Key Points
In familial WM, P/LP variants in highly penetrant genes constitute only a modest proportion of the deleterious variant load.
Each WM pedigree is largely unique in its genetic architecture; multiple genes and pathways are likely involved in the etiology of WM.
Visual Abstract
Waldenström macroglobulinemia (WM) is a rare hematological malignancy. Risk for WM is elevated 20-fold among first-degree relatives of patients with WM. However, the list of variants and genes that cause WM remains incomplete. In this study we analyzed exomes from 64 WM pedigrees for evidence of genetic susceptibility for this malignancy. We determined the frequency of pathogenic (P) or likely pathogenic (LP) variants among patients with WM; performed variant- and gene-level association analyses with the set of 166 WM cases and 681 unaffected controls; and examined the segregation pattern of deleterious variants among affected members in each pedigree. We identified P/LP variants in TREX1 and SAMHD1 (genes that function at the interface between innate immune response, genotoxic surveillance, and DNA repair) segregating in patients with WM from 2 pedigrees. There were additional P/LP variants in cancer-predisposing genes (eg, POT1, RECQL4, PTPN11, PMS2). In variant- and gene-level analyses, no associations were statistically significant after multiple testing correction. On a pathway level, we observed involvement of genes that play a role in telomere maintenance (q-value = 0.02), regulation of innate immune response (q-value = 0.05), and DNA repair (q-value = 0.08). Affected members of each pedigree shared multiple deleterious variants (median, n = 18), but the overlap between the families was modest. In summary, P/LP variants in highly penetrant genes constitute a modest proportion of the deleterious variants; each pedigree is largely unique in its genetic architecture, and multiple genes are likely involved in the etiology of WM.
Introduction
Waldenström macroglobulinemia (WM) is a rare hematological malignancy that belongs to the spectrum of plasma cell disorders and is a subtype of lymphoplasmacytic lymphoma (LPL).1-4 Together, WM and LPL account for ∼2% of newly diagnosed non-Hodgkin lymphoma cases in the United States.5 WM has an age-adjusted incidence of 0.36 per 100 000 in the United States, and incidence increases markedly with age.6 It is characterized by monoclonal immunoglobulin M (IgM) in serum and abnormal cells that share characteristics of lymphocytes and monoclonal plasma cells in the bone marrow. The lymph nodes, spleen, and other organs and tissues may be affected.7 The disease is indolent in most patients, often remaining asymptomatic for years.7,8 However, despite recent advances in treatment with the potential for long-term disease control, WM remains incurable.9
A genetic component for WM risk has long been suspected. Familial aggregation of WM has been observed for >60 years.10-12 Family history is the strongest risk factor in epidemiological studies,13 and a population-based registry study of first-degree relatives of patients with WM/LPL documented significantly elevated familial risk for WM (20-fold) as well as for related B-cell malignancies and monoclonal gammopathy of undetermined significance (MGUS).14 Early attempts to identify specific genetic loci, including a linkage study and a candidate gene association approach conducted in high-risk families, were promising and notable in suggesting genetic heterogeneity. Linkage analysis of 11 families with WM at high risk found significant evidence of linkage on chromosomes 1q and 4q and suggestive evidence for chromosomes 3 and 6, providing the first conclusive evidence that IgM MGUS is part of this disease spectrum.15 The study by Liang et al on candidate gene association in 165 unrelated familial cases with WM or related B-cell tumor vs 107 spouse controls reinforced the idea of WM genetic heterogeneity based on identification of associations with multiple genes.16 More recently, a genome-wide association study (GWAS) performed in 530 unrelated WM/LPL cases and 4362 controls of European ancestry17 identified 2 high-risk single-nucleotide variants (SNV) at 6p25.3 and 14q32.13 that together explained 4% of the familial risk. Despite these early promising results, next-generation sequencing efforts to identify rare predisposing genetic variants have been limited. Roccaro et al used exome sequencing to identify potential predisposition alleles in LAPTM5 and HCLS1 that segregated in 3 affected members of a multiplex family with WM.18 Follow-up screening of additional unrelated 246 WM cases identified significantly elevated frequency of these variants in familial cases compared with nonfamilial cases or unaffected controls. Another study identified a novel missense substitution in FHL2 in identical twins, 1 of whom was affected with WM and the other with IgM MGUS but not in their unaffected siblings.19 The FHL2 messenger RNA and protein expression levels were significantly lower in the peripheral blood cells of the patient with WM than in their healthy siblings, suggesting a role for the gene in the WM etiology. Somatic genome sequencing studies revealed an MYD88 c.794C>T (p.L265P) substitution affecting the NF-κB pathway in 91% of patients with WM,20 as well as inactivating variants in ARID1A (17% patients with WM) and CXCR4 (27% of patients with WM).21,22 Multiple highly recurrent copy-number variants were also reported.21
