Key Points
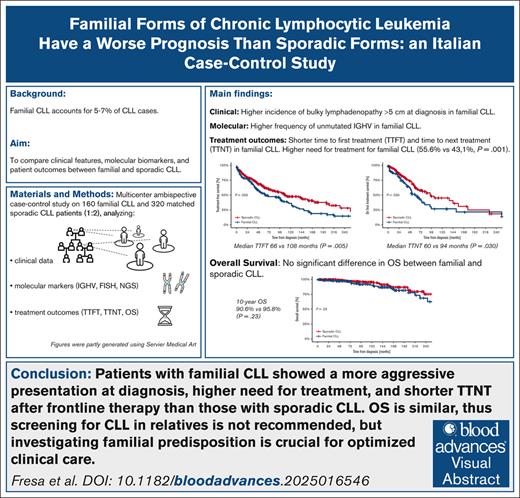
Familial CLL shows more frequent unmutated immunoglobulin heavy chain variable gene status but similar genetic features as sporadic CLL.
Familial CLL has a more aggressive presentation at diagnosis, higher need for treatment, and shorter TTNT but similar OS as sporadic CLL.
Visual Abstract
Familial chronic lymphocytic leukemia (CLL) constitutes 5% to 7% of CLL and has previously shown a more aggressive pattern of evolution than sporadic CLL, even if no difference in overall survival (OS) has been observed. This multicenter case-control study aimed to compare clinical features, molecular biomarkers, and patient outcomes of familial and sporadic CLL. Adult patients with CLL were enrolled from 18 Italian centers, with familial CLL defined as having at least 1 first-degree relative affected by CLL. Patients with sporadic CLL were matched for biological sex and age at diagnosis within 4 years (1:2 ratio). The Kaplan-Meier method was used to evaluate time-to-event outcomes. Of 480 enrolled patients, 160 had familial CLL and 320 sporadic CLL. Significant clinical and molecular differences between familial and sporadic CLL included the presence of lymphadenopathies at diagnosis >5 cm (7.2% vs 2.8%; P = .027) and unmutated immunoglobulin heavy chain variable gene (55.5% vs 36.2%; P < .001). First- and second-line treatments were required in 55.6% and 46.1% of familial CLL and 43.1% (P = .001) and 29.6% of sporadic CLL (P = .034), respectively. Both time to first treatment (TTFT) and time to next treatment (TTNT) were shorter for familial CLL than for sporadic CLL (median TTFT, 66 months vs 108 months; P = .005; median TTNT, 60 months vs 94 months; P = .030, respectively), although familiarity has not emerged as an independent prognostic factor. No difference in OS was observed. Considering the more aggressive course of familial CLL but similar OS, CLL screening in relatives is not recommended.
Introduction
There is evidence that having relatives with chronic lymphocytic leukemia (CLL) or non-Hodgkin lymphomas is a significant risk factor for developing CLL, with an odds ratio ranging from 3.0 to 8.5, compared with individuals without familiarity.1-3 The prevalence of monoclonal B-cell lymphocytosis (MBL) in first-degree relatives of patients with CLL is higher than in the general population (13%-22% vs 5%-12%), and the prevalence of hematological malignancies and CLL in relatives of patients with CLL has been estimated to be 8.6% to 13% and 5.2% to 7%, respectively.4-7
Regarding prognosis, the familial forms of CLL tend to manifest more aggressively than sporadic ones, even if no difference in overall survival (OS) has been observed. A recent prospective study evaluating the natural history of MBL in relatives of patients with CLL8 reported that family members of patients with CLL have a relatively high rate of progression from low-count MBL (clonal lymphocytes in peripheral blood <500/μL) to high-count MBL (clonal lymphocytes in peripheral blood >500/μL but <5000/μL) and from high-count MBL to CLL. In contrast, no evidence of progression from low-count MBL to CLL was observed in an Italian9 and a Spanish cohort10 of nonfamilial patients, indicating the existence of an inherited genetic component of low-count MBL progression in family members of patients with CLL.8
The prognostic impact of a familial trait was evaluated in an Italian retrospective study on 1449 consecutive patients with CLL.5 Familial and sporadic patients showed nonsignificantly different proportions of advanced stages (10.8% vs 7.1%), need of treatment (55% vs 60%), and OS at 10 years (67% vs 66%).5 However, both first- and second-degree relatives were considered as family patients in this study, and no data on molecular characteristics were provided.
Genetic anticipation, known as the phenomenon of earlier onset or increased severity of a disease when it is passed on to the next generation, has been observed in familial CLL, although evidence is mixed. Some studies have reported an earlier disease onset of ∼10 to 20 years and a more severe disease phenotype in younger generations,4,7,11,12 whereas in other studies this phenomenon has not been observed.5
Regarding the biological characteristics of familial CLL, previous studies have reported a higher frequency of mutated immunoglobulin heavy chain variable (IGHV) genes in familial CLL, along with intrafamilial concordance in mutation status.13 Conversely, our previous monocentric study showed a higher prevalence of unmutated IGHV genes in familial (65%) than sporadic CLL (35%).7 In addition, a high prevalence of chromosome 13q deletion (86%) has been observed in familial patients14 compared with a rate of 50% to 60% in sporadic patients.15,16 However, no data regarding the prevalence of somatic mutations in frequently affected CLL driver genes have been reported thus far, and our monocentric experience failed to identify a significant difference between the 2 groups.7
This case-control study, matching patients instead of considering population cohorts, aimed to further characterize the clinical features, molecular biomarkers, and patient outcomes of familial CLL and establish differences with respect to sporadic CLL.
