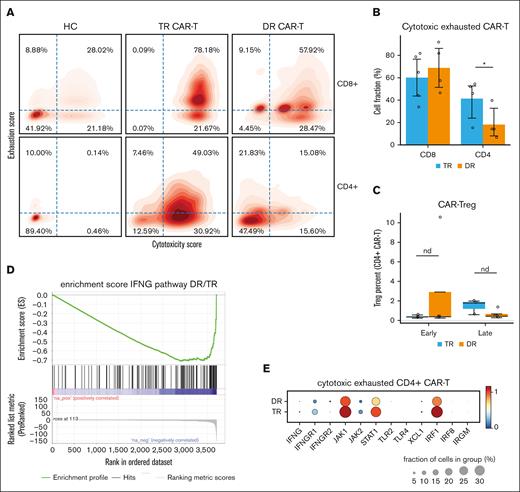
To elucidate the nature of cytotoxic and exhaustion genes of the CAR T cells, we calculated the cytotoxicity score and exhaustion score for every annotated CAR T cell (see “Methods”). Plotting single CAR T-cell scores in density maps for the 2 groups of patients reveals that in TR patients early after infusion (14-30 days), there was an enrichment of CD4+ cytotoxic exhausted cells compared with that of DR patients (Wilcoxon rank-sum test, P = .0303), unlike the CD8+ compartment (Wilcoxon rank-sum test, P = .196; Figure 4A-B). There was no difference in the exhaustion or cytotoxicity profile of non-CAR resident CD4+ (P = .392) or CD8+ T cells (P = .26; supplemental Figure 2).
Cytotoxic and exhausted CD4+ CAR T cells harboring interferon gamma signature are identified preferentially in patients with transient response to therapy. (A) Density map of cytotoxicity and exhaustion gene signature scores for CD4+ and CD8+ CAR T cells, by group. Cell fractions (%) are annotated in each quadrant. (B) Bar plot showing CD4+ CAR T-cell fraction (mean % ± SE) of “double positive” cytotoxic and exhausted cells derived from the top right quadrant in panel B, by group. Statistical significance is marked by asterisk and determined by Wilcoxon rank-sum test (P = .03). (C) Box plot showing CAR-Treg fraction (median % ± 1.5 interquartile range), by group. (D) Enrichment score (see “Methods”) for interferon gamma (IFNG) pathway of CD4+ CAR T cells from DR group vs TR group. (E) Dot plots showing normalized expression of genes related to interferon (IFNG) pathway in CD4+ CAR T cells, by group. Dot size is proportional to fraction of cells.
Cytotoxic and exhausted CD4+ CAR T cells harboring interferon gamma signature are identified preferentially in patients with transient response to therapy. (A) Density map of cytotoxicity and exhaustion gene signature scores for CD4+ and CD8+ CAR T cells, by group. Cell fractions (%) are annotated in each quadrant. (B) Bar plot showing CD4+ CAR T-cell fraction (mean % ± SE) of “double positive” cytotoxic and exhausted cells derived from the top right quadrant in panel B, by group. Statistical significance is marked by asterisk and determined by Wilcoxon rank-sum test (P = .03). (C) Box plot showing CAR-Treg fraction (median % ± 1.5 interquartile range), by group. (D) Enrichment score (see “Methods”) for interferon gamma (IFNG) pathway of CD4+ CAR T cells from DR group vs TR group. (E) Dot plots showing normalized expression of genes related to interferon (IFNG) pathway in CD4+ CAR T cells, by group. Dot size is proportional to fraction of cells.
CAR T-regulatory cells (CAR-Tregs) in the infusion products were recently reported to be associated with relapse of large B-cell lymphoma after anti-CD19 CAR T-cell therapy.39,40 Because CAR-Tregs are CD4+, we wondered whether the association between CD4+ CAR T cells and early relapse in our myeloma cohort could be attributed to CAR-Tregs. We found that CAR-Treg proportion was not increased in the TR group (Figure 4C). We further identified the exhausted cytotoxic CD4+ CAR T cells as either naïve or effector memory cells, enriched in IFNG signature (normalized enrichment score, –1.33; P < .001; FDR = 0.082; Figure 4D). DGE analysis confirmed the enrichment of specific IFNG pathway genes (IFNGR1, IFNGR2, JAK1, STAT1, and IRF1) in the TR group (Figure 4E).
