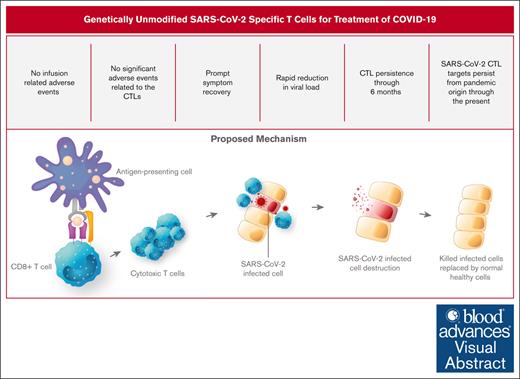
Key Points
SARS-CoV-2–specific CTLs were well tolerated in all 4 doses tested in high-risk ambulatory adults.
≥88% viral elimination in 92% of patients by day +4 and >99% viral elimination in everyone by day +14 on nasal swab testing.
Visual Abstract
Cytotoxic T lymphocytes (CTLs) destroy virally infected cells and are critical for the elimination of viral infections such as those caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Delayed and dysfunctional adaptive immune responses to SARS-CoV-2 are associated with poor outcomes. Treatment with allogeneic SARS-CoV-2–specific CTLs may enhance cellular immunity in high-risk patients providing a safe, direct mechanism of treatment. Thirty high-risk ambulatory patients with COVID-19 were enrolled in a phase 1 trial assessing the safety of third party, SARS-CoV-2–specific CTLs. Twelve interventional patients, 6 of whom were immunocompromised, matched the HLA-A∗02:01 restriction of the CTLs and received a single infusion of 1 of 4 escalating doses of a product containing 68.5% SARS-CoV-2–specific CD8+ CTLs/total cells. Symptom improvement and resolution in these patients was compared with an observational group of 18 patients lacking HLA-A∗02:01 who could receive standard of care. No dose-limiting toxicities were observed at any dosing level. Nasal swab polymerase chain reaction testing showed ≥88% and >99% viral elimination from baseline in all patients at 4 and 14 days after infusion, respectively. The CTLs did not interfere with the development of endogenous anti–SARS-CoV-2 humoral or cellular responses. T-cell receptor β analysis showed persistence of donor-derived SARS-CoV-2-specific CTLs through the end of the 6-month follow-up period. Interventional patients consistently reported symptomatic improvement 2 to 3 days after infusion, whereas improvement was more variable in observational patients. SARS-CoV-2–specific CTLs are a potentially feasible cellular therapy for COVID-19 illness. This trial was registered at www.clinicaltrials.gov as #NCT04765449.
Introduction
Early in the pandemic, individuals infected with COVID-19, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), who were older or had certain comorbidities, were identified as being at higher risk for more serious courses.1 Dysregulation of the angiotensin-converting enzyme 2 and the renin-angiotensin-aldosterone pathways by the virus has been linked to exacerbation of diabetic,2 pulmonary,3 cardiovascular,4 and renal disease5 as well as other comorbid conditions,6 which can, in turn, worsen the infectious outcome. Additional contributors to poor outcomes include obesity, a proinflammatory state and key dysregulator of immune function,7,8 as well as cancer and autoimmune diseases due to multiple factors including disease-related immune dysfunction,9 the immunosuppressant effects of therapy for these diseases,10,11 coexisting medical conditions,10 and delays in planned treatments due to SARS-CoV-2 infection.12 In older patients, immune dysregulation,13 predisposition to inflammation,14 and infirmity15 increase COVID-19 mortality. In the postvaccine era, although hospitalization and mortality have significantly decreased for most individuals, there is a continued need for novel therapies for specific groups. Comorbid conditions and advanced age remain associated with higher mortality due to COVID-19,16 and in subsets of immune-compromised patients, SARS-CoV-2 infection is associated with prolonged time to viral clearance and resistance to treatment,17 as well as increased rates of severe disease18 and poor outcomes.19
SARS-CoV-2 interferes with the kinetics of type I interferon induction, disrupting the complex interplay between the innate and adaptive immune systems necessary to effectively contain the virus. Interruptions in this pathway result in delayed signaling of the adaptive immune system,20 resulting in increased viral progression21 and deleterious inflammation.22 CD8+ cytotoxic T lymphocytes (CTLs) constitute a critical arm of the adaptive immune system, with the primary function of clearing viral pathogens through the elimination of virally infected cells. Rapid activation of bystander CD8+ T cells23 and early24 and robust25 COVID-19–specific CTL responses are associated with milder courses of the infection, whereas T-cell lymphopenia26 and T-cell exhaustion27 are hallmarks of more serious illness. These findings highlight the critical role that rapid and coordinated mobilization of the adaptive immune system, particularly CTLs, play in controlling COVID-19. We conducted a phase 1 study to determine the maximum tolerated dose of off-the-shelf genetically unmodified COVID-19–specific CTL therapy (CTLs), with the goal of rapidly providing cellular immunity to high-risk ambulatory patients with a newly diagnosed SARS-CoV-2 infection.
Methods
Study objectives, design, and patients
The primary objective of this phase 1 single-institution study, performed at Thomas Jefferson University, was to identify the maximum tolerated dose of an HLA-A∗02:01–restricted SARS-CoV-2–specific CTL product using a traditional phase one 3 + 3 study design.28 Additional objectives were to assess the pace of COVID-19 resolution by nasal polymerase chain reaction (PCR) testing, to determine whether the CTLs interfered with endogenous humoral and cellular immune responses to the virus, to identify the duration of CTL persistence after infusion, and to test for patient alloimmunization to the CTL donor.
Adult ambulatory patients with newly diagnosed COVID-19 and at least 1 of the Centers for Disease Control and Prevention’s high-risk COVID-19 features29 were eligible to participate. Patients had to be clinically stable without virus-induced hypoxia or evidence of COVID-19–related cytokine release syndrome (CRS). Patients were required to match the CTL donor at HLA-A∗02:01 only. To avoid inadvertent third-party engraftment, patients with significant pancytopenia or who matched the CTL donor at ≥5 of 6 HLA class I alleles were excluded. Full eligibility criteria are listed in supplemental Table 1.
Alternative treatments for COVID-19, such as steroids and monoclonal antibodies, were not permitted in patients receiving the CTLs, although remdesivir was allowed per protocol. In the event of COVID-19 progression, patients would be taken off the study and treated per institutional guidelines.
Study procedures
Upon enrollment, patients underwent rapid HLA typing. Patients possessing an HLA-A∗02:01 allele (interventional group) were to be treated with the CTLs within 96 hours of initial COVID-19 diagnosis based on home or PCR-based nasal swab testing. All home tests were confirmed by hospital-laboratory PCR analysis. Patients were admitted to the hospital and treated with a single infusion of 1 of 4 escalating doses of CTLs at 1 × 105/kg, 3 × 105/kg, 1 × 106/kg, or 3 × 106/kg of adjusted body weight (ideal weight plus 40% the difference between ideal and actual body weight). Day 0 was the day of CTL infusion, and patients were monitored in the hospital for 4 days after infusion (days +1 to +4). After discharge, interim histories were obtained daily by phone or in person through day +14. After infusion, nasal swab specimens for viral load by PCR were obtained twice weekly through day +14 or earlier if negative. HLA antibody screens to assess for alloimmunization were collected on or after day +28. Interventional patients were assessed in person at day +28 and 2, 3, and 6 months after CTL infusion. Studies for SARS-CoV-2–specific humoral and cellular responses were obtained at those times.