Methods
Study design
This is a retrospective, prospective multicenter study. The observation period was January 1987 to December 2021 for retrospective data and January 2022 to May 2024 for prospective data. The study was performed according to the Declaration of Helsinki, Good Clinical Practice, and applicable national regulations and approved by the ethical committee of Fondazione Policlinico Universitario Agostino Gemelli IRCCS (ID 4858). All the patients provided a written informed consent. Familial CLL was defined as the presence of the diagnosis of CLL and the presence of at least 1 first-degree relative with CLL. Upon referral to the center where patients were monitored and treated, each patient’s familiarity with CLL was ascertained. If patients confirmed familiarity, they were provided with a detailed questionnaire, developed with the assistance of a geneticist, to document the complete family pedigree. The variation in the number of patients across centers corresponds to the catchment area each serves, with a higher number of patients from tertiary and secondary centers. To minimize the risk of selection bias, sporadic patients were exclusively selected from tertiary centers, again by checking with patients for the presence of any known familiarity. For each patient with familial CLL, 2 patients with sporadic CLL were enrolled in the control cohort. The control cohort was matched for sex and age at diagnosis within 4 years. After diagnosis, patients underwent regular comprehensive diagnostic workup, follow-up, and treatment, if necessary, in accordance with their treating physicians’ choices.
A peripheral blood sample and a buccal swab were taken from each patient with familial CLL to be stored for subsequent DNA analysis.
Clinical and biological data from retrospective patients were collected anonymously, except for the information needed to reconstruct the patients’ pedigrees to avoid overlapping patients from the same family. Data collection included demographic information and clinical and laboratory characteristics that were extracted from the patients’ medical records. Data were also collected for a control cohort, which was necessary for comparison from an epidemiological (eg, prevalence of molecular biomarkers) and clinical perspective (eg, aggressiveness of the disease, prognostic impact of molecular biomarkers). In particular, markers of potential prognostic significance, including clinical features, complete blood count, lactate dehydrogenase, kidney and liver function, chromosomal aberrations detected by fluorescence in situ hybridization [del(13q14), tris12, del(11q22), and del(17p13)], recurrent genomic mutations studied by next-generation sequencing (TP53, NOTCH1, SF3B1, and BIRC3), IGHV gene mutational status, and β2-microglobulin level, were recorded.
Statistical analysis
Patient characteristics were described by frequency tables for qualitative variables and position indicators for quantitative variables. The associations between clinical-biological parameters and familiarity were analyzed using χ2 or Fisher exact test for qualitative variables and Mann-Whitney test for quantitative variables. Response rates were calculated based on the number of patients enrolled according to the treatment scheme received. Time to first treatment (TTFT) was calculated from the date of diagnosis to the date of first treatment or death from any cause; patients alive without treatment at the last follow-up were censored. Time to next treatment (TTNT) was calculated from the date of treatment start to the date of subsequent treatment or death from any cause; patients alive without subsequent treatment at the last follow-up were censored. OS was calculated from the date of treatment start to the date of death from any cause; patients alive at the last follow-up were censored. The choice to use TTNT instead of progression-free survival as a time-dependent variable is related to the fact that, because CLL is a chronic disease requiring treatment only in the case of symptomatic progression, the need for retreatment rather than asymptomatic clinical progression might be more representative of the true aggressiveness of the disease. Probabilities of TTFT, TTNT, and OS were estimated by the Kaplan-Meier method; the log-rank test was used to compare expected treatment regimens. Variables associated to prolonged survival in univariate analysis (P < .1) were included in a Cox model for multivariable comparisons. A stepwise forward selection procedure based on Wald statistics was used, setting 0.05 and 0.10 as the entry and removal limits, respectively. Confidence intervals (CIs) were calculated at 95%, all tests are 2 tailed, and differences with P < .05 are considered statistically significant. For the calculations, the software R 4.4.1 (https://www.R-project.org/), the survival (https://CRAN.R-project.org/package=survival), and the survminer packages (https://CRAN.R-project.org/package=survminer) were used together with the IBM SPSS version 28.0.
Results
The present analysis includes 480 patients with CLL diagnosed between January 1987 and December 2021 for retrospective data and between January 2022 and May 2024 for prospective data. The study involved 15 Italian centers (Table 1). Of the 480 patients enrolled, 160 had familial CLL and 320 had sporadic CLL. In both cohorts, 53.7% were male and 46.3% were female. Median age at diagnosis was 63 years for familial CLL (interquartile range [IQR], 52-68) and 62 years for sporadic CLL (IQR, 52-69). Median age at first treatment was 65 years in both groups (IQR, 55-72), and the median observation time of the entire population was 87 months (IQR, 42-155).
Italian centers involved in the study and patients with familial CLL enrolled for each center
| Centers . | Familial CLL, n . |
|---|---|
| Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Rome | 59 |
| A.O.U. Città della Salute e della Scienza, Torino | 26 |
| A.O.U. Careggi, Firenze | 19 |
| ASST Grande Ospedale Metropolitano Niguarda, Milan | 10 |
| Università di Padova, Padova | 9 |
| Università di Bari “Aldo Moro,” Bari | 7 |
| Ospedale Santa Maria Goretti, Latina | 7 |
| Università Sapienza, Rome | 7 |
| Università di Perugia, Perugia | 4 |
| Ospedale Oncologico Armando Businco, ARNAS G. Brotzu, Cagliari | 4 |
| Università del Piemonte Orientale, Novara | 2 |
| Azienda Ospedaliera Santa Maria di Terni, Terni | 2 |
| Ospedale Universitario Sant’Andrea, Rome | 2 |
| Azienda Sanitaria Universitaria Friuli Centrale, Udine | 1 |
| Immunoematologia e Ematologia, P.O. “San Luca,” Salerno | 1 |
| Centers . | Familial CLL, n . |
|---|---|
| Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Rome | 59 |
| A.O.U. Città della Salute e della Scienza, Torino | 26 |
| A.O.U. Careggi, Firenze | 19 |
| ASST Grande Ospedale Metropolitano Niguarda, Milan | 10 |
| Università di Padova, Padova | 9 |
| Università di Bari “Aldo Moro,” Bari | 7 |
| Ospedale Santa Maria Goretti, Latina | 7 |
| Università Sapienza, Rome | 7 |
| Università di Perugia, Perugia | 4 |
| Ospedale Oncologico Armando Businco, ARNAS G. Brotzu, Cagliari | 4 |
| Università del Piemonte Orientale, Novara | 2 |
| Azienda Ospedaliera Santa Maria di Terni, Terni | 2 |
| Ospedale Universitario Sant’Andrea, Rome | 2 |
| Azienda Sanitaria Universitaria Friuli Centrale, Udine | 1 |
| Immunoematologia e Ematologia, P.O. “San Luca,” Salerno | 1 |
A.O.U., Azienda Ospedaliero-Universitaria; ASST, Azienda Socio Sanitaria Territoriale.