TGF-β contributes to CD4+ CAR T-cell exhaustion
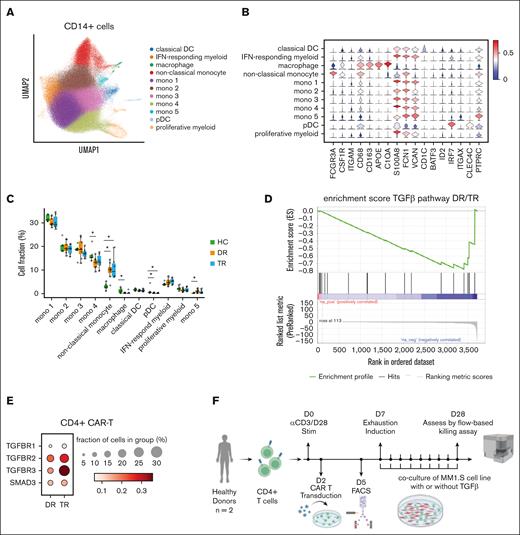
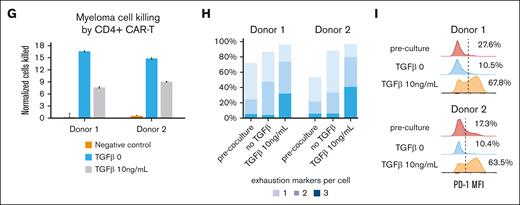
To understand whether other immune cells in the myeloma niche affect CAR T-cell state or their ability to kill myeloma cells, we performed fresh sorting of CD45+ CD19– CD3– cells from the BM of participants who underwent hip replacement and patients with myeloma undergoing CAR T-cell therapy. We performed in silico sorting of CD14+ cells (supplemental Figure 2) and clustered the cells after filtering and batch correction (see “Methods”). CD14+ cells in the BM comprised 11 clusters, including classical and plasmacytoid dendritic cells, CD163+ macrophages, 5 different monocyte subtypes expressing VCAN, FCN1, and S100A8, along with CD68+ CD16+ nonclassical monocytes (Figure 5A-B). Nonclassical monocytes were enriched in patients with myeloma receiving CAR T-cell therapy compared with participants who underwent hip replacement (P = .0052 for DR; P = .0041 for TR; Figure 5C). There was no difference in the frequency of CD14+ cell clusters between the TR and DR groups of patients. Because nonclassical monocytes were preferentially present in the BM samples of patients with myeloma, we simulated their interaction with annotated CAR T cells28 (see “Methods”). Interaction maps of CD4+ CAR T cells with nonclassical monocytes in the TR and DR groups reveal a possible role for the TGF-β axis, with interaction of TGFB1 in MM–associated nonclassical monocytes with TGFBR3 in CD4+ CAR T cells (supplemental Figure 2). We then computed the enrichment score of TGF-β pathway in CD4+ CAR T cells and found that the CD4+ CAR T cells from the TR group were enriched for TGF-β–related genes (normalized enrichment score, –1.39; P = .01; FDR = 0.055; Figure 5D). Additionally, TGFBR2 and TGFBR3 were preferentially expressed at the single-cell level in CD4+ CAR T cells in the TR group (Figure 5E). To assess the role of TGF-β in tumor cell killing of CD4+ CAR T cells, we performed a killing assay with home grown anti-BCMA CAR T cells (4-1BB CD3ζ) from 2 healthy donors, after CD4 selection (see “Methods”). We used a repetitive stimulation assay with a myeloma cell line to induce CAR T-cell exhaustion in vitro (Figure 5F; see “Methods”). We found a significantly decreased cytotoxicity of the CD4+ CAR T cells compared with the control after 3-week coculture (mean decrease in killing capacity for donor 1 and donor 2 was 54% and 39%, respectively; P < .001, Student t test; Figure 5G). TGF-β–treated CAR T cells expressed multiple exhaustion markers (PD1, TIM3, and CD39) compared with control at the end of the restimulation (mean fraction of cells, 36.3% vs 4.7%; P = .016, Student t test; Figure 5H). The CAR T cells expressing exhaustion markers were functionally exhausted, with hampered secretion of key cytokines interferon gamma and tumor necrosis factor α (supplemental Figure 2J). In addition, we find that TGF-β significantly reduces proliferation (assessed by Ki-67 flow cytometry staining) at 1 week of coculture and persists at 3 weeks after coculture (supplemental Figure 2K).
