UCK2, a pyrimidine salvage enzyme, was overexpressed in HTLV-1–infected T cells and supported cell proliferation in ATL leukemogenesis.
Azacitidine-resistant cells with UCK2 loss adapted to pyrimidine salvage dysfunction by increasing de novo pyrimidine biosynthesis.
Visual Abstract
Adult T-cell leukemia/lymphoma (ATL) is triggered by infection with human T-cell lymphotropic virus-1 (HTLV-1). Here, we describe the reprogramming of pyrimidine biosynthesis in both normal T cells and ATL cells through regulation of uridine-cytidine kinase 2 (UCK2), which supports vigorous proliferation. UCK2 catalyzes the monophosphorylation of cytidine/uridine and their analogues during pyrimidine biosynthesis and drug metabolism. We found that UCK2 was overexpressed aberrantly in HTLV-1–infected T cells but not in normal T cells. T-cell activation via T-cell receptor (TCR) signaling induced expression of UCK2 in normal T cells. Somatic alterations and epigenetic modifications in ATL cells activate TCR signaling. Therefore, we believe that expression of UCK2 in HTLV-1–infected cells is induced by dysregulated TCR signaling. Recently, we established azacitidine-resistant (AZA-R) cells showing absent expression of UCK2. AZA-R cells proliferated normally in vitro, whereas UCK2 knockdown inhibited ATL cell growth. Although uridine and cytidine accumulated in AZA-R cells, possibly because of dysfunction of pyrimidine salvage biosynthesis induced by loss of UCK2 expression, the amount of UTP and CTP was almost the same as in parental cells. Furthermore, AZA-R cells were more susceptible to an inhibitor of dihydroorotic acid dehydrogenase, which performs the rate-limiting enzyme of de novo pyrimidine nucleotide biosynthesis, and more resistant to dipyridamole, an inhibitor of pyrimidine salvage biosynthesis, suggesting that AZA-R cells adapt to UCK2 loss by increasing de novo pyrimidine nucleotide biosynthesis. Taken together, the data suggest that fine-tuning pyrimidine biosynthesis supports vigorous cell proliferation of both normal T cells and ATL cells.
Introduction
Adult T-cell leukemia/lymphoma (ATL) is a T-cell lymphoproliferative neoplasm caused by infection by human T-cell lymphotropic virus type 1 (HTLV-1).1,2 It takes several decades for HTLV-1 carriers to develop ATL. During ATL leukemogenesis, HTLV-1–infected T cells (ATL cells) acquire growth advantages through several mechanisms, including induction of viral oncoproteins3 and accumulation of genetic4,5 or epigenetic abnormalities,6 leading to disease development and progression from indolent ATL (consisting of smoldering and chronic types) to aggressive ATL (consisting of acute and lymphoma types). Several proteins are differentially expressed by HTLV-1–infected cells and ATL cells and are thought to be molecular targets for the treatment and diagnosis of ATL. For example, CCR47 and EZH28-10 are known to be therapeutic targets, and molecular diagnostics using CADM111,12 and TNFR213 as biomarkers are under development.
Previously, we reported that the accumulation of regional DNA hypermethylation is related to the development and progression of ATL and that DNA hypomethylating agents show efficacy against ATL.14 Furthermore, inactivation of uridine-cytidine kinase 2 (UCK2) and deoxycytidne kinase makes cells resistant to azacitidine (AZA) and decitabine, respectively.15 UCK proteins (UCK1 and UCK2 in humans) phosphorylate uridine and cytidine to yield monophosphorylated forms during pyrimidine nucleotide salvage synthesis.16 Although UCK1 is expressed ubiquitously in several normal human tissues, such as the liver, kidney, muscle, lung, and heart, the expression of UCK2 is limited to the human placenta and testis.17 Because UCK2 also phosphorylates AZA, which is then incorporated into DNA after sequential phosphorylation into AZA-dCTP,18 it is reasonable to suppose that inactivation of UCK2 makes cells resistant to AZA.
There is a correlation between aberrant overexpression of UCK2 and a poor prognosis in patients with lung cancer,19 breast cancer,20 and liver cancer.21 Knockdown of UCK2 inhibits growth and migration of lung cancer19 cells and hepatocellular carcinoma21 cell lines. However, it remains unclear how UCK2 overexpression contributes to tumorigenesis. Although UCK2 interacts with EGFR in hepatocellular carcinoma cell lines, leading to activation of the EGFR-AKT pathway, this interaction is independent of its kinase activity.22
Here, we found that UCK2 was overexpressed in HTLV-1–infected T cells and ATL cells when compared with HTLV-1–uninfected normal T cells. Therefore, the aim of this study was to better understand the role of UCK2 in ATL leukemogenesis.
Materials and methods
Human samples
All studies using human samples were performed in accordance with the Declaration of Helsinki and were approved by the institutional review board of Saga University (A2022-027-0). Primary peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll separation as described previously.14 All HTLV-1 carriers, patients with ATL, and healthy volunteers provided written informed consent to participate.
Mice
Animal studies were conducted in accordance with guidelines and animal protocols at Saga University (A2022-027-0), and in accordance with the German Animal Welfare Act. Immunodeficient Balb/c Rag-2−/− Jak3−/− mice, which lack mature T and B lymphocytes and natural killer cells,23 were maintained under a 12-hour light/dark cycle with free access to food and water.
Cell culture
MT-2 cells, derived from umbilical cord blood cells, which were transformed by coculture with PBMCs from a patient with ATL,24 were purchased from the JCRB Cell Bank (Osaka, Japan). TL-Om1 cells, derived from PBMCs from a patient with ATL, were provided by Masao Matsuoka (Kumamoto University). AZA-resistant (AZA-R) cells were established previously by long-term exposure to the drug.15 The parental cell lines, as well as the AZA-R subclones, were authenticated previously using short tandem repeat profiling.15 Cells were cultured in RPMI-1640 (Sigma-Aldrich, St. Louis, MO) containing 10% fetal bovine serum. All experiments were performed using mycoplasma-free cells.
Public database analyses
Isolation of cell subpopulations from PBMCs
Primary PBMCs were stained with anti-CD3 (BD Biosciences), anti-CD4 (BD Biosciences), anti-CD7 (BD Biosciences), and anti-CADM1 (MBL) antibodies and then sorted on a FACSAriaII (BD Biosciences) to isolate CD7+/CADM1− CD4 T cells (HTLV-1–uninfected T-cell fraction) and CADM1+ CD4 T cells (HTLV-1–infected T-cell fraction). Flow cytometry data were analyzed using FlowJo software (BD Biosciences). CD4+ T cells from healthy volunteers were isolated from PBMCs using a CD4+ T Cell Isolation Kit (Miltenyi Biotec).