Enrolled patients not possessing an HLA-A∗02:01 allele (observational group) were followed for interim history and outcomes. Interim histories (but no laboratory testing after initial eligibility studies) were collected using the same symptom list at the same time points as treated patients to compare outcomes between the 2 groups. This observational group could receive any type of treatment for COVID-19 as prescribed by their medical caregivers. The observational group patients were assigned a “day 0” based on when the HLA typing was resulted (day of consent or day after consent in all patients). Day 0 assignment had to be within 96 hours of the initial COVID-19 diagnosis based on home or PCR-based nasal swab testing. All home tests were confirmed by hospital-laboratory PCR analysis.
Assessment of which day patients first felt definitely improved (most reported symptoms better and/or patients states feeling better) and which day the patients felt all COVID-19–related symptoms resolved (performance status at or near 100% with no or minimal symptoms) was performed independently by 2 different study team members. Symptoms assessed were cough, presence and degree of shortness of breath, fever, chills, aches, sore throat, congestion/runny nose, headache, loss of taste or smell, nausea, vomiting, diarrhea, fatigue, and performance status. Observational patients were accrued until the treatment enrollment was complete. The follow-up period for patients in both groups was 6 months.
CTL manufacturing and administration
The CTLs were generated toward 7 HLA-A∗02:01 restricted SARS-CoV-2 peptides (from spike, nucleocapsid, nonstructural proteins 3, 7, and 8, and 2 from open reading frame [ORF] 3a proteins) using 1 apheresis product from 1 healthy donor who had COVID-19 illness ∼1 year earlier. The peptides were selected using data from the Wuhan strain of SARS-CoV-2, and the HLA-A∗02:01 restriction was chosen because it is the most common HLA allele worldwide. Manufacturing information and characteristics of the final product are shown in Table 1. After manufacture, CTLs were cryopreserved for off-the-shelf use. The CTLs were infused IV within 10 minutes of thawing. Patients were premedicated with acetaminophen and diphenhydramine as infusion reaction prophylaxis.
SARS-CoV-2 CTL manufacture and final characteristics
| CD3+ cells . | CD3+CD8+ cells . | CD3+CD4+ cells . | Tetramer positive out of CD3+CD8+ cells . | Tetramer positive out of total cells . | Cytotoxicity∗ . | Naïve T-cell content . | Monocyte content . | Natural killer cell content . | B-cell content . |
|---|---|---|---|---|---|---|---|---|---|
| 96.5% | 87.6% | 4% | 76.7% | 68.5% | 82% | 0% | 0.2% | 0.7% | 0.2% |
| CD3+ cells . | CD3+CD8+ cells . | CD3+CD4+ cells . | Tetramer positive out of CD3+CD8+ cells . | Tetramer positive out of total cells . | Cytotoxicity∗ . | Naïve T-cell content . | Monocyte content . | Natural killer cell content . | B-cell content . |
|---|---|---|---|---|---|---|---|---|---|
| 96.5% | 87.6% | 4% | 76.7% | 68.5% | 82% | 0% | 0.2% | 0.7% | 0.2% |
Target cells were pulsed with the specific SARS-CoV-2 target peptides to allow for binding to HLA-A∗02:01 before performing the assay. The CTLs were generated toward 7 HLA-A∗02:01–restricted SARS-CoV-2 peptides (from spike, nucleocapsid, nonstructural proteins 3, 7, and 8, and 2 from ORF3a proteins) using 1 apheresis product from 1 healthy donor who had COVID-19 illness ∼1 year earlier. The apheresis product was separated into monocytes and lymphocytes via elutriation. Dendritic cells were generated from monocytes and pulsed with the SARS-CoV-2 peptides and used for the first ex vivo resensitization. Thereafter, T cells were serially restimulated with peptide pulsed monocytes and enriched based on adherence of the target T-cell population to the monocytes. The contribution of each peptide to overall cytotoxicity was tested using HLA-A∗02:01 tetramers. Characteristics of the final product are shown above.
Cytotoxicity is shown at an effector-to-target cell ratio (effector cells = SARS-CoV-2 CTLs) of 10:1.
Trial safety and oversight
The study was approved by the Institutional Review Board of Thomas Jefferson University and performed in accordance with the Declaration of Helsinki and the International Conference on Harmonization E6 Guidelines for Good Clinical Practice. All patients provided written informed consent before enrollment. The trial was registered at www.clinicaltrials.gov as #NCT04765449.
The dose-limiting toxicity (DLT) monitoring period was 14 days. DLTs were defined as (1) grade ≥3 infusion reaction within 48 hours of CTL infusion (Common Terminology Criteria for Adverse Events, version 5); (2) modified American Society of Transplantation and Cellular Therapy Consensus Grading for cytokine release syndrome and neurologic toxicity associated with immune effector cells30 for hypoxia, hypotension, and neurotoxicity; (3) pancytopenia/marrow aplasia within 14 days of CTL infusion; and (4) any manifestation of acute (grades 2-4)31 or chronic graft-versus-host disease (GVHD).32 Supplemental Table 2 contains detailed criteria for DLT monitoring.
Within a dosing cohort, patients could not be treated until the prior patient was ≥4 days after CTL infusion. Between cohorts, escalation to a higher dose level was not permitted until the last patient on the previous cohort was at least 14 days after CTL infusion, and the Institutional Data Safety Monitoring Committee approved dose escalation. An internal medical monitor from Thomas Jefferson University and an external medical monitor from an outside institution, neither of whom were participants in the study, also reviewed the data and approved each dose escalation as well as the final cohort’s safety assessment.
Specimen testing
Low-resolution class I HLA genotyping was performed by real-time PCR using sequence-specific primer amplification (One Lambda). For HLA-A∗02–positive patients, identification of the HLA-A∗02 allele was achieved using high-resolution standard sequence-specific primer amplification (Olerup/CareDx). SARS-CoV-2 infection was made or confirmed by hospital-based PCR analysis of nasal swabs. The Roche cobas SARS-CoV-2 test on the cobas 6800 platform was used to determine cycle thresholds using envelope (E) and ORF genes as targets. The Illumina COVIDSeq test was used to sequence the entire SARS-CoV-2 genome (all coding regions and noncoding regions) to identify the particular SARS-CoV-2 variant present. Qualitative SARS-CoV-2 anti-nucleocapsid antibody and qualitative and quantitative SARS-CoV-2 antispike glycoprotein antibody analyses were performed using the Roche Diagnostics Elecsys Anti-SARS-CoV-2 assays. HLA antibody screens were performed by single-antigen bead testing and reported as calculated panel reactive antibodies. All of the above testing was performed at the Thomas Jefferson University clinical laboratories.
Posttreatment evaluation of endogenous SARS-CoV-2–specific T-cell responses and SARS-CoV-2–specific donor-derived T-cell persistence was performed by T-cell sequencing of the CDR3 regions of human T-cell receptor β (TCR-β) chains (at the nucleotide level) using Adaptive Immunosequencing (Adaptive Biotechnologies, Seattle, WA) in patients’ peripheral mononuclear cell samples as well as those from the CTL donor product for comparison purposes. Samples were to be chosen from a subset of patients representing each dosing level.