Interaction of nonclassical monocyes in the myeloma niche with CD4+ CAR T cells is linked to TGFb, blunting their killing capacity. (A) UMAP of CD14+ myeloid cells with annotated cell states. (B) Violin plots showing normalized expression of selected genes for each cell state. (C) Box plot showing the proportions of CD14+ myeloid cells (median % ± 1.5 interquartile range) of each cell state, by group. Statistical significance is marked by asterisk and determined by Wilcoxon rank-sum test (P < .001). (D) Enrichment score (see "Methods”) for TGF-β pathway of CD4+ CAR T cells from DR group vs TR group. (E) Dot plots showing normalized expression of genes related to TGF-β pathway in CD4+ CAR T cells, by group. Dot size is proportional to fraction of cells. (F) Scheme depicting experimental design of CD4+ CAR T cells in vitro killing assay performed by coculture with myeloma cell line MM1.S, with or without TGF-β. (G) Bar plot showing killing efficacy of CD4+ CAR T cells (number of cancer cells killed per CD4+ CAR T cell) after 3 weeks of coculture with myeloma cell line MM1.S, with or without TGF-β. (H) Frequency of CD4+ CAR T cells expressing 3 exhaustion markers (programmed cell death protein, TIM3, and CD39) after repetitive stimulation. (I) Flow cytometry histogram of programmed cell death protein mean fluorescence intensity (MFI) of CD4+ CAR T cells from 2 donors after 3 weeks of coculture with myeloma cell line MM1.S, with or without TGF-β. Mono, monocyte; pDC, plasmacytoid dendritic cell.
Interaction of nonclassical monocyes in the myeloma niche with CD4+ CAR T cells is linked to TGFb, blunting their killing capacity. (A) UMAP of CD14+ myeloid cells with annotated cell states. (B) Violin plots showing normalized expression of selected genes for each cell state. (C) Box plot showing the proportions of CD14+ myeloid cells (median % ± 1.5 interquartile range) of each cell state, by group. Statistical significance is marked by asterisk and determined by Wilcoxon rank-sum test (P < .001). (D) Enrichment score (see "Methods”) for TGF-β pathway of CD4+ CAR T cells from DR group vs TR group. (E) Dot plots showing normalized expression of genes related to TGF-β pathway in CD4+ CAR T cells, by group. Dot size is proportional to fraction of cells. (F) Scheme depicting experimental design of CD4+ CAR T cells in vitro killing assay performed by coculture with myeloma cell line MM1.S, with or without TGF-β. (G) Bar plot showing killing efficacy of CD4+ CAR T cells (number of cancer cells killed per CD4+ CAR T cell) after 3 weeks of coculture with myeloma cell line MM1.S, with or without TGF-β. (H) Frequency of CD4+ CAR T cells expressing 3 exhaustion markers (programmed cell death protein, TIM3, and CD39) after repetitive stimulation. (I) Flow cytometry histogram of programmed cell death protein mean fluorescence intensity (MFI) of CD4+ CAR T cells from 2 donors after 3 weeks of coculture with myeloma cell line MM1.S, with or without TGF-β. Mono, monocyte; pDC, plasmacytoid dendritic cell.