Western blot analysis
Whole cell lysates were prepared using RIPA buffer (Santa Cruz Biotechnology), and total protein content was measured using a protein assay (Bio-Rad). Equal amounts of each cell lysate were resolved in 4% to 12% Nu-polyacrylamide gels (Invitrogen), and proteins were detected using antibodies specific for UCK2 (Proteintech), beta-actin (Cell Signaling Technology), beta-tubulin (Cell Signaling Technology), UCK1 (Proteintech), phosphor-ZAP70 (Tyr319) (Cell Signaling Technology), Zap70 (Cell Signaling Technology), dihydroorotate dehydrogenase (DHODH) (Proteintech), and DNA methyltransferase1 (DNMT1) (Abcam).
T-cell activation and expansion
CD4+ T cells isolated from healthy volunteers were stimulated using anti-CD3/CD28 antibody-conjugated magnetic beads (Thermo Scientific). The cells were cultured in Advanced RPMI Medium 1640 (Thermo Scientific) containing 2% human AB serum (Thermo Scientific), 2 mM L-glutamin (Thermo Scientific), 100 U/mL human interleukin-2 (R & D Systems), and 100 U/mL penicillin/streptomycin.
Quantitative real-time polymerase chain reaction
RNA extraction, synthesis of complementary DNA, and quantification of UCK1 and UCK2 gene expression were performed as described previously.15
Lentivirus preparation and infection
The lentiviral particles used for transduction of shRNA targeting human UCK2 (pLV[shRNA]-EGFPU6>hUCK2[shRNA#1]) and scramble control shRNA (pLV[shRNA]-EGFP/Puro-U6>Scramble_shRNA) were prepared by VectorBuilder. Cells were infected with lentiviral particles using RetroNectin (Takara) and then used for further experiments.
Cell cycle analysis
The cell cycle was analyzed by staining DNA with propidium iodide, followed by flow cytometry analysis as described previously.26
SA-β-Gal staining
Senescence-associated β-galactosidase (SA-β-Gal) was detected using a staining kit (Cell Signaling Technology). Images were taken with a Zeiss Axio Imager. M2 is fitted with a Zeiss Axiocam 512 microscope color camera.
RNA sequencing and analysis
RNA sequencing (RNA-seq) was performed by Rhelixa Co, Ltd. Briefly, RNA-seq libraries were prepared using a NEBNext Ultra II Directional RNA Library Prep Kit (New England BioLabs). The purified libraries were sequenced on an Illumina NovaSeq 6000, with 150 × 2 paired-end reads. The quality of the raw paired-end sequence reads was assessed by FastQC (version 0.11.7; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and low-quality (<20) bases and adapter sequences were trimmed using Trimmomatic software (version 0.38) with the following parameters: ILLUMINACLIP: path/to/adapter.fa:two:30:10 LEADING:20 TRAILING:20 SLIDINGWINDOW:four:15 MINLEN:36. The trimmed reads were aligned to the reference genome (hg38) using RNA-seq aligner HISAT2 (version 2.1.0). The raw read counts were normalized to transcripts per million. Principal component analysis of the normalized counts was conducted, and each sample was projected onto the 2D plane of the first and second principal component analysis axes using stats (version 3.6.1) and gplots (version 3.0.1.1) in R. Gene set enrichment analysis27,28 using the Curated (C2) gene sets (reactome subset of CP) from the Molecular Signatures Database29,30 was used to identify enriched pathways.
Cell migration assay
Cell migration was assayed using a Transwell cell culture chamber (Corning). Cells in serum-free RPMI 1640 were seeded onto the upper insert well with a 5 μm-pore filter. Complete medium was added to a lower chamber and maintained for 24 hours at 37°C. The number of cells that migrated to the lower chamber was counted.
Xenograft mouse model
Cells were inoculated subcutaneously into 8-week-old female Balb/c Rag-2−/− Jak3−/− mice, and tumor volume, defined as (short axis)2 × (long axis)/2, was measured twice per week.
Metabolome analysis
Metabolites were extracted with methanol and then analyzed using the ω-Scan package at Human Metabolome Technologies, as described previously.31
Quantification of oxidized and reduced CoQ10
Cells were washed in phosphate-buffered saline and then resuspended in 2-propanol. Quantification of oxidized and reduced CoQ10 was performed by Kaneka Techno Research Co Ltd, as described previously.32
Measurement of cellular oxygen consumption rate (OCR)
OCR was measured using a Seahorse XF Pro Extracellular Flux analyzer (Agilent Technologies) and an Agilent Seahorse XF Cell Mito Stress Test Kit.
Statistical analysis
Data are expressed as the mean ± standard deviation. Differences between groups were tested using Dunnett test and student t test. Results were considered significant when a value of P < .05 was obtained by a 2-sided test. All calculations were performed using R v.3.4.4 (The R Foundation for Statistical Computing) or EZR33 (a graphical user interface for R; Saitama Medical Center, Jichi Medical University, Saitama, Japan).
Results
Aberrant induction of UCK2 expression triggered by HTLV-1
First, we used gene expression data sets (GSE55851 and GSE33615), which have been reported previously,11,25 to analyze changes in expression of the UCK2 gene in HTLV-1–infected cells. UCK2 expression in HTLV-1–infected T cells and/or ATL cells isolated from asymptomatic HTLV-1 carriers and patients with ATL was higher than that of their uninfected counterparts (HTLV-1–uninfected T cells) (Figure 1A). However, expression of a homologous gene, UCK1, was not higher in HTLV-1–infected T cells (Figure 1B). Another data set showed that expression of UCK2 but not UCK1, increased significantly in PBMCs from patients with chronic and acute ATL but not smoldering ATL (Figure 1C-D).
Aberrant overexpression of UCK2 during ATL leukemogenesis. (A-B) Expression of the UCK2 and UCK1 genes in HTLV-1–infected cells and normal T cells was analyzed using GSE55851 data set. HTLV-1–infected T cells (CADM1+/CD7 dimly+ [yellow], and CADM1+/CD7− [red]), and normal T cells (CADM1−/CD+ [blue]), were isolated from healthy volunteers (normal), asymptomatic carriers (AC), and patients with smoldering (S), chronic (C), and acute (A) ATL. (C-D) Expression of the UCK2 and UCK1 genes by PBMCs from healthy volunteers and patients with ATL was analyzed using GSE33615 data set. Normal CD4+ T cells were isolated from healthy volunteers (n = 21). PBMCs were isolated from patients with S (n = 4), C (n = 20), and A (n = 26) ATL. Differences between normal cells and each ATL subtype were assessed using Dunnett test. ∗P < .05. (E) HTLV-1–infected T cells (CADM1+ T cells) and their normal counterparts (CADM1−/CD7+ T cells) were isolated from healthy volunteers (N, n = 3), HTLV-1 carriers (AC, n = 1), and patients with smoldering ATL (S, n = 2), chronic (C, n = 2) and acute ATL (A, n = 2). UCK2 expression was measured by western blotting. (F) UCK2 expression by ATL cell lines and CD3+CD4+CD7+CADM1− T cells (CD4+ T cell) from 3 volunteers was compared by western blotting.