To assess endogenous T-cell responses after infusion, the analysis focused on TCR-β CDR3 sequences that were present in the patient after infusion but that were undetectable in both the patient before infusion or the CTL donor product. Newly arising sequences in these patient samples that were present in the ImmuneCode database, a compendium of 160 000 COVID-reactive TCR-β sequences,33,34 were taken as evidence of SARS-CoV-2 endogenous T-cell responses not abrogated by CTL infusion.
To assess donor-derived T-cell persistence, T-cell clones found in the CTL donor product but not in the patient’s pretreatment samples were analyzed to quantify signals most attributable to the CTL donor product. These sequences were analyzed at the DNA, not amino acid, level.
Responses were then quantified by the number and/or frequency of SARS-CoV-2–specific TCRs. Specifically, clonal breadth (the proportion of distinct TCRs that are SARS-CoV-2 specific divided by the number of unique TCRs sequenced in the sample) and clonal depth (the sum frequency of the SARS-CoV-2–specific TCRs in the repertoire) were determined for each sample. Additional information regarding this method is contained in the supplemental Materials.
Results
Patient characteristics
Patient characteristics are shown in Table 2. From 7 October 2021 to 14 July 2022, a total of 30 patients were enrolled on trial, and the 6-month follow-up for all patients was completed on 19 January 2023. Twelve of the patients matched the HLA-A∗02:01 restriction of the CTLs, which were infused at a median of 2 days (range, 1-4) after SARS-CoV-2 diagnosis. Total and virus-specific CTL doses for this group are listed in Table 3. Patient 1 was lost to follow-up after 2½ months. Patient 5 died of preexisting, progressive lymphoma 4½ months after receiving CTLs. Eighteen patients were HLA-A∗02:01 negative and were followed in the observational group. The interventional group had a higher median number of comorbidities (3 vs 2.5), more individuals who were unvaccinated or unresponsive to vaccine (4/12 [25%] vs 1/18 [6%]), and a higher number of immunocompromised patients due to treatment for cancer or autoimmune disease (6/12 [50%] vs 1/18 [6%]) than the observational group. Remdesivir was initiated on day +2 in interventional patient 1 but was discontinued before completing the planned 4-day course due to clinical improvement.
Patient characteristics
| Interventional group (received CTLs) . | Observation group (did not receive CTLs) . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient number . | Age-race/ethnicity . | No. of prior vaccine/booster . | Day∗ symptoms definitely better . | Day∗ symptoms resolved . | Comorbid conditions . | CTL dosing level . | Variant† . | Patient number . | Age-race/ethnicity . | No. of prior vaccine/boosters . | Day∗ symptoms definitely better . | Day∗ symptoms resolved . | Comorbid conditions . | Alternate therapies . |
| Fall 2021 | ||||||||||||||
| 1♂ | 24-AA | None | 3 | 8 | DM, HTN, lung disease, obesity | 1 | Delta | 1♂ | 77-Cau | 2 | 23 | 180 | Age, HTN, obesity | Moab day +2, ICU/ventilator |
| 2♀ | 73-Cau | 2 | 3 | 13 | Age, HTN, Obesity | 1 | Delta | 2♀ | 65-Cau | 2 | 5 | 60 | Age, HTN, obesity | |
| 3♀‡ | 47-His | 1 | 2 | 6 | CAD, CVA, DM, HTN, SLE | 1 | Delta | 3♀ | 59-AA | 2 | 2 | 11 | DM, MI, obesity | Moab day 0 (After HLA typing was known) |
| 4♀ | 44-His | 2 | 3 | 7 | Obesity | Moab day +1 | ||||||||
| 5♂ | 53-AA | 2 | 7 | 9 | DM, HTN, obesity | Moab day +1 | ||||||||
| 6♀ | 51-Cau | None | 4 | 8 | DM, HTN, obesity | Moab day +1 | ||||||||
| End December 2021/January-March 2022 | ||||||||||||||
| 4♂‡ | 56-Cau | 2 | 2 | 10 | Colon Ca, obesity | 2 | Omicron BA.1 | 7♂ | 65-Cau | 3 | 4 | 14 | Age, HTN, obesity | |
| 5♀‡ | 44-His | None | 2 | 4 | Lymphoma, obesity | 2 | Omicron BA.1 | 8♂ | 61-AA | 2 | 6 | 14 | DM, HTN | |
| 6♀‡ | 73-Cau | 1 | 2 | 6 | Age, HTN, pancreatic Ca | 2 | Omicron BA.1 | 9♀ | 28-AA | 2 | 3 | 5 | HTN | |
| April-June 2022 | ||||||||||||||
| 7♂ | 58-Cau | 3 | 2 | 10 | HTN, DM | 3 | Omicron BA.2 | 10♀ | 68-Cau | 3 | 4 | 12 | Age | Nirmatrelvir/ritonavir day +2 |
| 8♀‡ | 83-Cau | 3-NMR | 2 | 9 | Age, HTN, lymphoma | 3 | Omicron BA.2 | 11♂ | 85-Cau | 3 | 5 | 11 | Age, DM, HTN | |
| 9♀‡ | 63-AA | 3-NMR | 2 | 7 | HTN, obesity, lymphoma, | 3 | Omicron BA.2 | 12♀ | 55-Cau | 4 | 3 | 9 | DM, HTN, obesity | |
| 10♂ | 67-Cau | 3 | 2 | 6 | Age, DM, HTN, AFib, obesity | 4 | Omicron BA.2 | 13♂ | 63-Cau | 2 | 5 | 9 | HTN, obesity | |
| 14♂ | 68-AA | 3 | 1 | 12 | Age, CVA, heart disease | |||||||||
| July 2022 | ||||||||||||||
| 11♀ | 60-Cau | None-COVID 6 mo prior | 3 | 11 | DM, HTN, obesity | 4 | Omicron BA.5 (presumed) | 15♀ | 40-Cau | 3 | 3 | 28 | HTN, obesity | |
| 12♂ | 49-Cau | 3 | 2 | 8 | DM, HTN, obesity | 4 | Omicron BA.5 | 16♀‡ | 26-Cau | 4 | 11 | 90 | Obesity, Behcet’s | |
| 17♂ | 55-Cau | 4 | 3 | 11 | HTN, obesity | |||||||||
| 18♀ | 54-Cau | 2 | 2 | 11 | obesity | |||||||||
| Interventional group (received CTLs) . | Observation group (did not receive CTLs) . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient number . | Age-race/ethnicity . | No. of prior vaccine/booster . | Day∗ symptoms definitely better . | Day∗ symptoms resolved . | Comorbid conditions . | CTL dosing level . | Variant† . | Patient number . | Age-race/ethnicity . | No. of prior vaccine/boosters . | Day∗ symptoms definitely better . | Day∗ symptoms resolved . | Comorbid conditions . | Alternate therapies . |
| Fall 2021 | ||||||||||||||
| 1♂ | 24-AA | None | 3 | 8 | DM, HTN, lung disease, obesity | 1 | Delta | 1♂ | 77-Cau | 2 | 23 | 180 | Age, HTN, obesity | Moab day +2, ICU/ventilator |
| 2♀ | 73-Cau | 2 | 3 | 13 | Age, HTN, Obesity | 1 | Delta | 2♀ | 65-Cau | 2 | 5 | 60 | Age, HTN, obesity | |
| 3♀‡ | 47-His | 1 | 2 | 6 | CAD, CVA, DM, HTN, SLE | 1 | Delta | 3♀ | 59-AA | 2 | 2 | 11 | DM, MI, obesity | Moab day 0 (After HLA typing was known) |
| 4♀ | 44-His | 2 | 3 | 7 | Obesity | Moab day +1 | ||||||||