Clinically, patients with familial CLL more frequently exhibited bulky lymphadenopathy (>5 cm) at diagnosis than those with sporadic CLL (7.2% [11/152] vs 2.8% [8/290], respectively; P = .027). No differences were observed between the 2 groups at diagnosis regarding disease stage or presence of splenomegaly or hypogammaglobulinemia (Table 2). IGHV gene mutational status was unmutated in 55.5% of patients with familial CLL (61/110) and 36.2% of patients with sporadic CLL (77/213; P < .001). No differences were detected with respect to the presence of common genetic abnormalities, including del(17p), del(11q), tris12, and del(13q) and TP53, NOTCH1, SF3B1, and BIRC3 mutations (Table 2). To note, familial CLL harbored del(17p) (11.3% vs 6.4%), del(11q) (10.4% vs 5.6%), and del(13q) (53% vs 44.6%) more frequently than sporadic CLL, although no statistically significant difference was found. No relationships between lymphadenopathies >5 cm and del(11q) were found in both cohorts.
Clinical and molecular characteristics of patients with familial and sporadic CLL
| . | Sporadic CLL (n = 320) . | Familial CLL (n = 160) . | P value . |
|---|---|---|---|
| Age at diagnosis, median (IQR), y | 62 (52-69) | 63 (52-68) | n.e. |
| Sex, n (%) | n.e. | ||
| M | 172 (53.8) | 86 (53.8) | |
| F | 148 (46.3) | 74 (46.3) | |
| Rai stage at diagnosis, n (%) | .56 | ||
| 0 | 156 (48.8) | 67 (41.9) | |
| I | 61 (19.1) | 38 (23.8) | |
| II | 75 (23.4) | 39 (24.4) | |
| III | 6 (1.9) | 6 (3.8) | |
| IV | 8 (2.5) | 3 (1.9) | |
| Unknown | 14 (4.4) | 7 (4.4) | |
| Binet stage at diagnosis, n (%) | .72 | ||
| A | 223 (69.7) | 111 (69.4) | |
| B | 75 (23.4) | 36 (22.5) | |
| C | 8 (2.5) | 7 (4.4) | |
| Unknown | 14 (4.4) | 6 (3.8) | |
| Splenomegaly, n (%) | 64/283 (22.6) | 41/151 (27.2) | .29 |
| Lymph nodes at diagnosis >5 cm, n (%) | 8/290 (2.8) | 11/152 (7.2) | .027 |
| IgG <400 mg/dL, n (%) | 6/178 (3.4) | 4/95 (4.2) | .72 |
| del(17p), n (%) | 15/235 (6.4) | 13/115 (11.3) | .11 |
| del(13q), n (%) | 104/233 (44.6) | 61/115 (53.0) | .14 |
| del(11q), n (%) | 13/234 (5.6) | 12/115 (10.4) | .10 |
| Trisomy 12, n (%) | 32/234 (13.7) | 9/115 (7.8) | .11 |
| Unmutated IGHV gene, n (%) | 77/213 (36.2) | 61/110 (55.5) | <.001 |
| Mutated TP53, n (%) | 23/166 (13.9) | 13/96 (13.5) | .94 |
| Mutated NOTCH1, n (%) | 30/131 (22.9) | 17/56 (30.4) | .28 |
| Mutated SF3B1, n (%) | 22/127 (17.3) | 4/38 (10.5) | .31 |
| Mutated BIRC3, n (%) | 15/127 (11.8) | 4/37 (10.8) | .87 |
| . | Sporadic CLL (n = 320) . | Familial CLL (n = 160) . | P value . |
|---|---|---|---|
| Age at diagnosis, median (IQR), y | 62 (52-69) | 63 (52-68) | n.e. |
| Sex, n (%) | n.e. | ||
| M | 172 (53.8) | 86 (53.8) | |
| F | 148 (46.3) | 74 (46.3) | |
| Rai stage at diagnosis, n (%) | .56 | ||
| 0 | 156 (48.8) | 67 (41.9) | |
| I | 61 (19.1) | 38 (23.8) | |
| II | 75 (23.4) | 39 (24.4) | |
| III | 6 (1.9) | 6 (3.8) | |
| IV | 8 (2.5) | 3 (1.9) | |
| Unknown | 14 (4.4) | 7 (4.4) | |
| Binet stage at diagnosis, n (%) | .72 | ||
| A | 223 (69.7) | 111 (69.4) | |
| B | 75 (23.4) | 36 (22.5) | |
| C | 8 (2.5) | 7 (4.4) | |
| Unknown | 14 (4.4) | 6 (3.8) | |
| Splenomegaly, n (%) | 64/283 (22.6) | 41/151 (27.2) | .29 |
| Lymph nodes at diagnosis >5 cm, n (%) | 8/290 (2.8) | 11/152 (7.2) | .027 |
| IgG <400 mg/dL, n (%) | 6/178 (3.4) | 4/95 (4.2) | .72 |
| del(17p), n (%) | 15/235 (6.4) | 13/115 (11.3) | .11 |
| del(13q), n (%) | 104/233 (44.6) | 61/115 (53.0) | .14 |
| del(11q), n (%) | 13/234 (5.6) | 12/115 (10.4) | .10 |
| Trisomy 12, n (%) | 32/234 (13.7) | 9/115 (7.8) | .11 |
| Unmutated IGHV gene, n (%) | 77/213 (36.2) | 61/110 (55.5) | <.001 |
| Mutated TP53, n (%) | 23/166 (13.9) | 13/96 (13.5) | .94 |
| Mutated NOTCH1, n (%) | 30/131 (22.9) | 17/56 (30.4) | .28 |
| Mutated SF3B1, n (%) | 22/127 (17.3) | 4/38 (10.5) | .31 |
| Mutated BIRC3, n (%) | 15/127 (11.8) | 4/37 (10.8) | .87 |
Significant associations with familial forms of CLL are set in bold.