Aberrant overexpression of UCK2 during ATL leukemogenesis. (A-B) Expression of the UCK2 and UCK1 genes in HTLV-1–infected cells and normal T cells was analyzed using GSE55851 data set. HTLV-1–infected T cells (CADM1+/CD7 dimly+ [yellow], and CADM1+/CD7− [red]), and normal T cells (CADM1−/CD+ [blue]), were isolated from healthy volunteers (normal), asymptomatic carriers (AC), and patients with smoldering (S), chronic (C), and acute (A) ATL. (C-D) Expression of the UCK2 and UCK1 genes by PBMCs from healthy volunteers and patients with ATL was analyzed using GSE33615 data set. Normal CD4+ T cells were isolated from healthy volunteers (n = 21). PBMCs were isolated from patients with S (n = 4), C (n = 20), and A (n = 26) ATL. Differences between normal cells and each ATL subtype were assessed using Dunnett test. ∗P < .05. (E) HTLV-1–infected T cells (CADM1+ T cells) and their normal counterparts (CADM1−/CD7+ T cells) were isolated from healthy volunteers (N, n = 3), HTLV-1 carriers (AC, n = 1), and patients with smoldering ATL (S, n = 2), chronic (C, n = 2) and acute ATL (A, n = 2). UCK2 expression was measured by western blotting. (F) UCK2 expression by ATL cell lines and CD3+CD4+CD7+CADM1− T cells (CD4+ T cell) from 3 volunteers was compared by western blotting.
Next, we isolated HTLV-1–infected T cells and normal T cells from healthy volunteers, an asymptomatic HTLV-1 carrier, and patients with smoldering, chronic, and acute ATL based on CADM1 expression status. Western blot analysis revealed that expression of the UCK2 protein was upregulated significantly in HTLV-1–infected T cells and ATL cells (CADM1-positive cells) compared with normal T cells (CADM1-negative cells) (Figure 1E). Expression was also observed in all 7 HTLV-1–infected cell lines and ATL cell lines tested, as well as other cell lines established from other T-cell malignancies, including ALL and ALCL, but not in normal CD4+ T cells isolated from HTLV-1–uninfected healthy volunteers (Figure 1F; supplemental Figure 1A). Taken together, the data suggest that aberrant induction of UCK2 expression results in growth advantages in a portion of HTLV-1–infected T cells and ATL cells.
UCK2 expression in T cells is induced by activation of TCR signaling
Next, we tried to understand how expression of UCK2 is induced in HTLV-1–infected T cells. Although the amounts of UCK2 varied widely between cell lines, cellular characteristics, such as expression of the oncoviral protein Tax or dependency upon interleukin-2 for cell growth, did not reflect expression status (supplemental Figure 1B-C). Previously, we found that aberrant regional DNA methylation accumulates in HTLV-1–infected T cells and that DNA methylation status reflects disease status.14 However, treatment of MT-2 cells with DNA demethylating agents induced UCK2 gene expression slightly (supplemental Figure 1D). In addition, there was no significant difference in UCK2 promoter methylation status between HTLV-1–infected cells (or ATL cells) isolated from HTLV-1 carriers or patients with any disease type of ATL and that of their uninfected counterparts (supplemental Figure 1E). Thus, DNA methylation seems not to be involved in the aberrant expression of UCK2.
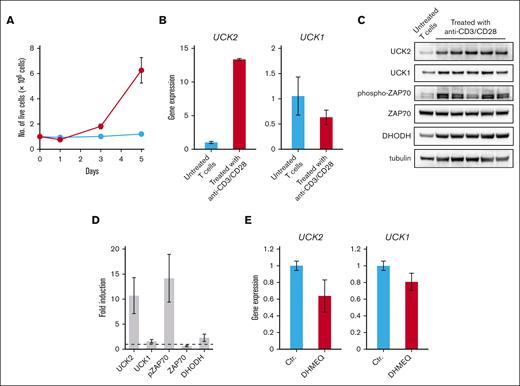
T cells undergo clonal expansion in response to the activation of TCR signaling, which is caused by antigen recognition. However, several activating mutations have been identified in components of the TCR signaling pathway, which might drive vigorous proliferation of ATL cells.4 In addition, we found that several regulators of TCR signaling in HTLV-1–infected cells and ATL cells are silenced by promoter hypermethylation.14 Therefore, we next focused on the involvement of TCR signaling in the induction of UCK2. As expected, analysis of a gene expression data set (GSE13738) revealed that expression of UCK2, but not UCK1, in normal T cells was induced by activation of TCR signaling triggered by antigen recognition (supplemental Figure 2A-B). We then isolated HTLV-1–uninfected CD4+ T cells from healthy volunteers and stimulated them with anti-CD3/CD28 antibody-conjugated magnetic beads. Although T cells started to expand 2 to 3 days after stimulation, expression of UCK2 increased significantly on day 1 (Figure 2A-B). Strong induction of UCK2 protein expression upon TCR activation, concomitant with phosphorylation of ZAP70, was observed 5 days after stimulation, whereas UCK1 was induced only slightly (Figure 2C-D).
Induction of UCK2 expression through the activation of the TCR signaling pathway. (A) CD4+ T cells isolated from 5 healthy volunteers were cultured in the presence (blue line) or absence (red line) of anti-CD3/CD28 antibodies conjugated beads. At 1, 3, and 5 days after seeding, cells were stained with trypan blue, and the number of live cells was counted. The results represent the mean of 5 biological replicates of CD4+ T cells isolated from 5 independent donors. (B) Real-time PCR conducted to measure expression of the UCK2 and UCK1 genes by T cells treated (or not) with anti-CD3/CD28 antibody-conjugated beads for 1 day. Graphs show relative expression of the UCK2 and UCK1 genes (compared with untreated T cells) after normalization to ACTB mRNA levels. The results are expressed as the mean ± standard deviation (SD) of 3 biological replicates. (C) Immunoblots showing upregulation of UCK2 expression 5 days after treatment with anti-CD3/CD28 antibody-conjugated beads. Although 5 biological replicates of T cells isolated from 5 different donors were prepared, untreated samples were pooled, and analyzed as a single sample because the cells were small and did not grow. (D) The graph shows relative expression relative to that in a pooled untreated sample. Results are expressed as the mean ± SD of 5 biological replicates of CD4+ T cells isolated from 5 independent donors. (E) Expression of UCK2 and UCK1 genes in TL-Om1 cells treated for 16 hours with DHMEQ. PCR, polymerase chain reaction.