Discussion
This study is, to our knowledge, the largest single-cell analysis of anti-BCMA CAR T cells in vivo, with >1.1 million single cells analyzed from 15 patients and 5 healthy donors. Rather than focusing on the infusion products, we assayed the CAR T cells and their microenvironment in the myeloma niche. Our results suggest that specific subtypes of CAR T-cell states early after infusion contribute to the variability in response to anti-BCMA CAR T-cell therapy in patients with R/R and functional high-risk MM. Unlike previous studies focusing on single-cell analysis of CD8+ CAR T cells,41,42 this extensive database of freshly sorted CAR T cells and immune niche cells sheds light on the role of cytotoxic-like CD4+ CAR T cells. Cytotoxic CD4+ tumor infiltration lymphocytes (TILs) have been described in the setting of solid tumors, interacting with other microenvironment cells.43 In the setting of CAR T cells, increased CD4:CD8 ratio in the infusion product is associated with better expansion and clinical outcomes.7 Surprisingly, we have found that across 3 different products, CD4+ CAR T cells expressing cytotoxic and activation markers were associated with a worse clinical outcome and early relapse. Unlike previous cases described for anti-CD19 CAR T cell in patients with chronic lymphocytic leukemia, our single-cell TCR analysis early after infusion suggests that anti-BCMA CD4+ CAR T cells are polyclonal.44 In vivo tracking of CAR T cells reveals that, at days 90 and beyond after infusion, there is no persistence in patients who have had transient response, compared with those with durable responses. The association between early expansion of exhausted cytotoxic and polyclonal CD4+ CAR T cells and transient response or early relapse requires prospective validation. However, our study introduces the importance of cell states in vivo as possible predictors of outcome after CAR T-cell therapy, which constitutes a clinical unmet need. In this context, the relevant exhaustion coexpressed markers are TIM3 and TIGIT (rather than other canonical markers of exhaustion such as programmed cell death protein or cytotoxic T-lymphocyte-associated antigen 4), which might inform future combination therapeutic approaches.
Our analysis of immune cells in the BM uncovers nonclassical monocytes, which are preferentially present in the myeloma niche compared with that of age-matched controls. Single-cell interaction analysis points to a possible role of the TGF-β axis in the interaction of CD4+ CAR T cells with these nonclassical monocytes. In vitro modeling of CAR T-cell exhaustion also demonstrates that TGF-β represents a T-cell–extrinsic mechanism contributing to T-cell exhaustion beyond repetitive antigen stimulation. This finding further corroborates the negative impact of TGF-β on immunotherapy in the tumor microenvironment and makes the case for future research of TGF-β–negating armoring strategies.45-47
Strengths of this study include (1) large data set comprising 1.1 million single cells from patients who received transfusion of anti-BCMA CAR T cells containing 4-1BB costimulatory domain; and (2) fresh tissue sampling with immediate processing and sorting to minimize stress-induced activation, in conjunction with clonal assessment and cell surface marker analysis. A limitation in our study is the inability to analyze the cell states of the infusion product and compare those with the ones found in vivo. However, because patient-to-patient variability can affect the expansion of CAR T cells and their cell states, the in vivo analysis early after infusion might be a predictor of long-term outcomes.
Acknowledgments
The authors thank the patients and their families, the Parnassus Flow Cytometry CoLab, and the Human Genetics CoLab. The authors thank Ajai Chari for critical review of the manuscript.
This work was supported by the Parker Institute for Cancer Immunotherapy and the National Institutes of Health ([NIH], National Cancer Institute grant R35CA253175). The authors also acknowledge support from the Parnassus Flow Cytometry CoLab (RRID:SCR_018206), which was supported in part by NIH grant P30 DK063720 and NIH S10 Instrumentation grant S10 1S10OD021822-01.
Authorship