F, female; IgG, immunoglobulin G; M, male; n.e., not evaluable.
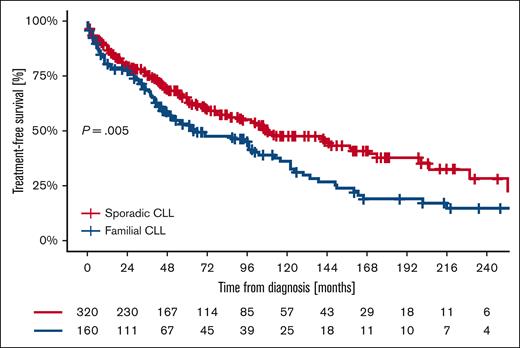
First-line treatment was required in 55.6% of patients with familial CLL (89/160) and 43.1% of patients with sporadic CLL (138/320; P = .001). The median TTFT was 66 months (95% CI, 36.7-95.2) in patients with familial CLL and 108 months (95% CI, 76.0-140.0) in patients with sporadic CLL (P = .005). For familial patients, the proportion of treatment-free patients was 52.4% at 5 years, 36.0% at 10 years, 19.4% at 15 years, and 14.9% at 20 years; for controls, the proportion of treatment-free patients was 65.4% at 5 years, 47.6% at 10 years, 37.8% at 15 years, and 28.7% at 20 years (Figure 1). The variables independently associated with a shorter TTFT in multivariate analysis were stage at diagnosis Rai I to II (hazard ratio [HR], 2.34; 95% CI, 1.55-3.52), stage Rai III to IV (HR, 28.58; 95% CI, 11.42-71.49), splenomegaly (HR, 1.64; 95% CI, 1.11-2.44), lymphadenopathy >5 cm (HR, 8.40; 95% CI, 4.08-17.29), presence of del(11q) (HR, 1.88; 95% CI, 1.05-3.37), and unmutated IGHV gene status (HR, 2.43; 95% CI, 1.68-3.50). Familiarity, despite correlating with shorter TTFT at univariate analysis (HR, 1.58; 95% CI, 1.04-2.39), was not significant in multivariate analysis. (Table 3).
Univariate and multivariable analysis for TTFT
| . | Univariate, HR (95% CI) . | Multivariable, HR (95% CI) . |
|---|---|---|
| Sex | ||
| M | 1.47 (1.13-1.91) | |
| F | Ref. | |
| Age, y | 0.99 (0.98-1.01) | |
| Rai stage at diagnosis | ||
| 0 | Ref. | Ref. |
| I-II | 3.36 (2.49-4.53) | 2.34 (1.55-3.52) |
| III-IV | 23.94 (14.15-40.53) | 28.58 (11.42-71.49) |
| Splenomegaly | ||
| Yes | 3.83 (2.84-5.15) | 1.64 (1.11-2.44) |
| No | Ref. | Ref. |
| Lymph nodes at diagnosis >5 cm | ||
| Yes | 8.70 (5.26-14.39) | 8.40 (4.08-17.29) |
| No | Ref. | Ref. |
| IgG <400 mg/dL | ||
| Yes | 2.23 (0.98-5.08) | |
| No | Ref. | |
| del(17p) | ||
| Yes | 3.00 (1.97-4.57) | |
| No | Ref. | |
| del(13q) | ||
| Yes | 1.15 (0.88-1.51) | |
| No | Ref. | |
| del(11q) | ||
| Yes | 3.17 (2.04-4.94) | 1.88 (1.05-3.37) |
| No | Ref. | Ref. |
| Trisomy 12 | ||
| Yes | 1.37 (0.88-2.12) | |
| No | Ref. | |
| IGHV gene | ||
| Unmutated | 3.36 (2.50-4.53) | 2.43 (1.68-3.50) |
| Mutated | Ref. | Ref. |
| TP53 | ||
| Mutated | 1.70 (1.15-2.51) | |
| WT | Ref. | |
| Familial CLL | ||
| Cases | 1.45 (1.12-1.89) | |
| Controls | Ref. |
| . | Univariate, HR (95% CI) . | Multivariable, HR (95% CI) . |
|---|---|---|
| Sex | ||
| M | 1.47 (1.13-1.91) | |
| F | Ref. | |
| Age, y | 0.99 (0.98-1.01) | |
| Rai stage at diagnosis | ||
| 0 | Ref. | Ref. |
| I-II | 3.36 (2.49-4.53) | 2.34 (1.55-3.52) |
| III-IV | 23.94 (14.15-40.53) | 28.58 (11.42-71.49) |
| Splenomegaly | ||
| Yes | 3.83 (2.84-5.15) | 1.64 (1.11-2.44) |
| No | Ref. | Ref. |
| Lymph nodes at diagnosis >5 cm | ||
| Yes | 8.70 (5.26-14.39) | 8.40 (4.08-17.29) |
| No | Ref. | Ref. |
| IgG <400 mg/dL | ||
| Yes | 2.23 (0.98-5.08) | |
| No | Ref. | |
| del(17p) | ||
| Yes | 3.00 (1.97-4.57) | |
| No | Ref. | |
| del(13q) | ||
| Yes | 1.15 (0.88-1.51) | |
| No | Ref. | |
| del(11q) | ||
| Yes | 3.17 (2.04-4.94) | 1.88 (1.05-3.37) |
| No | Ref. | Ref. |
| Trisomy 12 | ||
| Yes | 1.37 (0.88-2.12) | |
| No | Ref. | |
| IGHV gene | ||
| Unmutated | 3.36 (2.50-4.53) | 2.43 (1.68-3.50) |
| Mutated | Ref. | Ref. |
| TP53 | ||
| Mutated | 1.70 (1.15-2.51) | |
| WT | Ref. | |
| Familial CLL | ||
| Cases | 1.45 (1.12-1.89) | |
| Controls | Ref. |
Variables significantly associated with lower TTFT are set in bold.