Induction of UCK2 expression through the activation of the TCR signaling pathway. (A) CD4+ T cells isolated from 5 healthy volunteers were cultured in the presence (blue line) or absence (red line) of anti-CD3/CD28 antibodies conjugated beads. At 1, 3, and 5 days after seeding, cells were stained with trypan blue, and the number of live cells was counted. The results represent the mean of 5 biological replicates of CD4+ T cells isolated from 5 independent donors. (B) Real-time PCR conducted to measure expression of the UCK2 and UCK1 genes by T cells treated (or not) with anti-CD3/CD28 antibody-conjugated beads for 1 day. Graphs show relative expression of the UCK2 and UCK1 genes (compared with untreated T cells) after normalization to ACTB mRNA levels. The results are expressed as the mean ± standard deviation (SD) of 3 biological replicates. (C) Immunoblots showing upregulation of UCK2 expression 5 days after treatment with anti-CD3/CD28 antibody-conjugated beads. Although 5 biological replicates of T cells isolated from 5 different donors were prepared, untreated samples were pooled, and analyzed as a single sample because the cells were small and did not grow. (D) The graph shows relative expression relative to that in a pooled untreated sample. Results are expressed as the mean ± SD of 5 biological replicates of CD4+ T cells isolated from 5 independent donors. (E) Expression of UCK2 and UCK1 genes in TL-Om1 cells treated for 16 hours with DHMEQ. PCR, polymerase chain reaction.
Several mutations activating the TCR-nuclear factor κB (NF-κB) pathway have been identified in ATL cells4; indeed, this pathway is a druggable target in ATL. Dehydroxymethylepoxyquinomicin (DHMEQ) is a NF-κB inhibitor that shows efficacy in ATL cells in vitro and in vivo.34 Treatment of TL-Om1 cells with DHMEQ suppressed the expression of UCK2 and UCK1 (Figure 2E). These results suggest that UCK2 is overexpressed in HTLV-1–infected cells and ATL cells through activation of TCR signaling.
Knockdown of UCK2 in HTLV-1-infected cells and ATL cells inhibits cell growth
To reveal the biological significance of UCK2 during ATL leukemogenesis, TL-Om1 and MT-2 cell lines were transduced with a retroviral vector expressing shRNA targeting UCK2 (sh-UCK2), leading to a marked reduction in the amount of UCK2 protein (Figure 3A). Knockdown of UCK2 strongly suppressed the growth of both cell lines (Figure 3B). Although it partially induced cell death in TL-Om1 cells, we observed only cytostatic effects and unchanged viability in MT-2 cells until at least 5 days after processing (Figure 3C). Although knockdown of UCK2 did not induce apoptosis (supplemental Figure 3A), cells were arrested in the G1 phase, with enlarged cell morphology, 3 days after processing (Figures 3D; supplemental Figure 3B). As expected, SA-β-gal–positive MT-2 cells accumulated gradually after transduction with sh-UCK2 (Figure 3E). Principal component analysis plots of gene expression profiles clearly showed that UCK2 knockdown cells clustered separately from control cells (Figure 3F). Gene set enrichment analysis showed that genes involved in DNA synthesis were downregulated in UCK2 knockdown cells (Figure 3G).
Growth inhibition induced by knockdown of UCK2. (A) Immunoblots showing reduced expression of UCK2, but not UCK1, protein in TL-Om1 and MT-2 cells at 3 days after transduction with shRNA targeting UCK2 (sh-UCK2), or a scramble shRNA control (Con-sh). (B-C) Cells were stained with trypan blue stain 3 to 5 days after transduction of Con-sh [blue] or sh-UCK2 [red], and the number of live cells was counted. The results are expressed as the mean ± SD of 3 independent experiments. (D) Cell cycle distribution was examined 3 days after transduction. Blue columns, G1; red columns, S; green columns, G2/M. Results are expressed as the mean ± SD of 3 independent experiments. (E) TL-Om1 cells were stained for SA-β-gal at 3 to 5 days after transduction. (F) Principal component analysis plot in gene expression profiles of TL-Om1 [circle] and MT-2 [triangle] cells transduced for 3 days with Con-sh [blue] or sh-UCK2 [red]. (G) Gene set enrichment analysis of the Con-sh and sh-UCK2 group using the Curated (C2) gene sets (Reactome subset of CP) from the Molecular Signatures Database. FDR, False discovery rate; NES, Normalized enrichment score.
Growth inhibition induced by knockdown of UCK2. (A) Immunoblots showing reduced expression of UCK2, but not UCK1, protein in TL-Om1 and MT-2 cells at 3 days after transduction with shRNA targeting UCK2 (sh-UCK2), or a scramble shRNA control (Con-sh). (B-C) Cells were stained with trypan blue stain 3 to 5 days after transduction of Con-sh [blue] or sh-UCK2 [red], and the number of live cells was counted. The results are expressed as the mean ± SD of 3 independent experiments. (D) Cell cycle distribution was examined 3 days after transduction. Blue columns, G1; red columns, S; green columns, G2/M. Results are expressed as the mean ± SD of 3 independent experiments. (E) TL-Om1 cells were stained for SA-β-gal at 3 to 5 days after transduction. (F) Principal component analysis plot in gene expression profiles of TL-Om1 [circle] and MT-2 [triangle] cells transduced for 3 days with Con-sh [blue] or sh-UCK2 [red]. (G) Gene set enrichment analysis of the Con-sh and sh-UCK2 group using the Curated (C2) gene sets (Reactome subset of CP) from the Molecular Signatures Database. FDR, False discovery rate; NES, Normalized enrichment score.
Taken together, these data indicate that increased expression of UCK2 supplies pyrimidine nucleotides by activating pyrimidine salvage biosynthesis, which supports vigorous proliferation of HTLV-1–infected cells.