Contribution: G.L. conceived the study, designed and performed experiments, analyzed data, and wrote the manuscript; Z.F. analyzed single-cell RNA sequencing (scRNA-seq) data and wrote the manuscript; K.W. designed and performed experiments of targeted amplification and flow cytometry killing assays, analyzed data, and wrote the manuscript; E.M. analyzed scRNA-seq data; A.H.-W. designed and performed CAR T-cell transduction and killing assays; A.S. performed killing assays and flow cytometry experiments; H.C. performed scRNA-seq library preparation and flow cytometry experiments; M.B. analyzed scRNA-seq data; S.K. and A.C. designed flow cytometry panels; S.B., J.W., S.W., and N.S. recruited patients and provided deidentified clinical data; E.H. performed bone marrow collection from healthy donors undergoing surgery; K.T.R. designed CAR T-cell killing assays; C.J.Y. designed experiments and provided guidance on data analysis; and T.M. and L.F. conceived the study, designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: G.L. is employed by and holds stock in Caribou Biosciences. S.W. is employed by and holds stock in Bristol Myers Squibb (BMS). N.S. is employed by and owns stock in AstraZeneca; served as a consultant for Amgen, CareDx, CSL Behring, GlaxoSmithKline (GSK), Indapta Therapeutics, Karyopharm Therapeutics, Kite, Oncopeptides, and Sanofi; and received research funding from bluebird bio, BMS/Celgene, Janssen, Nektar, Poseida, Precision BioSciences, Sutro Biopharma, and TeneoBio. K.T.R. is a cofounder, consultant, scientific advisory board member, and stockholder of Arsenal Biosciences, Dispatch Therapeutics, and Moonlight Bio; is a consultant and stockholder in Cell Design Labs (now a Gilead company); holds stock in Gilead; and is an adviser to Venrock. C.J.Y. is a cofounder of Survey Genomics; a scientific advisory board member of Related Sciences and Immunai; a consultant for Maze Therapeutics, TRex Bio, ImYoo, and Santa Ana Bio; and received research support from the Chan Zuckerberg Initiative, Chan Zuckerberg Biohub, Genentech, BioLegend, ScaleBio, and Illumina. T.M. received research funding from Sanofi, BMS, Janssen, and Amgen, and is a consultant for GSK and Pfizer. L.F. received research funding from AbbVie, Bavarian Nordic, BMS, Dendreon, Janssen, Merck, Nektar, Roche/Genentech, and Parker Institute; served as a consultant to AbbVie, Actym Therapeutics, Amgen, AstraZeneca, Atreca, BioAtla, Bolt, BMS, Crescendo, Daiichi Sankyo, ImmunoGenesis, Innovent, Merck, NGM Bio, Nutcracker, RAPT, Senti, Sutro, and Roche/Genentech; and has ownership interests in Actym Therapeutics, Atreca, BioAtla, Bolt, ImmunoGenesis, Nutcracker, RAPT, and Senti, unrelated to the submitted work. The remaining authors declare no competing financial interests.
Correspondence: Lawrence Fong, Fred Hutchinson Cancer Center, 1100 Fairview Ave, Seattle, WA 98109; email: lawrence.fong@fredhutch.org.
References
Author notes
G.L., Z.F., K.W., T.M., and L.F. contributed equally to this study.
Single-cell transcriptomic data are available in Gene Expression Omnibus database (accession number GSE246342).
Flow cytometry data or additional unidentified patient data will be shared upon request from the corresponding author, Lawrence Fong (lawrence.fong@fredhutch.org).
The full-text version of this article contains a data supplement.

![Annotated CAR T cells from 15 patients with myeloma, their cell states, and TCR clonality. (A) Scheme showing annotation of CAR T cells by de novo transcriptome assembly and targeted amplification. (B) UMAP of 690 939 T cells with projection of annotated CAR T cells (n = 100 157). (C) Fraction of CD4+ and CD8+ CAR T cells over time (mean ± standard error [SE]). (D) Bar plot showing proportions of annotated CAR T-cell states. Significance level was determined using Wilcoxon rank-sum test (P < .001). (E) Gini coefficients (mean ± SE) for non-CAR T cells and CAR T cells, by CD4+ and CD8+, over time. Baseline represents before CAR T-cell infusion; early, day 14 and day 30 after infusion; and late, day ≥90 after infusion. (F) Pie charts of expanded CAR T-cell clonotypes (same TCR clonotype in >1 cell), by CD4+ and CD8+. MAIT, mucosal-associated invariant T cell; NKT, natural killer T cell.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/13/10.1182_bloodadvances.2023012416/3/m_blooda_adv-2023-012416-gr2.jpeg?Expires=1769085995&Signature=0f60TOZJ0rdbXBkVBq~cMWMCvm9sxXfXjmHAcpDBMz8llnzsXvhy-~N-ShMM6~GLvvSZ5xMdfa23-piXq1g-GYhuEGjZzCiprtjOyrZ1fzkmEHLy2Bo-T89XdZXDcVedwyHn1BugjOdGKgKCb7QR~am2R-8cAjG4qMquDOYHSJ61CGCRjdnuGGgBn31f0EivIr-5EZ8CNIG4Vgixj5ragG~mGD4yP01WaMCrLC25hMJBZCYMb9SctSKGo4AFtd9kdPgZQ~7yxQBFEXHULx9DxJKhDYBRuVofe6QUIwmQvFcgskJ4uh3Xe6ROeqa5n0Ns97RVzdMfwPTZ9TJonR4FYw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)