Ref., reference; WT, wild type.
The overall response rate to first-line treatment was 79.8% (71/89) in patients with familial CLL and 79.7% (110/138) in patients with sporadic CLL (P = .98). Even when stratifying patients according to the treatment received, there were no significant differences between the 2 cohorts in patients treated with chemotherapy (CT), chemoimmunotherapy (CIT), Bruton tyrosine kinase inhibitor (BTKi), and B-cell lymphoma 2 inhibitor (BCL2i; Table 4).
Overall response rate of patients stratified by the treatment received: CT, CIT, and inhibitors
| . | n . | ORR (%) . | P value . |
|---|---|---|---|
| CT | .47 | ||
| Controls | 15 | 6 (40.0) | |
| Cases | 11 | 6 (54.6) | |
| CIT | .93 | ||
| Controls | 61 | 51 (83.6) | |
| Cases | 41 | 34 (82.9) | |
| Inhibitors | .93 | ||
| Controls | 60 | 52 (86.7) | |
| Cases | 36 | 50 (86.1) |
| . | n . | ORR (%) . | P value . |
|---|---|---|---|
| CT | .47 | ||
| Controls | 15 | 6 (40.0) | |
| Cases | 11 | 6 (54.6) | |
| CIT | .93 | ||
| Controls | 61 | 51 (83.6) | |
| Cases | 41 | 34 (82.9) | |
| Inhibitors | .93 | ||
| Controls | 60 | 52 (86.7) | |
| Cases | 36 | 50 (86.1) |
ORR, overall response rate.
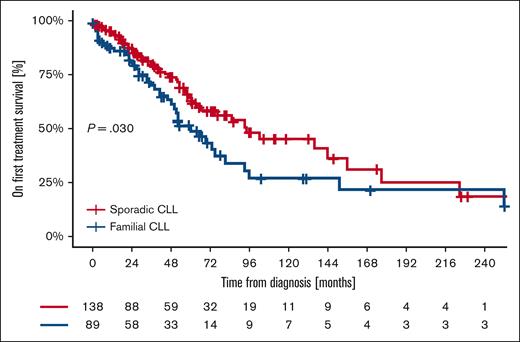
Second-line treatment was required in 46.1% of patients with familial CLL (41/89) vs 29.6% of patients with sporadic CLL (41/138; P = .034). TTNT after frontline therapy was shorter for patients with familial CLL (median TTNT of 60 months [95% CI, 42.9-77.1]) than sporadic CLL (median TTNT of 94 months [95% CI, 43.7-144.3]; P = .030). In particular, for familial patients, the TTNT was 48.9% at 5 years, 27.2% at 10 years, 21.7% at 15 years, and 21.7% at 20 years; for sporadic patients, the TTNT was 64.6% at 5 years, 46.8% at 10 years, 25.7% at 15 years, and 19.3% at 20 years (Figure 2). The only variable independently associated with a shorter TTNT in the multivariable analysis was unmutated IGHV gene status (HR, 2.86; 95% CI, 1.70-4.83; Table 5). Considering the impact of unmutated IGHV genes on outcome, we performed the analysis stratifying patients according to IGHV gene mutational status. The 5-year TTFT was superimposable between familial and sporadic CLL for both mutated (familial 64.7% vs sporadic 70.0%; P = .70) and unmutated IGHV genes (familial 27.1% vs sporadic 38.8%; P = .70). The 5-year TTNT was superimposable between familial and sporadic CLL for mutated IGHV genes (familial 78.1% vs sporadic 79.5%; P = .65), whereas, in patients with unmutated IGHV genes, familial CLL showed a significantly shorter 5-year TTNT than sporadic CLL (familial 31.3% vs sporadic 53.2%; P = .015).
Univariate and multivariable analysis for TTNT
| . | Univariate, HR (95% CI) . | Multivariable, HR (95% CI) . |
|---|---|---|
| Biological sex | ||
| M | 1.07 (0.69-1.64) | |
| F | Ref. | |
| Age, y | 1.01 (0.99-1.03) | |
| Rai stage at diagnosis | ||
| 0 | Ref. | |
| I-II | 1.21 (0.73-2.00) | |
| III-IV | 1.61 (0.78-3.34) | |
| Splenomegaly | ||
| Yes | 0.99 (0.60-1.64) | |
| No | Ref. | |
| Lymph nodes at diagnosis >5 cm | ||
| Yes | 0.78 (0.34-1.80) | |
| No | Ref. | |
| IgG <400 mg/dL | ||
| Yes | 1.70 (0.41-7.11) | |
| No | Ref. | |
| del(17p) | ||
| Yes | 1.71 (0.97-3.03) | |
| No | Ref. | |
| del(13q) | ||
| Yes | 0.98 (0.63-1.53) | |
| No | Ref. | |
| del(11q) | ||
| Yes | 1.70 (0.87-3.31) | |
| No | Ref. | |
| Trisomy 12 | ||
| Yes | 0.50 (0.20-1.25) | |
| No | Ref. | |
| IGHV gene | ||
| Unmutated | 2.86(1.70-4.83) | 2.86 (1.70-4.83) |
| Mutated | Ref. | Ref. |
| TP53 | ||
| Mutated | 1.51 (0.88-2.57) | |
| WT | Ref. | |
| Familial CLL | ||
| Cases | 1.58(1.04-2.39) | |
| Controls | Ref. |
| . | Univariate, HR (95% CI) . | Multivariable, HR (95% CI) . |
|---|---|---|
| Biological sex | ||
| M | 1.07 (0.69-1.64) | |
| F | Ref. | |
| Age, y | 1.01 (0.99-1.03) | |
| Rai stage at diagnosis | ||
| 0 | Ref. | |
| I-II | 1.21 (0.73-2.00) | |
| III-IV | 1.61 (0.78-3.34) | |
| Splenomegaly | ||
| Yes | 0.99 (0.60-1.64) | |
| No | Ref. | |
| Lymph nodes at diagnosis >5 cm | ||
| Yes | 0.78 (0.34-1.80) | |
| No | Ref. | |
| IgG <400 mg/dL | ||
| Yes | 1.70 (0.41-7.11) | |
| No | Ref. | |
| del(17p) | ||
| Yes | 1.71 (0.97-3.03) | |
| No | Ref. | |
| del(13q) | ||
| Yes | 0.98 (0.63-1.53) | |
| No | Ref. | |
| del(11q) | ||
| Yes | 1.70 (0.87-3.31) | |
| No | Ref. | |
| Trisomy 12 | ||
| Yes | 0.50 (0.20-1.25) | |
| No | Ref. | |
| IGHV gene | ||
| Unmutated | 2.86(1.70-4.83) | 2.86 (1.70-4.83) |
| Mutated | Ref. | Ref. |
| TP53 | ||
| Mutated | 1.51 (0.88-2.57) | |
| WT | Ref. | |
| Familial CLL | ||
| Cases | 1.58(1.04-2.39) | |
| Controls | Ref. |
Variables significantly associated with lower TTNT are set in bold.