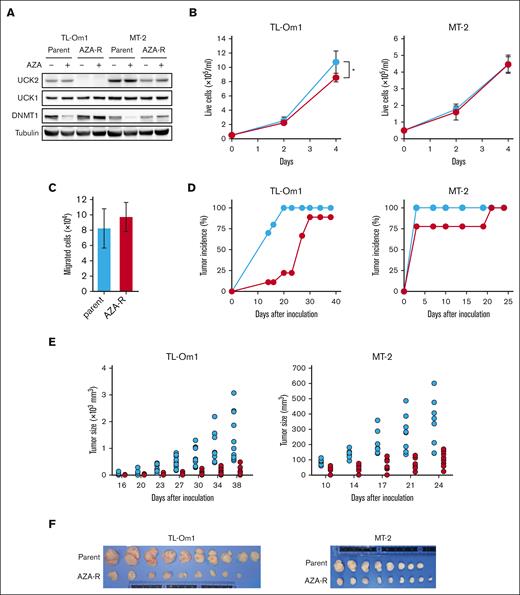
Decreased expression of UCK2 inhibits growth of xenograft tumors established from AZA-R cells
UCK proteins (UCK1 and UCK2) are involved not only in the pyrimidine salvage pathway but also in the monophosphorylation of AZA, which is a rate-limiting step during the activation of AZA (supplemental Figure 4). Recently, we established AZA-R cells from TL-Om1 and MT-2 cells through long-term exposure to the drug in vitro. As we reported previously,15 we did not detect UCK2 expression in AZA-R TL-Om1 cells, and it fell by almost half in AZA-R MT-2 cells (Figure 4A). AZA is incorporated into cells by nucleotide transporters and then converted into AZA-CTP or AZA-dCTP via continuous phosphorylation. AZA-dCTP is then incorporated into the growing daughter strand of DNA, leading to the depletion of DNMT1. Treatment with AZA decreased the amount of DNMT1 protein in both parental cell lines but not in their AZA-R derivatives (Figure 4A). AZA-R TL-Om1 cells showed a slight growth defect compared with parental cells, but AZA-R MT-2 cells did not (Figure 4B). These results appear inconsistent with those showing that knockdown of UCK2 led to strong inhibition of TL-Om1 and MT-2 growth (as shown in Figure 3B). Because most cells transduced with short hairpin-RNA targeting UCK2 died upon prolonged cultivation, we could not isolate stable UCK2 knockdown cells. Although UCK2 plays a role in migration of lung cancer cells,19 there was no difference in the migration between AZA-R TL-Om1 and parental cells (Figure 4C).
Adaptive growth of AZA-R cells showing decreased expression of UCK2 in vitro but not in vivo. (A) Immunoblots showing amounts of protein in each cell line after treatment for 4 days with 100 nM AZA. (B) The number of live cells was measured by trypan blue staining. Blue line, parental cells; red line, AZA-R cells. The results are expressed as the mean ± SD of 3 independent experiments. ∗P < .05. (C) The number of migrated cells was counted after staining with trypan blue. Migration was induced by incubation for 24 hours with FBS. The results are expressed as the mean ± SD of 3 independent experiments. (D-E) Tumor development and size in Balb/c Rag-2−/− Jak3−/− mice injected subcutaneously with parental cells (blue) or AZA-R (red) TL-Om1 and MT-2 cells. Tumors measuring more than 5 mm in diameter were counted. (F) TL-Om1 tumors were isolated from mice sacrificed on day 39, and MT-2 tumors were isolated from mice sacrificed on day 25. FBS, fetal bovine serum.
Adaptive growth of AZA-R cells showing decreased expression of UCK2 in vitro but not in vivo. (A) Immunoblots showing amounts of protein in each cell line after treatment for 4 days with 100 nM AZA. (B) The number of live cells was measured by trypan blue staining. Blue line, parental cells; red line, AZA-R cells. The results are expressed as the mean ± SD of 3 independent experiments. ∗P < .05. (C) The number of migrated cells was counted after staining with trypan blue. Migration was induced by incubation for 24 hours with FBS. The results are expressed as the mean ± SD of 3 independent experiments. (D-E) Tumor development and size in Balb/c Rag-2−/− Jak3−/− mice injected subcutaneously with parental cells (blue) or AZA-R (red) TL-Om1 and MT-2 cells. Tumors measuring more than 5 mm in diameter were counted. (F) TL-Om1 tumors were isolated from mice sacrificed on day 39, and MT-2 tumors were isolated from mice sacrificed on day 25. FBS, fetal bovine serum.
However, engraftment of AZA-R TL-Om1 cells transplanted subcutaneously into immunodeficient mice was delayed significantly compared with that of parental cells (Figure 4D). Furthermore, growth of both AZA-R cell tumor xenografts was suppressed when compared with that of the parental cells (Figure 4E-F).
Upregulation of de novo pyrimidine nucleotide synthesis rescues growth of AZA-R TL-Om1 cells
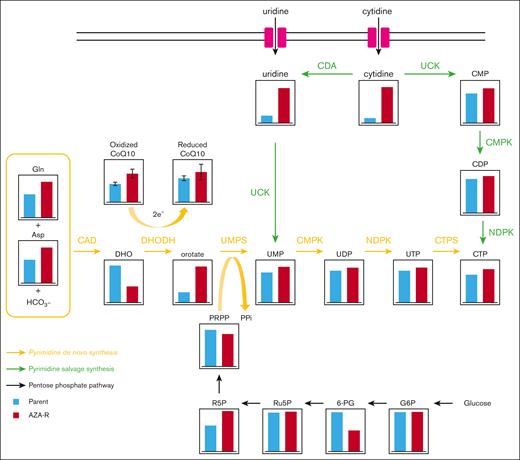
To better understand why AZA-R cells grew normally in vitro, we focused on pyrimidine nucleotide metabolism. Because AZA-R TL-Om1 cells showed no detectable expression of UCK2 upon western blot analysis, we compared the metabolomic profile of AZA-R TL-Om1 cells with that of parental cells. We noted markedly greater accumulation of intracellular uridine and cytidine in AZA-R cells than in parental cells, possibly due to UCK2 deficiency in the former (Figure 5). In addition, we observed decreased levels of dihydroorotate (DHO) and increased levels of orotate in AZA-R cells (Figure 5), suggesting activation of dihydroorotate dehydrogenase (DHODH), which is the rate-limiting enzyme of de novo pyrimidine nucleotide synthesis.
Reprogramming of pyrimidine nucleotide biosynthesis in AZA-R cells showing reduced expression of UCK2. Each metabolite (except for CoQ10) of TL-Om1 AZA-R cells (red) cultured under normal growth conditions was identified and compared with those of TL-Om1 parental cells (blue) using the ω-Scan package (HMT). Both oxidized and reduced CoQ10 present in whole cell extracts was measured as described in “Materials and methods.” CDA, Cytidine deaminase; CMP, Cytidine monophosphate; CMPK, Cytidine monophosphate kinase; NDPK, Nucleoside diphosphate kinase; UMP, Uridine monophosphate.