| 5♂ | 53-AA | 2 | 7 | 9 | DM, HTN, obesity | Moab day +1 | ||||||||
| 6♀ | 51-Cau | None | 4 | 8 | DM, HTN, obesity | Moab day +1 | ||||||||
| End December 2021/January-March 2022 | ||||||||||||||
| 4♂‡ | 56-Cau | 2 | 2 | 10 | Colon Ca, obesity | 2 | Omicron BA.1 | 7♂ | 65-Cau | 3 | 4 | 14 | Age, HTN, obesity | |
| 5♀‡ | 44-His | None | 2 | 4 | Lymphoma, obesity | 2 | Omicron BA.1 | 8♂ | 61-AA | 2 | 6 | 14 | DM, HTN | |
| 6♀‡ | 73-Cau | 1 | 2 | 6 | Age, HTN, pancreatic Ca | 2 | Omicron BA.1 | 9♀ | 28-AA | 2 | 3 | 5 | HTN | |
| April-June 2022 | ||||||||||||||
| 7♂ | 58-Cau | 3 | 2 | 10 | HTN, DM | 3 | Omicron BA.2 | 10♀ | 68-Cau | 3 | 4 | 12 | Age | Nirmatrelvir/ritonavir day +2 |
| 8♀‡ | 83-Cau | 3-NMR | 2 | 9 | Age, HTN, lymphoma | 3 | Omicron BA.2 | 11♂ | 85-Cau | 3 | 5 | 11 | Age, DM, HTN | |
| 9♀‡ | 63-AA | 3-NMR | 2 | 7 | HTN, obesity, lymphoma, | 3 | Omicron BA.2 | 12♀ | 55-Cau | 4 | 3 | 9 | DM, HTN, obesity | |
| 10♂ | 67-Cau | 3 | 2 | 6 | Age, DM, HTN, AFib, obesity | 4 | Omicron BA.2 | 13♂ | 63-Cau | 2 | 5 | 9 | HTN, obesity | |
| 14♂ | 68-AA | 3 | 1 | 12 | Age, CVA, heart disease | |||||||||
| July 2022 | ||||||||||||||
| 11♀ | 60-Cau | None-COVID 6 mo prior | 3 | 11 | DM, HTN, obesity | 4 | Omicron BA.5 (presumed) | 15♀ | 40-Cau | 3 | 3 | 28 | HTN, obesity | |
| 12♂ | 49-Cau | 3 | 2 | 8 | DM, HTN, obesity | 4 | Omicron BA.5 | 16♀‡ | 26-Cau | 4 | 11 | 90 | Obesity, Behcet’s | |
| 17♂ | 55-Cau | 4 | 3 | 11 | HTN, obesity | |||||||||
| 18♀ | 54-Cau | 2 | 2 | 11 | obesity | |||||||||
AA, African American; AFib, atrial fibrillation; Ca, cancer; Cau, Caucasian; CAD, coronary artery disease; CVA, cerebral vascular accident; DM, diabetes mellitus; His, Hispanic; HTN, hypertension; MI, myocardial infarction; Moab, monoclonal antibody; NMR, no measurable response to vaccine; SLE, systemic lupus erythematosus.
Days to symptoms “definitely better” and days to health “back to baseline” both start from day 0.
SARS-CoV-2 variants analyzed for interventional group only.
Immune compromised due to cancer or autoimmune disease.
Interventional patient dosing and immunity
| . | Patient 1 . | Patient 2 . | Patient 3 . | Patient 4 . | Patient 5∗ . | Patient 6 . | Patient 7 . | Patient 8∗ . | Patient 9∗ . | Patient 10 . | Patient 11 . | Patient 12 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA class I allele matches with CTL donor | A∗02:01 | A∗02:01 | A∗02:01 B∗18:01 | A∗02:01 C∗07:01 | A∗02:01 | A∗02:01 | A∗02:01 | A∗02:01 | A∗02:01 | A∗02:01 | A∗02:01 C∗07:01 | A∗02:01 C∗07:01 |
| Dose level | 1 × 105/kg | 1 × 105/kg | 1 × 105/kg | 3 × 105/kg | 3 × 105/kg | 3 × 105/kg | 1 × 106/kg | 1 × 106/kg | 1 × 106/kg | 3 × 106/kg | 3 × 106/kg | 3 × 106/kg |
| Adjusted weight (kg) | 94.9 | 55.1 | 57.7 | 92.7 | 73.04 | 46.4 | 90.3 | 53.52 | 74.16 | 97.94 | 81 | 104 |
| Total cell dose (10e6) | 9.5 | 5.51 | 5.80 | 27.80 | 21.91 | 13.92 | 90.30 | 53.52 | 74.16 | 294.0 | 243.0 | 312.0 |
| Virus-specific CTL dose (10e6) | 6.51 | 3.77 | 3.97 | 19.04 | 15.01 | 9.54 | 61.86 | 36.66 | 50.80 | 201.39 | 166.46 | 213.72 |
| Anti-HLA donor antibody detected at or after day +28 | None | None | None | None | None | None | None | None | None | None | A∗26, DRB1∗08 | None |
| Vaccine or history of COVID-19 infection before CTLs | No hx Vac No hx Cov | Vac × 2 No hx Cov | Vac × 1 No hx Cov | Vac × 2 No hx Cov | No Vac No hx Cov | Vac × 1 No hx Cov | Vac × 3 No hx Cov | NVR No hx Cov | NVR No hx Cov | Vac × 3 No hx Cov | No Vac Hx of Cov (6 mo prior) | Vac × 3 No hx Cov |
| Antibody status† 1 mo after CTLs | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >1.76 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | No anti-spike antibody | In Allo HSCT | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >184 U/mL |
| +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | No Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | No Anti-NC antibody | In Allo HSCT | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | |
| Antibody status† 2 mo after CTLs | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >1.31 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >1.04 U/mL | In Allo HSCT | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >172.5 U/mL |
| +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | No Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | No Anti-NC antibody | In Allo HSCT | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | |
| Antibody status† 6 mo after CTLs | N/A‡ | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | N/A‡ | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | After Allo HSCT | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >215 U/mL |
| N/A‡ | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | N/A‡ | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | After Allo HSCT | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody |
| . | Patient 1 . | Patient 2 . | Patient 3 . | Patient 4 . | Patient 5∗ . | Patient 6 . | Patient 7 . | Patient 8∗ . | Patient 9∗ . | Patient 10 . | Patient 11 . | Patient 12 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA class I allele matches with CTL donor | A∗02:01 | A∗02:01 | A∗02:01 B∗18:01 | A∗02:01 C∗07:01 | A∗02:01 | A∗02:01 | A∗02:01 | A∗02:01 | A∗02:01 | A∗02:01 | A∗02:01 C∗07:01 | A∗02:01 C∗07:01 |
| Dose level | 1 × 105/kg | 1 × 105/kg | 1 × 105/kg | 3 × 105/kg | 3 × 105/kg | 3 × 105/kg | 1 × 106/kg | 1 × 106/kg | 1 × 106/kg | 3 × 106/kg | 3 × 106/kg | 3 × 106/kg |
| Adjusted weight (kg) | 94.9 | 55.1 | 57.7 | 92.7 | 73.04 | 46.4 | 90.3 | 53.52 | 74.16 | 97.94 | 81 | 104 |
| Total cell dose (10e6) | 9.5 | 5.51 | 5.80 | 27.80 | 21.91 | 13.92 | 90.30 | 53.52 | 74.16 | 294.0 | 243.0 | 312.0 |
| Virus-specific CTL dose (10e6) | 6.51 | 3.77 | 3.97 | 19.04 | 15.01 | 9.54 | 61.86 | 36.66 | 50.80 | 201.39 | 166.46 | 213.72 |
| Anti-HLA donor antibody detected at or after day +28 | None | None | None | None | None | None | None | None | None | None | A∗26, DRB1∗08 | None |