Stratifying patients according to the treatment received, the median TTNT for patients treated with CT was 33 months (95% CI, 0-70.2) for familial and 67 months (95% CI, 0-158.4) for sporadic patients (P = .86). For patients treated with CIT, the median TTNT was 52 months (95% CI, 30.0-74.0) for familial patients and 94 months (95% CI, 64.7-123.2) for sporadic patients (P = .018). For patients treated with BTKi or BCL2i, the median TTNT was 65 months (95% CI, not evaluable) for familial patients, whereas the median was not reached for sporadic patients (P = .41).
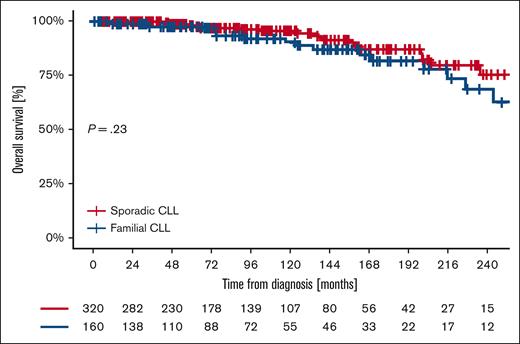
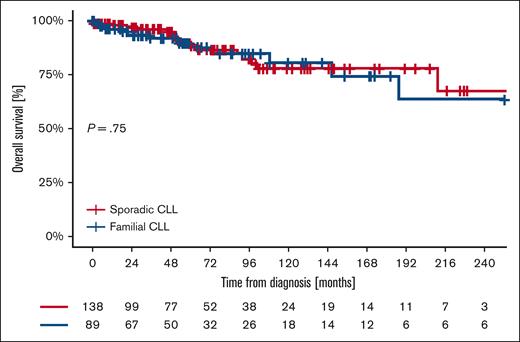
The OS from diagnosis at 5, 10, 15, and 20 years was 97.9%, 90.6%, 82.0%, and 68.6% for familial patients and 98.2%, 95.8%, 87.3%, and 75.4% for sporadic patients (P = .23) (Figure 3). OS since first treatment at 5, 10, 15, and 20 years was 87.8%, 80.8%, 75.0%, and 64.3% for familial patients and 89.7%, 77.1%, 77.1%, and 64.3% for sporadic controls (P = .75; Figure 4). The variables independently associated with a shorter OS in the multivariable analysis were nonmutated IGHV gene mutational status (HR, 3.81; 95% CI, 1.41-10.31) and age at diagnosis (HR, 1.09; 95% CI, 1.03-1.16; Table 6). Comparing OS in different “eras” stratified based on available treatment options, 5- and 10-year OS did not differ between cases and controls in the CT era (1987-2009), the CIT era (2010-2015), or the BTKi/BCL2i era (2016-2024) (Table 7).
Kaplan-Meier of OS from diagnosis of familial and sporadic CLL.
Kaplan-Meier of OS from diagnosis of familial and sporadic CLL.
Kaplan-Meier of OS from the first treatment received of familial and sporadic CLL.
Kaplan-Meier of OS from the first treatment received of familial and sporadic CLL.