Reprogramming of pyrimidine nucleotide biosynthesis in AZA-R cells showing reduced expression of UCK2. Each metabolite (except for CoQ10) of TL-Om1 AZA-R cells (red) cultured under normal growth conditions was identified and compared with those of TL-Om1 parental cells (blue) using the ω-Scan package (HMT). Both oxidized and reduced CoQ10 present in whole cell extracts was measured as described in “Materials and methods.” CDA, Cytidine deaminase; CMP, Cytidine monophosphate; CMPK, Cytidine monophosphate kinase; NDPK, Nucleoside diphosphate kinase; UMP, Uridine monophosphate.
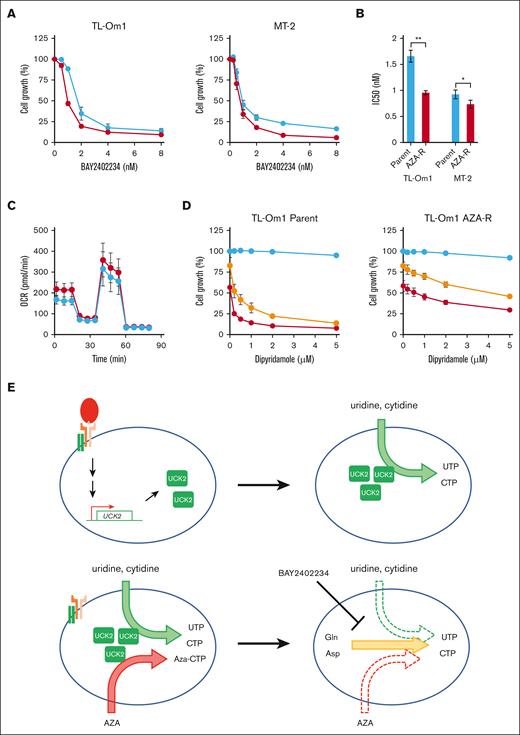
As expected, AZA-R cells were more sensitive to BAY2402234, a selective DHODH inhibitor, than the parental cells (Figure 6A-B). We also succeeded in establishing another AZA-R cell line from an MDS cell line, SKM-1, by long-term exposure to the drug (supplemental Figure 5A). The cells showed reduced expression of UCK2 and increased susceptibility to BAY2402234, as seen for AZA-R cells derived from ATL cell lines (supplemental Figure 5B-C). Although several reports show that DHODH activity is linked to respiratory chain function,35 we did not observe any significant differences in OCR between AZA-R TL-Om1 cells and parental cells (Figure 6C). Dipyridamole inhibits transporting nucleosides,36 leading to dysfunction of pyrimidine salvage biosynthesis. Although treatment with dipyridamole alone did not suppress cell growth, combination with small amounts of BAY2402234 (IC20 and IC40 doses) dramatically enhanced the susceptibility of parental TL-Om1 cells to dipyridamole but not AZA-R cells (Figure 6D).
Inhibition of de novo pyrimidine biosynthesis in AZA-R cells showing reduced expression of UCK2. (A) AZA-R cells (red) derived from TL-Om1 and MT-2 cells, and each parental cell line (blue) were treated for 96 hours with BAY2402234, a DHIODH-specific inhibitor. Cell growth was measured using a CCK-8 kit. Absorbance of nontreated cells was set as 100%. The results are expressed as the mean ± SD of 3 independent experiments. (B) Fifty percent inhibitory concentration (IC) values of BAY2402234. Data are expressed as the mean ± SD of 3 independent experiments. ∗P < .05; ∗∗P < .01. (C) An extracellular flux assay was used to measure the OCR in AZA-R cells (red) derived from TL-Om1 cells, as well as parental cells (blue), cultivated under normal growth conditions. Oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, rotenone, and antimycin A were added to the assay. (D) AZA-R cells derived from TL-Om1 and parental cell line were treated for 96 hours with dipyridamole alone (blue) or in the presence of IC20 (yellow; 1.2 nM for parental cells and 0.75 nM for AZA-R cells) and IC 40 (red; 1.6 nM for parental cells and 1 nM for AZA-R cells) of BAY2402234. Cell growth was measured using a CCK-8 kit. Absorbance of nontreated cells was set as 100%. The results are expressed as the mean ± SD of 3 independent experiments. (E) Graphical schema showing reprogramming of pyrimidine biosynthesis in normal T cells upon T-cell activation (upper panel), and in malignant T cells upon downregulation of UCK2 (which induces resistance to AZA) (bottom panel).
Inhibition of de novo pyrimidine biosynthesis in AZA-R cells showing reduced expression of UCK2. (A) AZA-R cells (red) derived from TL-Om1 and MT-2 cells, and each parental cell line (blue) were treated for 96 hours with BAY2402234, a DHIODH-specific inhibitor. Cell growth was measured using a CCK-8 kit. Absorbance of nontreated cells was set as 100%. The results are expressed as the mean ± SD of 3 independent experiments. (B) Fifty percent inhibitory concentration (IC) values of BAY2402234. Data are expressed as the mean ± SD of 3 independent experiments. ∗P < .05; ∗∗P < .01. (C) An extracellular flux assay was used to measure the OCR in AZA-R cells (red) derived from TL-Om1 cells, as well as parental cells (blue), cultivated under normal growth conditions. Oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, rotenone, and antimycin A were added to the assay. (D) AZA-R cells derived from TL-Om1 and parental cell line were treated for 96 hours with dipyridamole alone (blue) or in the presence of IC20 (yellow; 1.2 nM for parental cells and 0.75 nM for AZA-R cells) and IC 40 (red; 1.6 nM for parental cells and 1 nM for AZA-R cells) of BAY2402234. Cell growth was measured using a CCK-8 kit. Absorbance of nontreated cells was set as 100%. The results are expressed as the mean ± SD of 3 independent experiments. (E) Graphical schema showing reprogramming of pyrimidine biosynthesis in normal T cells upon T-cell activation (upper panel), and in malignant T cells upon downregulation of UCK2 (which induces resistance to AZA) (bottom panel).