Etiologic heterogeneity for WM is also supported by epidemiological studies that have identified host and environmental WM risk factors, including personal history of autoimmune conditions (Sjögren syndrome and systemic lupus erythematosus); adult weight; hay fever and infections; and occupational exposure and exposures to tobacco smoking, pesticides, wood dust, and organic solvents.13,23
In this study, to better understand the genetic etiology of WM, we analyzed exomes of 64 WM pedigrees with 1 to 8 affected members in each for evidence of genetic susceptibility for this malignancy. Considering the strong familial clustering and the inability of the previous studies to identify highly penetrant variants associated with WM, we hypothesized that multiple low-penetrance variants specific to each family are likely to be involved in the predisposition to this malignancy. We determined the frequency of pathogenic (P) or likely pathogenic (LP) variants among affected members across all families. We performed variant- and gene-level association analyses in WM cases vs unaffected controls. In addition, we examined the segregation pattern of deleterious variants among all affected members in each pedigree. To our knowledge, this is the largest set of WM families reported to date.
Materials and methods
The full version of “Materials and methods” can be found in supplemental Methods.
Patients and sample collection
This study included patients diagnosed with WM, together with their consenting family members, enrolled in the US National Cancer Institute study of individuals and families at high risk of hematolymphoid cancers (ClinicalTrials.gov identifier: NCT00039676), as previously described.15 In addition, families containing at least 2 members with WM or MGUS were enrolled in a study of familial plasma cell dyscrasias (HCL/P 2007.460/3) conducted at the Hospices Civils de Lyon. Each protocol was approved by its respective institutional review board, and all participants provided written informed consent for sample collection and analysis. We included WM, MGUS, and related B-cell tumors as cases for this analysis based on their known coaggregation. For simplicity, in the text we refer to patients with WM and related B-cell disorders as “WM cases.” Patients’ diagnoses and other relevant clinical information are shown in supplemental Table 1. WM cases (n = 173) included all known affected family members. Controls free of cancer (n = 681) were selected from the Prostate, Lung, Colorectal, and Ovarian24 cancer screening trial samples. All pedigrees but 1, and all controls were of European descent.
Exome sequencing
Genomic DNA was extracted from blood using standard methods. DNA was captured with NimbleGen SeqCap EZ Exome Library and sequenced on the Illumina platform (HiSeq 2000/2500/4000 and NextSeq500 instruments).
Exome sequence data processing
The human reference genome and the “known gene” transcript annotation were downloaded from the University of California Santa Cruz database (http://genome.ucsc.edu/), version hg19 (corresponding to Genome Reference Consortium assembly GRCh37). Data processing was performed as described previously.25
Data filtering and variant prioritization
For rare variant analyses, variants with a population frequency of >1% in gnomAD, version 2.1.1, and variants present in this study’s controls at a frequency of >10% were filtered out. Variants with population frequency of >1% and ≤5% in gnomAD, version 2.1.1 were analyzed in common variant analyses. To filter out putative somatic variants, only the variants with variant allele frequencies (VAF) of ≥0.35 and ≤0.70 were considered in the analyses. Remaining variants were further prioritized: (1) variants classified in ClinVar as P or LP; (2) loss-of-function variants including frameshifting deletions and insertions, nonsense, start loss, and canonical splice-sites; (3) missense variants with CADD_phred_score≥25, REVEL_score of ≥0.5 and MetaSVM_score = D (deleterious); (4) variants with deleterious splicing effects as determined by an in silico prediction tool spliceAI, delta_score of ≥0.5 for either acceptor or donor gain or loss; (5) nonframeshifting deletions/insertions and stop-loss variants; and (6) all remaining variants of uncertain significance (VUS).