Univariate and multivariable analysis for OS
| . | Univariate . | Multivariable . |
|---|---|---|
| Biological sex | ||
| M | 1.24 (0.66-2.33) | |
| F | Ref. | |
| Age | 1.08(1.04-1.12) | 1.09(1.03-1.16) |
| Rai stage at diagnosis | ||
| 0 | Ref. | |
| I-II | 1.23(0.59-2.55) | |
| III-IV | 4.56(1.66-12.51) | |
| Splenomegaly | ||
| Yes | 1.44 (0.64-3.25) | |
| No | Ref. | |
| Lymph nodes at diagnosis >5 cm | ||
| Yes | 0.88 (0.11-7.02) | |
| No | Ref. | |
| IgG <400 mg/dl | ||
| Yes | 5.67 (0.69-46.35) | |
| No | Ref. | |
| del(17p) | ||
| Yes | 0.51 (0.12-2.20) | |
| No | Ref. | |
| del(13q) | ||
| Yes | 0.57 (0.28-1.17) | |
| No | Ref. | |
| del(11q) | ||
| Yes | 2.50 (0.75-8.40) | |
| No | Ref. | |
| Trisomy 12 | ||
| Yes | 0.04 (0-36.83) | |
| No | Ref. | |
| IGHV gene | ||
| Unmutated | 3.86(1.76-8.44) | 3.81 (1.41-10.31) |
| Mutated | Ref. | Ref. |
| TP53 | ||
| Mutated | 2.83(1.20-6.68) | |
| WT | Ref. | |
| Familial CLL | ||
| Cases | 1.46(0.78-2.76) | |
| Controls | Ref. |
| . | Univariate . | Multivariable . |
|---|---|---|
| Biological sex | ||
| M | 1.24 (0.66-2.33) | |
| F | Ref. | |
| Age | 1.08(1.04-1.12) | 1.09(1.03-1.16) |
| Rai stage at diagnosis | ||
| 0 | Ref. | |
| I-II | 1.23(0.59-2.55) | |
| III-IV | 4.56(1.66-12.51) | |
| Splenomegaly | ||
| Yes | 1.44 (0.64-3.25) | |
| No | Ref. | |
| Lymph nodes at diagnosis >5 cm | ||
| Yes | 0.88 (0.11-7.02) | |
| No | Ref. | |
| IgG <400 mg/dl | ||
| Yes | 5.67 (0.69-46.35) | |
| No | Ref. | |
| del(17p) | ||
| Yes | 0.51 (0.12-2.20) | |
| No | Ref. | |
| del(13q) | ||
| Yes | 0.57 (0.28-1.17) | |
| No | Ref. | |
| del(11q) | ||
| Yes | 2.50 (0.75-8.40) | |
| No | Ref. | |
| Trisomy 12 | ||
| Yes | 0.04 (0-36.83) | |
| No | Ref. | |
| IGHV gene | ||
| Unmutated | 3.86(1.76-8.44) | 3.81 (1.41-10.31) |
| Mutated | Ref. | Ref. |
| TP53 | ||
| Mutated | 2.83(1.20-6.68) | |
| WT | Ref. | |
| Familial CLL | ||
| Cases | 1.46(0.78-2.76) | |
| Controls | Ref. |
Variables significantly associated with lower OS are set in bold.
OS stratified by the different “eras” of treatment
| . | n . | OS from the first treatment . | P value . | |
|---|---|---|---|---|
| % at 5 years . | % at 10 years . | |||
| Prerituximab era (1987-2009) | .58 | |||
| Controls | 14 | 92.9 | 85.7 | |
| Cases | 11 | 90.9 | 90.9 | |
| Preinhibitors era (2010-2015) | .89 | |||
| Controls | 28 | 92.9 | 80.9 | |
| Cases | 15 | 86.7 | 78.0 | |
| Inhibitors era (2016-2024) | .98 | |||
| Controls | 94 | 86.4 | n.a. | |
| Cases | 62 | 84.6 | n.a. | |
| . | n . | OS from the first treatment . | P value . | |
|---|---|---|---|---|
| % at 5 years . | % at 10 years . | |||
| Prerituximab era (1987-2009) | .58 | |||
| Controls | 14 | 92.9 | 85.7 | |
| Cases | 11 | 90.9 | 90.9 | |
| Preinhibitors era (2010-2015) | .89 | |||
| Controls | 28 | 92.9 | 80.9 | |
| Cases | 15 | 86.7 | 78.0 | |
| Inhibitors era (2016-2024) | .98 | |||
| Controls | 94 | 86.4 | n.a. | |
| Cases | 62 | 84.6 | n.a. | |
n.a., not available.
Secondary neoplasms occurred in 18.8% of patients with familial CLL (30/160) and 15.9% of patients with sporadic CLL (51/320; P = .44). Richter transformation occurred in 2.0% of familial patients (3/148) and 2.5% of sporadic patients (8/317; P = .74).
Discussion
In this multicenter study, we enrolled 160 patients with familial CLL. To date, this is the study with the largest number of patients with familial CLL that compares their clinical outcome with a cohort of patients with sporadic CLL.
In contrast to previous studies4,5,7 that showed no differences in clinical presentation between consecutive patients with familial and sporadic CLL, this case-control study showed a higher incidence of lymphadenopathy >5 cm at diagnosis in familial than sporadic CLL. The proportion of patients requiring treatment was also higher in familial than in sporadic CLL, as was the time to treatment. These results suggest a more aggressive clinical presentation in familial forms of CLL, previously reported through the phenomenon of anticipation in population studies involving consecutive nonmatched patients.5 This phenomenon was also observed in our previous monocentric study on consecutive patients with CLL at the Fondazione Policlinico Universitario Agostino Gemelli IRCCS, which also showed an earlier onset of the disease in subsequent generations.7
Literature data regarding IGHV gene mutational status present conflicting findings, with both lower13 and higher7 prevalence of unmutated IGHV genes in familial CLL reported. Our analysis revealed a higher proportion of patients presenting with unmutated IGHV genes in familial than in sporadic CLL, consistent with the more aggressive clinical phenotype in patients with familial CLL. Furthermore, although no statistically significant difference was found, the fluorescence in situ hybridization analysis showed a numerically higher percentage of patients with del(17p) and del(11q) in familial than in sporadic CLL (11.3% vs 6.4% and 10.4% vs 5.6%, respectively), which may have also contributed to the more aggressive course in the familial patients. We also observed a higher but statistically not significant percentage of patients with del(13q) among the familial patients (familial CLL, 53% vs sporadic CLL, 44.6%). A higher incidence of del(13q) in familial CLL had also been observed in a previous study reporting 85% of patients with familial CLL harboring del(13q) (11/13), but mostly with an intrafamilial pattern (10/11 patients with the deletion belonged to a total of 3 families).14
The higher aggressiveness of familial CLL also emerged from the survival analysis. Not only did patients with familial CLL require a second treatment more frequently, but the median TTNT was also significantly shorter than in patients with sporadic CLL, although the only factor independently associated with a lower TTNT was the unmutated IGHV gene. The role of the unmutated IGHV gene, which emerged as the only risk factor affecting TTNT in our cohort, probably underlines the fact that its influence on progression outweighs the familiarity, rather than excluding familiarity as a risk factor. This consideration is further corroborated by the analysis conducted through stratification of patients based on IGHV gene mutational status. In particular, in patients with unmutated IGHV genes, familial CLL exhibited a significantly shorter TTNT than sporadic CLL. This finding underscores the potentially greater aggressiveness of familial CLL, even when the IGHV gene variable is controlled for.