Discussion
Here, we demonstrate overexpression of UCK2 but not UCK1 in HTLV-1–infected T cells and ATL cells isolated from primary PBMC samples (Figure 1). UCK proteins catalyze phosphorylation of uridine and cytidine during pyrimidine nucleotide salvage biosynthesis. We also showed that knockdown of UCK2 using shRNA strongly inhibited tumor cell growth, despite intact expression of UCK1 (Figure 3A-B). Although UCK1 and UCK2 show 70% sequence homology, the activity of UCK2 is 15 to 20 times higher than that of UCK1.37 Thus, UCK2 is important specifically for ATL leukemogenesis. As reported previously, expression of UCK2 in normal human tissues is limited to the placenta and testis,17 and was not detected in HTLV-1–uninfected CD4+ T cells (Figure 1E-F).
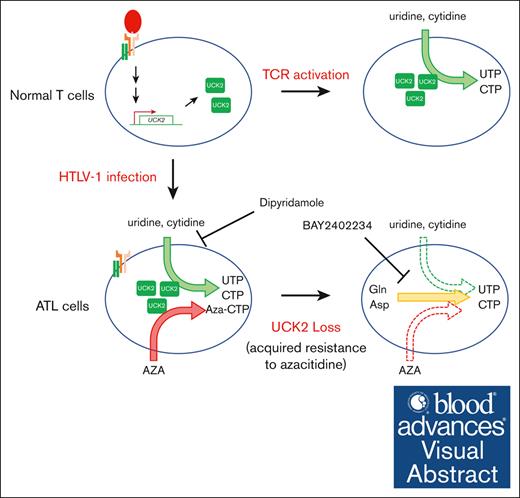
It is well-known that the metabolic profile of T cells is reprogrammed dramatically during differentiation and expansion.38 Here, we found that UCK2 was strongly upregulated in HTLV-1–uninfected CD4+ cells activated via TCR signaling stimulated by anti-CD3/CD28 antibodies (Figure 2C-D). It is important to note that induction of DHODH, a rate-limiting enzyme for de novo synthesis of pyrimidine nucleotides,16 was not as pronounced (Figure 2C-D). Although ATP and amino acids are required for de novo biosynthesis of pyrimidine nucleotides,39 T cells might switch to the more energy-efficient nucleotide salvage pathway to support vigorous proliferation (Figure 6D). Several genetic and epigenetic abnormalities, which activate the TCR-NF-κB signaling pathway in the absence of antigen recognition, accumulate in HTLV-1–infected cells and ATL cells, and this signaling axis is a known driver of ATL leukemogenesis.4,14 Treatment with the NF-κB inhibitor DHMEQ suppressed expression of UCK2 in ATL cells (Figure 2E). Taken together, the data suggest that dysregulated TCR-NF-κB signaling causes overexpression of UCK2 in HTLV-1–infected cells and ATL cells.
Because UCK2 also phosphorylates AZA, it is reasonable that inactivation of UCK2 makes cells resistant to AZA15 (Figure 4A). However, AZA-R cells showing reduced expression of UCK2 grew almost as well as the parental cells (Figure 4B), whereas knockdown of UCK2 by shRNA rapidly and strongly suppressed cell growth in the same background cell lines (Figure 3B). Metabolome analysis revealed that although AZA-R cells accumulated uridine and cytidine, UMP levels were almost identical to those in parental cells. By contrast, the level of DHO decreased, and that of orotate increased, in AZA-R cells (Figure 5). Based on these results, we think that inactivation of UCK2 makes cells resistant to AZA, leading to dysfunction of salvage pyrimidine nucleotide synthesis. Therefore, AZA-R cells showing reduced expression of UCK2 need to increase the de novo synthesis of pyrimidine nucleotides to drive vigorous cell proliferation (Figure 6E). We think that the in vitro culture conditions are nutritionally rich, meaning that cells may still be supplied with pyrimidine nucleotides through de novo pyrimidine nucleotide synthesis even they lack UCK2 expression. By contrast, the subcutaneous microenvironment (in the mouse model) might be nutritionally poor, meaning de novo pyrimidine nucleotide synthesis might not be sufficient to supply the pyrimidine nucleotides needed to drive growth. This may be why AZA-R cells proliferated normally in vitro (Figure 4B) but not in vivo (Figure 4E).
Because inhibition of DHODH shows efficacy against AML cells through induction of differentiation, several specific inhibitors are under development.40 It is important to note that UCK2-downregulated AZA-R cells were more susceptible to the DHODH inhibitor, BAY2402234 (Figure 6A-B). A clinical trial results synopsis of BAY2402234 (NCT03404726) in both patients with AML and MDS/CMML patients reported no objective responses, presumably because inhibition of pyrimidine biosynthesis is compensated by the salvage pathway; therefore, the study was terminated because of the lack of sufficient clinical benefit. It has been reported that bone marrow mononuclear cells from patients with AML who relapsed on AZA treatment show reduced expression of UCK2,41 similar to our in vitro data. Therefore, DHODH inhibitors may show higher efficacy in patients whose tumor cells have lower levels of UCK2 protein. We think that patient stratification may improve the clinical outcome of DHODH inhibitors and that UCK2 may be a useful biomarker for stratification.
It is also important to note that dipyridamole, which inhibits pyrimidine salvage biosynthesis, suppressed tumor cell growth when used in combination with a low-dose BAY2402234 (Figure 6D). Although dipyridamole has been reported to have multiple target proteins, including phosphodiesterases,42 HSP90,43 nucleoside transporters,36 and so on, the efficacy was less in AZA-R TL-Om1 cells (UCK2-deficient cells) compared with parental cells (Figure 6D). We think that the differences in efficacy are due to the differences in dependence on pyrimidine biosynthesis pathways. Although shRNA-mediated knockdown of UCK2 strongly inhibits ATL cell growth (Figure 3C), UCK2-targeted therapy is a potent approach for the treatment of tumor cells, which enhance pyrimidine salvage biosynthesis, such as ATL cells. Although development of DHODH-targeted therapy is ongoing, only a few selective small molecular inhibitors of UCK2 have been identified.44,45 Dipyridamole is an old antiplatelet agent introduced in the early 1960s.46 Thus, we believe that combination therapy with dipyridamole and a low-dose DHODH inhibitor is a potent alternative approach to targeting pyrimidine salvage biosynthesis.
Because UCK2 expression was also upregulated in normal T cells stimulated by treatment with anti-CD3/CD28 antibodies (Figure 2A-D), regulation of pyrimidine nucleotide synthesis through UCK2 might be important to maintain homeostasis within the normal T cell. Therefore, we think that UCK2 is also a potential target for autoimmune diseases and graft-versus-host disease, and that activation of UCK2 could support effective T-cell proliferation in immunotherapy.
In summary, both normal and malignant T cells adapt to the environmental conditions and maintain growth by fine-tuning pyrimidine nucleotide biosynthesis. UCK2 is a key regulator of pyrimidine nucleotide metabolism that supports ATL cell proliferation in ATL leukemogenesis.