| Vaccine or history of COVID-19 infection before CTLs | No hx Vac No hx Cov | Vac × 2 No hx Cov | Vac × 1 No hx Cov | Vac × 2 No hx Cov | No Vac No hx Cov | Vac × 1 No hx Cov | Vac × 3 No hx Cov | NVR No hx Cov | NVR No hx Cov | Vac × 3 No hx Cov | No Vac Hx of Cov (6 mo prior) | Vac × 3 No hx Cov |
| Antibody status† 1 mo after CTLs | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >1.76 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | No anti-spike antibody | In Allo HSCT | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >184 U/mL |
| +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | No Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | No Anti-NC antibody | In Allo HSCT | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | |
| Antibody status† 2 mo after CTLs | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >1.31 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >1.04 U/mL | In Allo HSCT | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >172.5 U/mL |
| +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | No Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | No Anti-NC antibody | In Allo HSCT | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | |
| Antibody status† 6 mo after CTLs | N/A‡ | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | N/A‡ | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | After Allo HSCT | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >250 U/mL | +Anti-spike antibody >215 U/mL |
| N/A‡ | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | N/A‡ | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody | After Allo HSCT | +Anti-NC antibody | +Anti-NC antibody | +Anti-NC antibody |
Allo HSCT, allogeneic hematopoietic stem cell transplantation; Cov, COVID-19; NC, Nucleocapsid; No hx Cov, no history of COVID-19; No hx Vac, no history of SARS-CoV-2 vaccine; NVR, No vaccine response; Vac, Vaccine.
Patients 5, 8, and 9 under treatment for lymphoma at time of COVID-19. Patient 5 admitted for autologous HSCT 28 days after CTL infusion, underwent HSCT without complication, but died of preexisting, progressive lymphoma 4½ months after CTLs. Patient 9 admitted for successful allogeneic HSCT 16 days after CTL infusion.
Reference range for a positive spike antibody titer is >0.80 U/mL to >250.00 U/mL.
Six-month sample not available due to lost to follow-up after 2½ months, (patient 1) and death at 4½ months, (patient 5).
Safety of the CTLs
There were no DLTs, infusion-related reactions, evidence of CRS, or GVHD at any dosing level. No unexpected side effects were observed in any patient through the 6-month follow-up period. There was 1 grade 3 significant adverse event associated with a preexisting elevation in aspartame aminotransferase level, which worsened with acetaminophen use. All other adverse events were grade ≤2 (supplemental Table 3). Three patients had brief episodes of grade 2 hypoxia, with the lowest saturation of peripheral oxygen (SPO2) being 89% before initiating supplemental oxygen. Patient 1 on dosing level 1 with multifocal pneumonia (Delta variant) received 1 to 2 L of oxygen for <2 hours on day +2. Patient 2 on dosing level 1 (Delta variant) was treated with 1 to 2 L of oxygen for 6½ hours on day +1. Patient 8 treated on dosing level 3 (Omicron variant) received 1 L of oxygen for ∼1 hour on day +1. These limited events were likely due to infection with COVID-19 in the first 2 instances and the sedating effects of diphenhydramine in an 83-year-old patient in the third.
Alloimmunization
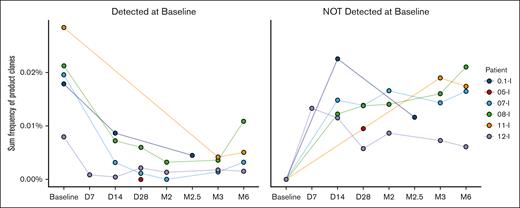
Eleven of 12 patients had no evidence of alloimmunization to the CTL donor. Two HLA antibodies against CTL donor antigens were detected in a previously transfused, multiparous patient who also had multiple other unrelated anti-HLA antibodies. This patient’s baseline specimen could not be processed to assess whether these antibodies were new or preexisting. Donor-derived CTLs were detected through the 6-month time period in this patient (patient 11 in Figure 1) on TCR-β testing, suggesting that there was no rejection of the CTLs due to antidonor antibodies.
CTL product clones are detected in patients through 6 months after infusion. The figure on the left shows the frequencies of the subset of SARS-CoV-2–specific CTL donor sequences present in both the patients’ baseline samples and the CTL donor product (CTL donor-patient overlap). The figure on the right excludes all these overlap clones detected at baseline and shows only donor CTL clones that were not present in the patients’ preinfusion samples. These were used to assess the frequency of donor CTLs over time. Sequences were tracked at the DNA, not amino acid, level. All four 6-month samples exhibited low but nonzero frequencies of CTL product clones undetected preinfusion. Clones most consistent with donor-derived CTLs were detected in patient 5 on dosing level 2 at day +28, but lack of baseline sample in this patient precludes definite confirmation. Testing performed by Adaptive Biotechnologies. D, day; M, month.
CTL product clones are detected in patients through 6 months after infusion. The figure on the left shows the frequencies of the subset of SARS-CoV-2–specific CTL donor sequences present in both the patients’ baseline samples and the CTL donor product (CTL donor-patient overlap). The figure on the right excludes all these overlap clones detected at baseline and shows only donor CTL clones that were not present in the patients’ preinfusion samples. These were used to assess the frequency of donor CTLs over time. Sequences were tracked at the DNA, not amino acid, level. All four 6-month samples exhibited low but nonzero frequencies of CTL product clones undetected preinfusion. Clones most consistent with donor-derived CTLs were detected in patient 5 on dosing level 2 at day +28, but lack of baseline sample in this patient precludes definite confirmation. Testing performed by Adaptive Biotechnologies. D, day; M, month.