Stratifying TTNT based on the treatment received, a statistically significant difference emerged between patients with familial and sporadic CLL treated with CIT (52 vs 94 months; P = .018). To date, this finding has never been reported by any study and further confirms an increased clinical risk in patients with familial CLL. In contrast, no difference was observed in median TTNT between the familial and sporadic cohorts treated with BTKi or BCL2i therapy (65 months vs not reached, respectively; P = .41). Considering the relatively recent introduction of the inhibitors, a longer observation time will be needed to assess the impact of familial CLL on the outcome of patients treated with these drugs.
With regard to OS, no difference was observed between familial and sporadic forms of CLL, which is in line with previously published findings.5 An Italian retrospective study reported a 67% 10-year OS for sporadic and 61% for familial patients.5 The higher 10-year OS observed in our study (90.6% and 95.8% for sporadic and familial patients, respectively) is probably caused by the different “era” at which enrollment was performed, which for the earlier study ended in 2000,5 before the development of anti-CD20 monoclonal antibodies and inhibitors. Given the similar OS between familial and sporadic CLL, we obviously do not recommend screening for familial forms of CLL, but, considering the increased aggressiveness from a clinical point of view, it is important to investigate a possible familial predisposition of patients with CLL to optimize clinical care.
The rate of Richter transformation was not significantly different between familial and sporadic patients (2.0% vs 2.5%), consistent with data from the literature.17 We also did not detect a difference in the rate of secondary malignancies between the 2 populations, in contrast to a previous study reporting more frequent secondary malignancies among the familial CLL patients.17 However, in that study, the comparison data for sporadic CLL were extracted from the Surveillance, Epidemiology, and End Results registry, not allowing direct comparisons from a statistical point of view and not taking into account the inhomogeneity of the analyzed populations. Historically, assessing the risk of developing secondary malignancies in patients already having CLL has always been controversial, owing to the risk of bias in an assessment that must consider multiple possible risk factors. Only recently, a large population study performed on 19 705 consecutive patients showed that the only risk factor significantly associated with the development of secondary malignancies is the use of fludarabine in the treatment of patients with CLL.18 Therefore, the discrepancy between the result observed in our study and that previously reported could be related to a different proportion of patients receiving CT, thus exposing patients to different risk factors for the development of secondary neoplasms.
The main strengths of this study are the large number of familial CLL patients and the homogeneity between the 2 cohorts, achieved by performing a case-control match with patients with sporadic CLL. Limitations of the study are the partly retrospective nature of the data collection and, because no screening for clonality on peripheral blood was performed in relatives of patients with sporadic CLL, the possibility that some patients from the sporadic CLL group may actually have familial CLL.
Whole-exome sequencing analysis to identify possible recurrent gene mutations in patients with familial CLL is currently ongoing, and the results could further help shed light on possible risk factors for familial CLL susceptibility and its increased aggressiveness and identify novel therapeutic targets for this subgroup of patients.
Acknowledgments
The authors thank all of the patients enrolled in this study, their family members and caregivers, and the Ministry of Health-Ricerca Corrente 2024.
Authorship
Contribution: A.F., I.I., and L.L. performed the research; A.F., A. Tomasso, C.V., A.S., A.M.F., A.V., A.R., P.S., A. Giordano, F.P., R. Murru, F.R.M., A.M., F.M., R.L., A. Galitzia, I.A., E.C., R. Moia, G.D., R.P., F.A., J.O., L.S., G.B., A.C., M.I.D.P., A. Tedeschi, M.C., and E.S. collected data; D.G.E. and A.F. performed data analysis; A.F. and L.L. wrote the manuscript; D.G.E., E.S., and L.L. supervised the study; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: A.F. reports honoraria from AstraZeneca, BeiGene, and AbbVie. C.V. reports honoraria from AstraZeneca, AbbVie, and Johnson & Johnson (J&J), and support for attending meetings from AstraZeneca and Takeda. A.M.F. reports travel, accommodations, and expenses from AbbVie and BeiGene, and serves as a consultant for AbbVie, BeiGene, AstraZeneca, and Janssen. A.V. reports membership on an entity's board of directors or advisory committees of AbbVie, J&J, Takeda, and AstraZeneca SpA; research funding from AbbVie, BeiGene, J&J, and AstraZeneca SpA; serves as a consultant for BeiGene and AstraZeneca SpA; and reports participation on speakers' bureaus for BeiGene, J&J, and AstraZeneca SpA. P.S. reports honoraria from and membership on the board of directors or advisory committees of Janssen, AstraZeneca, AbbVie, and BeiGene. F.P. reports honoraria from AstraZeneca and Takeda. F.R.M. reports consultancy for, membership on the board of directors or advisory committees of, and participation in the speakers' bureaus of, AstraZeneca SpA. F. Autore reports honoraria from BeiGene, AstraZeneca, AbbVie, and Janssen. A. Tedeschi reports membership on the board of directors or advisory committees and being on the speakers bureau of AstraZeneca, AbbVie, BeiGene, Janssen, and Lilly. M.C. reports honoraria from AbbVie, AstraZeneca, and Janssen; membership on the board of directors or advisory committees of AbbVie, AstraZeneca, and Janssen; and support for attending meetings from AbbVie, AstraZeneca, and Janssen. L.L. reports research funding from AstraZeneca and AbbVie; honoraria from AstraZeneca, AbbVie, J&J, BeiGene, and Lilly; and membership on the board of directors or advisory committees of AstraZeneca, AbbVie, J&J, BeiGene, and Lilly. The remaining authors declare no competing financial interests.
The current affiliation for F.M. is BeiGene, Milan, Italy.
Correspondence: Luca Laurenti, Fondazione Policlinico Universitario A. Gemelli IRCCS, Istituto di Ematologia, Largo A. Gemelli 8, 00168 Rome, Italy; email: luca.laurenti@unicatt.it.
References
Author notes
A.F. and I.I. contributed equally to this study.
Original data are available on request from the corresponding author, Luca Laurenti (luca.laurenti@unicatt.it).