Acknowledgments
The authors thank Masao Matsuoka at Kumamoto University and Yasuaki Yamada at Nagasaki University Graduate School of Biomedical Sciences for providing HTLV-1–infected human T-cell lines. Flow cytometry analyses, pyrosequencing, and real-time polymerase chain reaction were conducted at the Analytical Research Center for Experimental Sciences, Saga University.
This work was supported by JSPS KAKENHI, grant numbers JP17H06956 and JP20K07593, by Nippon Shinyaku, by The Shinnihon Foundation of Advanced Medical Treatment Research, and by OHARA Pharmaceutical Co.
Authorship
Contribution: T.W. and S.K. designed the experiments; T.W. conducted most experiments; Y.Y., Y.F.-K., K. Kawasoe, K. Kidoguchi, H.U., K. Kamachi, N.Y.-S., and S.O. contributed to animal experiments and a part of in vitro experiments; T.W., H.N., and E.S. performed experiments using human clinical samples; T.W. and Y.K. performed bioinformatics analyses; T.W. and S.K. obtained funding for this work and T.W. and S.K. wrote the manuscript, with input from all authors.
Conflict-of-interest disclosure: Y.K. and Y.F.-K. are full-time employees of OHARA Pharmaceutical Co. T.W. received grants from Nippon Shinyaku. S.K. has received honoraria from Bristol Myers Squibb, Novartis, Pfizer, and Otsuka Pharmaceuticals; and research funding from Bristol Myers Squibb, Novartis, Pfizer, and Otsuka Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Tatsuro Watanabe, Department of Drug Discovery and Biomedical Sciences, Faculty of Medicine, Saga University, 5-1-1 Nabeshima, Saga, Japan; email: sn6538@cc.saga-u.ac.jp.
References
Author notes
RNA sequencing data are available from DDBJ under accession number DRA016349. Other data sets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
The full-text version of this article contains a data supplement.

![Aberrant overexpression of UCK2 during ATL leukemogenesis. (A-B) Expression of the UCK2 and UCK1 genes in HTLV-1–infected cells and normal T cells was analyzed using GSE55851 data set. HTLV-1–infected T cells (CADM1+/CD7 dimly+ [yellow], and CADM1+/CD7− [red]), and normal T cells (CADM1−/CD+ [blue]), were isolated from healthy volunteers (normal), asymptomatic carriers (AC), and patients with smoldering (S), chronic (C), and acute (A) ATL. (C-D) Expression of the UCK2 and UCK1 genes by PBMCs from healthy volunteers and patients with ATL was analyzed using GSE33615 data set. Normal CD4+ T cells were isolated from healthy volunteers (n = 21). PBMCs were isolated from patients with S (n = 4), C (n = 20), and A (n = 26) ATL. Differences between normal cells and each ATL subtype were assessed using Dunnett test. ∗P < .05. (E) HTLV-1–infected T cells (CADM1+ T cells) and their normal counterparts (CADM1−/CD7+ T cells) were isolated from healthy volunteers (N, n = 3), HTLV-1 carriers (AC, n = 1), and patients with smoldering ATL (S, n = 2), chronic (C, n = 2) and acute ATL (A, n = 2). UCK2 expression was measured by western blotting. (F) UCK2 expression by ATL cell lines and CD3+CD4+CD7+CADM1− T cells (CD4+ T cell) from 3 volunteers was compared by western blotting.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/6/10.1182_bloodadvances.2023011131/2/m_blooda_adv-2023-011131-gr1.jpeg?Expires=1769200349&Signature=QNtuBPY7Lp1AdpYtq4jYvIvGNyvWzpzMZIc0bmguPpAvpGcfpqc3-Z0QYqHcq6-e4UkqxJQBn0zpoPYtPz4tepMeO-SoAotFt0CvlNh7UBDHg-8h3yU2g4uv5zoEf-DUno~ANzlF1hVQ-3WgqiuIIEfV7TzbfCuphWWC5Lj8seqbKdfpwXXktGmz-VBfZAR9pZ0zcZhzU2P~CTk5nMoeuAy1lwJmaNJqJfIsGEccy7tTfOu2ngyBUCohg248Wuj7Zw4VsB3Qf99S~hlsEY1FOTKUKmA9UfajlPK4~ZVdP~A3ZdMz-bv~ok8zbcZlPlhmMkwq6G2v4Yf4kQwGf0jPFg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Growth inhibition induced by knockdown of UCK2. (A) Immunoblots showing reduced expression of UCK2, but not UCK1, protein in TL-Om1 and MT-2 cells at 3 days after transduction with shRNA targeting UCK2 (sh-UCK2), or a scramble shRNA control (Con-sh). (B-C) Cells were stained with trypan blue stain 3 to 5 days after transduction of Con-sh [blue] or sh-UCK2 [red], and the number of live cells was counted. The results are expressed as the mean ± SD of 3 independent experiments. (D) Cell cycle distribution was examined 3 days after transduction. Blue columns, G1; red columns, S; green columns, G2/M. Results are expressed as the mean ± SD of 3 independent experiments. (E) TL-Om1 cells were stained for SA-β-gal at 3 to 5 days after transduction. (F) Principal component analysis plot in gene expression profiles of TL-Om1 [circle] and MT-2 [triangle] cells transduced for 3 days with Con-sh [blue] or sh-UCK2 [red]. (G) Gene set enrichment analysis of the Con-sh and sh-UCK2 group using the Curated (C2) gene sets (Reactome subset of CP) from the Molecular Signatures Database. FDR, False discovery rate; NES, Normalized enrichment score.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/6/10.1182_bloodadvances.2023011131/2/m_blooda_adv-2023-011131-gr3.jpeg?Expires=1769200349&Signature=leieP0pcA13~4gclbve3bnFA1wlbs0h3Vft7HxLHQM8a68XpK7wFWEhGg~UdsY5gQ~yFbSYiXOnSwV3P3SSdZkEJKOvJIdlu3Ccde15B2p4ngFzW1XgQkTbJ4K~BHIOR2C9xACeOooQqLX302O07fS6sGxjxCO6sIQ1b6~HDWc4jRkrpdHu3b6YQaqIwJp0IAl4uDVnRjObdGqoeRBBoK2VDte1xTWCJgFWKUz57PbtJHEVh1XZe8VCEcyP9UvMYUedeMrkRwokG3Ndk5K1xDo~syxbVhnemUNgy0GOJdfQ8g5kL6jsBK0Y4lLB5qWIHm2tdzcqRm-aoL6Av-eFKZQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)