Nasal specimen results
PCR detection of SARS-CoV-2 RNA using 2 targets (ORF1a and E) yielded concordant results, consistent with good internal reproducibility. Results were given as Ct values, with lower values reflecting a higher viral load (supplemental Table 4). For E, the median Ct value at baseline was 21.3 (range, 15.63-30.64). By day +14 after treatment, 83% of patients were PCR negative for COVID-19, with 25% of patients reaching PCR negativity by day +4. Reduction of viral burden, calculated based on the Ct data (Table 4), showed that 88% of the virus had been eliminated in 11 of 12 patients (92%) by day +4, and >99% viral elimination was achieved in all patients by day +14. Two humorally immunocompromised patients with a recent history of anti-CD20 monoclonal antibody treatment did not achieve complete PCR negativity by day +14, although both had >99% reduction in PCR-assessed viral load at this time.
COVID-19 nasal swab Ct values and percent reduction from baseline specimen after CTL infusion
| Interventional patient . | Pre-CTL baseline . | Day +4 . | Day +7 . | Day +10∗ . | Day +14 . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Target 1 (E) . | Target 2 (ORF1a) . | Target 1 (E) . | Target 2 (ORF1a) . | Target 1 (E) . | Target 2 (ORF1a) . | Target 1 (E) . | Target 2 (ORF1a) . | Target 1 (E) . | Target 2 (ORF1a) . | |||||||||
| Ct . | Ct . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | |
| 1 | 23.18 | 24.11 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 |
| 2 | 20.16 | 20.54 | 28.18 | 99.61 | 28.65 | 99.64 | 27.58 | 99.42 | 27.77 | 99.33 | 20.41 | 15.91 | 20.91 | 22.62 | NLP | 100 | NLP | 100 |
| 3 | 29.39 | 29.91 | 32.41 | 87.67 | 33.21 | 89.85 | 31.3 | 73.39 | 32.82 | 86.70 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 |
| 4 | 19.58 | 19.86 | 25.36 | 98.18 | 25.96 | 98.54 | 33.02 | 99.99 | 34.98 | 99.99 | 33.34 | 99.99 | 36.02 | 99.99 | NLP | 100 | NLP | 100 |
| 5 | 24.5 | 24.88 | 20.43 | −1579 | 20.71 | −1700 | 22.63 | −265 | 23.04 | −258 | ND | ND | 31.74 | 99.34 | 32.76 | 99.58 | ||
| 6 | 30.64 | 32.7 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 |
| 7 | 18.96 | 19.18 | 28.35 | 99.85 | 28.45 | 99.84 | 33.4 | 99.99 | 34.1 | 99.99 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 |
| 8 | 20.01 | 20.11 | 25.29 | 97.43 | 25.29 | 97.24 | 29.65 | 99.87 | 29.86 | 99.88 | 31.67 | 99.97 | 32.6 | 99.98 | 27.51 | 99.45 | 27.73 | 99.49 |
| 9 | 23.92 | 19.3 | 27.56 | 91.98 | 24.13 | 96.48 | 34.03 | 99.91 | 31.79 | 99.98 | NLP | 100 | 37.02 | 99.99 | NLP | 100 | NLP | 100 |
| 10 | 15.63 | 15.74 | 27.14 | 99.97 | 27.21 | 99.96 | 27.53 | 99.97 | 27.7 | 99.97 | ND | ND | ND | ND | NLP | 100 | NLP | 100 |
| 11 | 20.47 | 20.48 | NLP | 100 | NLP | 100 | TNR | TNR | TNR | TNR | TNR | TNR | TNR | TNR | TNR | TNR | TNR | TNR |
| 12 | 22.2 | 22.17 | 30.08 | 99.58 | 30.2 | 99.62 | 36.91 | 99.99 | NLP | 100 | NLP | 100 | NLP | 100 | TNR | TNR | TNR | TNR |
| Interventional patient . | Pre-CTL baseline . | Day +4 . | Day +7 . | Day +10∗ . | Day +14 . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Target 1 (E) . | Target 2 (ORF1a) . | Target 1 (E) . | Target 2 (ORF1a) . | Target 1 (E) . | Target 2 (ORF1a) . | Target 1 (E) . | Target 2 (ORF1a) . | Target 1 (E) . | Target 2 (ORF1a) . | |||||||||
| Ct . | Ct . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | Ct . | % Red . | |
| 1 | 23.18 | 24.11 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 |
| 2 | 20.16 | 20.54 | 28.18 | 99.61 | 28.65 | 99.64 | 27.58 | 99.42 | 27.77 | 99.33 | 20.41 | 15.91 | 20.91 | 22.62 | NLP | 100 | NLP | 100 |
| 3 | 29.39 | 29.91 | 32.41 | 87.67 | 33.21 | 89.85 | 31.3 | 73.39 | 32.82 | 86.70 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 |
| 4 | 19.58 | 19.86 | 25.36 | 98.18 | 25.96 | 98.54 | 33.02 | 99.99 | 34.98 | 99.99 | 33.34 | 99.99 | 36.02 | 99.99 | NLP | 100 | NLP | 100 |
| 5 | 24.5 | 24.88 | 20.43 | −1579 | 20.71 | −1700 | 22.63 | −265 | 23.04 | −258 | ND | ND | 31.74 | 99.34 | 32.76 | 99.58 | ||
| 6 | 30.64 | 32.7 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 |
| 7 | 18.96 | 19.18 | 28.35 | 99.85 | 28.45 | 99.84 | 33.4 | 99.99 | 34.1 | 99.99 | NLP | 100 | NLP | 100 | NLP | 100 | NLP | 100 |
| 8 | 20.01 | 20.11 | 25.29 | 97.43 | 25.29 | 97.24 | 29.65 | 99.87 | 29.86 | 99.88 | 31.67 | 99.97 | 32.6 | 99.98 | 27.51 | 99.45 | 27.73 | 99.49 |
| 9 | 23.92 | 19.3 | 27.56 | 91.98 | 24.13 | 96.48 | 34.03 | 99.91 | 31.79 | 99.98 | NLP | 100 | 37.02 | 99.99 | NLP | 100 | NLP | 100 |
| 10 | 15.63 | 15.74 | 27.14 | 99.97 | 27.21 | 99.96 | 27.53 | 99.97 | 27.7 | 99.97 | ND | ND | ND | ND | NLP | 100 | NLP | 100 |
| 11 | 20.47 | 20.48 | NLP | 100 | NLP | 100 | TNR | TNR | TNR | TNR | TNR | TNR | TNR | TNR | TNR | TNR | TNR | TNR |
| 12 | 22.2 | 22.17 | 30.08 | 99.58 | 30.2 | 99.62 | 36.91 | 99.99 | NLP | 100 | NLP | 100 | NLP | 100 | TNR | TNR | TNR | TNR |
To determine viral load reduction for each patient, the difference in Ct on each follow-up day from day 0 baseline was determined. Next, the percentage of SARS-CoV-2 burden remaining was calculated by converting the exponent difference to a percentage (ie, 2 to the power of the difference between baseline and remaining). If it took 1 more cycle, 50% of the viral burden would be left; if 2 more cycles, 25 %; 3 cycles, 12.5%, etc. Finally, we converted the percentage remaining to a percentage eliminated by subtracting the percentage remaining from 100%. Each patient in this way was compared with their original Ct (ie, their initial burden).
% Red, Percentage of reduction in viral load from baseline sample; ND, not done; NLP, No longer positive; TNR, testing was not repeated because the patient had previously achieved PCR negativity.
The day +10 testing was not mandatory but was obtained in most patients. PCR testing could be stopped once a negative specimen was obtained. Patients 2 and 3 became PCR negative after treatment for bacterial sinus infection. Patient 2 reported resolution of maxillary sinus pressure at the time her PCR data rebounded, potentially because of opening of a reservoir of virus in her maxillary sinus mucous, which was secluded from CTLs. Patients 5, 8, and 9 were under treatment for lymphoma at the time of COVID-19 diagnosis.
Humoral immunity
The CTLs did not interfere in the development of endogenous humoral responses to COVID-19 nucleocapsid protein in patients capable of humoral responses, and antispike titers showed no pattern of decline over the 6-month follow-up period (Table 3). Treated patients with solid tumor or autoimmune diagnoses produced COVID-19 antibodies after infection, whereas 2 patients with lymphoma and recent anti-CD20 monoclonal antibody therapy produced no detectable antibodies.
Cellular immunity
The development of endogenous cellular anti–SARS-CoV-2 responses and the persistence of infused CTLs in the recipients was assessed by TCR-β analysis in 5 interventional patients; 1 from dose level 1; 2 from dose level 3; and 2 from dose level 4. Preinfusion samples from all the patients on dosing level 2 were unsuitable for analysis precluding formal analysis. An average of 279 154 (range, 39 128-750 304) T cells were profiled from the patient samples. All tested patients developed COVID-19–reactive T-cell depth and breadth in both CD4 and CD8 compartments across multiple ORFs, consistent with an endogenous anti–COVID-19 T-cell response.
Regarding persistence, SARS-CoV-2–specific donor TCR-β clones, specifically documented as not detected in the patient before CTL infusion but present in the CTL product, were identified in all tested postinfusion patient samples. The frequencies of CTL donor clones were on average 6.6 times higher than the frequencies of endogenous COVID-19–reactive T cells documented in the patients at baseline or produced thereafter, suggestive of ex vivo expansion. The CTL donor clones were identified at 2½ months after infusion (the latest sample tested) from patient 1 on the first dose level and in all 4 remaining patients on dosing levels 3 and 4 through the 6-month follow-up time period (Figure 1). Three of the four 6-month samples met Adaptive Biotechnologies’ COVID classifier criteria for a positive result, whereas 1 sample from patient 11 (fourth dosing level), while detectable, just missed the criteria for being classified as positive.
In a separate laboratory study, CTL products from 2 other HLA-A∗02:01–positive donors recognizing the same 7 COVID-19 peptides were found to be largely distinct in TCR-β DNA sequences from those in the product used for the study and from each other (supplemental Figure 1). This result demonstrates the unlikelihood that multiple patients produced endogenous COVID-19 responses bearing the same TCR-β sequences as found in the clinical product. Patient 8, with recently treated lymphoma and serologic vaccine failure, exhibited the highest continual increase in CTL product TCR clonal depth and breadth through 6 months. After the initial analysis, a day +28 sample from patient 5 treated on the second dosing level was analyzed. An average of 210 298 T cells were profiled, and the sample was classified as positive for CTL donor cells, although the lack of a preinfusion sample for comparison purposes may have confounded this result.
Symptoms comparison
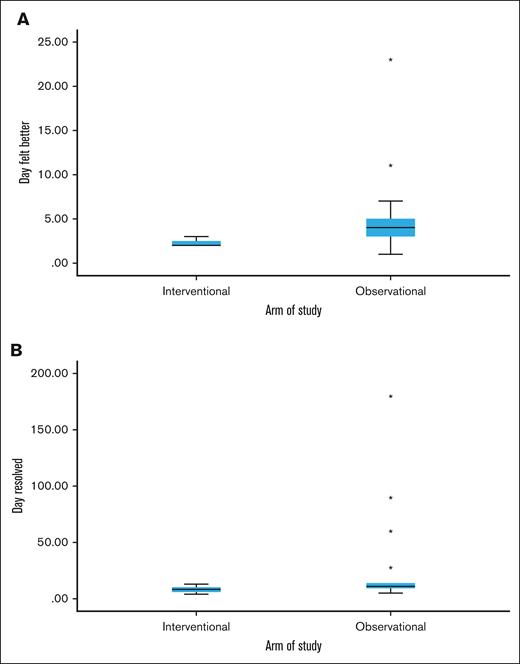
Every patient in the interventional group reported definite symptom improvement at day +2 or +3 after CTL infusion, consistent with a treatment effect. Patients in the observational group had more variable improvement in symptoms, which ranged from days +1 to +23 (median, day +4; Figure 2). Complete resolution of symptoms was also quicker and more consistent in the interventional vs observational group, at 8 days (range, 4-13) vs 11 days (range, 5-180), respectively. No patients in the interventional group experienced progression of COVID-19, postacute sequelae SARS-CoV-2 (PASC), or recurrent COVID-19 through the 6-month follow-up period. Patients 5 and 9 were admitted for autologous and allogeneic hematopoietic stem cell transplants (HSCT) at days +28 and +16, respectively, and had no COVID-19–related events throughout their HSCT courses. In the observational group, 1 patient had progression of COVID-19 requiring mechanical ventilation in an intensive care unit, a second developed PASC with significant symptoms persisting through the 6-month follow-up period, and a third had persistent fatigue due to possible PASC vs deconditioning.
Symptoms comparison between interventional vs observational groups. Box and whisker plots showing the median and range of days when COVID-19 symptoms were definitely better (A) and when symptoms resolved (B) between the interventional and observational groups. In the interventional group, all patients definitely felt better in either 2 or 3 days, narrowing the range. Outliers (each star denotes an outlier) occurred in the observational group with delayed symptom improvement in 2 patients and delayed resolution of symptoms in 4 patients. One of these patients developed PASC and remained symptomatic through the end of the study at day 180, widening the time to resolution range in the observational group.
Symptoms comparison between interventional vs observational groups. Box and whisker plots showing the median and range of days when COVID-19 symptoms were definitely better (A) and when symptoms resolved (B) between the interventional and observational groups. In the interventional group, all patients definitely felt better in either 2 or 3 days, narrowing the range. Outliers (each star denotes an outlier) occurred in the observational group with delayed symptom improvement in 2 patients and delayed resolution of symptoms in 4 patients. One of these patients developed PASC and remained symptomatic through the end of the study at day 180, widening the time to resolution range in the observational group.
Discussion
The primary finding in this trial was that SARS-CoV-2–specific CTL therapy was well tolerated at all tested dosing levels. The side-effect profiles of 2 types of T-cell immunotherapy, off-the-shelf virus-specific T cells (VSTs) used in postallogeneic HSCT recipients and chimeric antigen receptor (CAR) T cells for the treatment of malignancies, were used to predict potential adverse reactions to the CTLs used in the trial. Over decades of use, VSTs have not been associated with significant infusion reactions, CRS, or problematic GVHD.35 Although the manufacturing approach and higher content of virus-specific CD8+ cells in the product used in this study are different than the majority of VSTs used in HSCT recipients, we anticipated that these 2 therapies would be more alike regarding side-effect profile vs CAR T cells, which are genetically modified, target both normal and malignant cells, and may be associated with tumor lysis syndrome. The results of this trial support a CTL safety profile more analogous to VSTs than CAR T cells.
The CTLs did not interfere with endogenous anti–SARS-CoV-2 humoral or cellular immunity. The analysis of postinfusion cellular immunity showed a more extended persistence of the CTLs than anticipated from the HSCT experience. Based on the high degree of HLA mismatch and the lack of preexisting immune compromise in 4 of 6 patients tested for CTL persistence, we anticipated that the single dose of CTLs used in this trial would be more quickly eliminated than the VSTs administered to immune-compromised HSCT recipients. In the post-HSCT setting, although VSTs may last for up to 12 weeks,36-38 persistence is more transient in many patients,39,40 highlighted by the need for additional infusions to gain viral response.38,41,42 In contrast, CTL donor TCR-β clones were detected after a single dose in all patients tested at the 6-month end of follow-up testing point. This prolonged persistence may be due to a higher virus-specific CTL content per dose vs post-HSCT products,38,41,42 the use of more sensitive tests to detect CTLs, or the existence of a viral reservoir after COVID-19 infection43 that provides continued CTL stimulation. Expansion and persistence of allogeneic T cells have been associated with disease control in many settings. Whether the prolonged persistence of the CTLs used in this study or their ability to facilitate endogenous immunity, as has been postulated for the durable effects of VSTs in HSCT patients,44 is of benefit in the treatment of COVID-19, PASC, or alternate future viral or oncologic targets for these CTLs merits further examination.
The patients in the interventional group had several higher risk characteristics than those in the observational group, including active therapy for cancer in 5 of 12 patients before developing COVID-19 infection. Despite that, patients treated with CTLs recovered as promptly and in some cases more quickly than those on the observational arm. The consistency of symptom relief in the days +2 to +3 after CTL time frame, together with a decrease in viral loads in nasal swabs by day +4 in 11 of 12 patients, suggests a treatment effect. These findings warrant follow-up studies examining CTL efficacy in populations remaining at risk for severe COVID-19 disease including patients with humoral immunodeficiency45 in which cellular immunity against SARS-CoV-2 is critical for disease recovery.46 To this point, interventional patient 8, with lymphoma and failure to seroconvert after several vaccine attempts, had the highest frequency of anti-COVID TCR-β CTLs and returned to baseline health as promptly as her non–immune-compromised counterparts.
We did not identify SARS-CoV-2 mutations affecting the study CTLs’ target peptides from the time of peptide selection in 2020 through the end of the Omicron surge and conclusion of study follow-up in early 2023. Unlike antibodies, T cells target small peptides and are not affected by conformational changes from mutations that fall elsewhere in the parent protein. The ability to individually assess CTL reactivity to a precisely known group of peptides restricted by a single HLA allele allows for rapid assessment regarding the impact of new variants on efficacy and “tuning” of the peptide set if needed, thus allowing for consistent dosing of virus-specific CD8+ CTLs. This precision and the high percentage of SARS-CoV-2–reactive CTLs would be compromised if additional HLA restrictions, less well-defined immunizing pools, and multiple viral targets were used to generate CTL products, although wider applicability, such as in post-HSCT VST treatment,41 would be achieved per batch with these multitarget sensitization approaches.
We acknowledge the study limitations. The number of treated patients was small, in part due to a lack of DLTs. CTL tolerability, reduction in viral load and symptoms, and CTL persistence did not appear to be influenced by CTL dose, thus additional testing will be necessary to determine optimal dosing. In addition, because the treatment was limited to HLA-A∗02:01 individuals, which potentially introduced an HLA-specific bias, future studies will require confirmation of these findings in patients with different HLA types. Comparison of nasal PCR data between interventional and observational patients may have provided additional data regarding potential efficacy signals had these samples been obtained in the observational group. Per protocol, SARS-CoV-2 testing via nasal swab sampling was not required once an individual tested negative. This may have prevented the detection of later viral rebound, especially in patients who rapidly converted to negative very early after treatment. However, there was no clinical evidence of rebound in any of the treated patients.
The data support the safety of these SARS-CoV-2–specific CTLs in this tested cohort. SARS-CoV-2 mutations have not affected the CTL targets to date, suggesting that this type of therapy may have some advantages over humorally based treatments for COVID-19 illness. Future studies examining the efficacy and optimal dose of the CTLs, the durability of the CTLs in recipients beyond 6 months, and the implications of this persistence are needed.
Acknowledgments
The authors sincerely thank all the patients participating in this trial for their invaluable contribution to research regarding SARS-CoV-2. The authors also thank the staff of 10 Thompson, Thomas Jefferson University Hospital in Philadelphia, who have been courageously caring for patients with COVID-19 from the onset of the pandemic, including those patients participating in this trial. Finally, the authors acknowledge Dennis Glass as well as study coordinators Samantha Matusiak and Brenda Grande for their support of this trial.
The study was funded by Tevogen Bio.
Authorship
Contribution: All authors had access to primary clinical trial data; D.G. and N.F. analyzed the clinical outcome data which included collaboration with Adaptive Biotechnologies regarding analysis of TCRβ results; N.F., J.L.W., and A.O. analyzed and confirmed CTL dosing data; H.M. and Y.H. analyzed HLA typing results and Z.-X.W. analyzed and confirmed SARS-CoV-2 variant and viral load data; N.F., D.G., and B.L. provided the overall study design; D.G., N.F., U.G., M.M., P.F., J.K., N.N., and A.P. planned and conducted the clinical trial; D.G., N.F., Y.H., H.M., and Z.-X.W. analyzed clinical and laboratory data; N.F., J.L.W., A.O., and K.K. developed and manufactured the CTL product; D.G. was the primary writer of the manuscript; and all other authors provided input and contributed to manuscript revisions, approved the final draft, and vouched for the completeness and accuracy of the data and analyses.
Conflict-of-interest disclosure: D.G. and J.L.W. were unpaid consultants for Tevogen Bio during the conduct of the study. N.F. also served as an unpaid consultant and scientific advisor to Tevogen Bio. The remaining authors declare no competing financial interests.
The current affiliation for D.G. is Tevogen Bio, Philadelphia, PA.
The current affiliation for N.F. is Tevogen Bio, Philadelphia, PA.
The current affiliation for A.O. is Tevogen Bio, Philadelphia, PA.
The current affiliation for J.L.W. is University of Virginia Health, Charlottesville, VA.
The current affiliation for M.M. is Philadelphia College of Osteopathic Medicine, Philadelphia, PA.
The current affiliation for A.P. is Children’s Hospital of Philadelphia, Philadelphia, PA.
Correspondence: Dolores Grosso, Department of Medical Oncology, Sidney Kimmel Cancer Center, Thomas Jefferson University, 834 Chestnut St, Suite 308, Philadelphia, PA 19107; email: dolores.grosso@tevogen.com.
References
Author notes
Sequencing data of T-cell receptor β analysis of patient and cytotoxic T lymphocyte donor samples are available at https://doi.org/10.21417/DG2023NC. Access to deidentified patient data and supporting clinical documents may be available upon request and subject to review by the study sponsor. Requests should be made to shannon.rudolph@tevogen.com. A material transfer and/or data access agreement with the sponsor will be required to access the data, which would be provided electronically. Access will be available for 6 months after publication.
The full-text version of this article contains a data supplement.