Statistical tests
Variant- and gene-level analyses were performed on rare coding variants. To address relatedness between the cases, Scalable and Accurate Implementation of Generalized mixed model (SAIGE) and SAIGE-GENE+ (version 1.0.4) were used for association analyses.26,27 Age and sex were used as covariates in the association analyses. Principal component analysis was performed to cluster individuals with shared ancestry (supplemental Figure 1). Multiple testing was adjusted by false discovery rate computation in variant- and gene-based analyses (with the cutoff q-value ≤ 0.05).
Frequency of P/LP variants among affected members across all families (variant frequency visualization with ONCOPRINT)
Variants classified as P/LP in ClinVar were summarized in an oncoprint using associated clinical and genomic characteristics. The oncoprint was generated by using R library “Complex Heatmap.”
Variant segregation pattern analysis in pedigrees
In total, 64 pedigrees that included 1 to 8 affected individuals were analyzed. Prioritized variants present in all affected individuals within each pedigree were further considered.
Candidate gene list compilation
A list of genes that may be involved in the etiology of WM was compiled using results of previously published linkage and GWAS studies, somatic sequencing studies of WM, and studies of germ line variation in tumor predisposition syndromes.28,29 Genes involved in lymphocyte biology were included in the list as well (supplemental Table 2).
Pathway and ontological analysis of genes
Pathway and ontological analyses of gene sets were performed with Enrichr (Gene Ontology, Biological Process, 2021 database).
Results
Overview
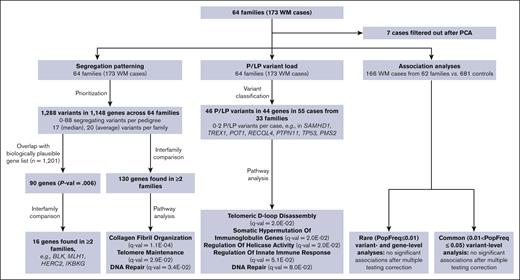
To identify variants and/or genes involved in the etiology of WM we examined the frequency of P or LP variants in the WM cases across 64 pedigrees, determined the segregation pattern of variants among WM affected individuals in each pedigree, and performed association analyses with the exome sequencing data obtained from the germ line DNA of 166 WM cases and 681 unrelated unaffected controls. The data sets and analyses performed in the study are schematically represented in Figure 1.
Schematic representation of analyses and sample sets used in the study. PCA, principal component analysis.
Schematic representation of analyses and sample sets used in the study. PCA, principal component analysis.
P/LP variants in WM cases
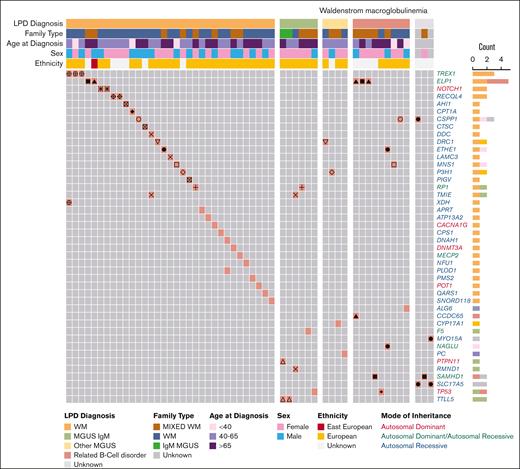
First, we examined the P/LP variant profile in WM cases (Figure 2; Table 1; supplemental Table 3A). We identified 46 P/LP variants in 44 genes in 55 WM cases from 35 pedigrees. Of these, 15 P/LP variants in 24 WM cases from 15 families resided in 14 genes with autosomal dominant (AD), autosomal dominant/autosomal recessive (AD/AR), or X-linked mode of inheritance clinical traits (Tier 1 variants in Table 1; supplemental Table 3A). The remainder of the variants, all of which were heterozygous, were observed in AR genes (Tier 2). There were at most 2 P/LP variants per affected individual. Pathway analysis of the 44 genes with P/LP variants using the Gene Ontology database identified several statistically significant (q-value ≤ 0.05) functional categories including “Telomeric D-loop Disassembly” (RECQL4, POT1, q-value = 2.0E−02), “Somatic Hypermutation Of Immunoglobulin Genes” (PMS2, SAMHD1, q-value = 2.0E−02), “Regulation Of Helicase Activity” (POT1, TP53, q-value = 2.0E−02); and suggestively significant (q-value ≤ 0.10) categories, including “Regulation Of Innate Immune Response” (TREX1, PTPN11, SAMHD1, q-value = 5.1E−02) and “DNA Repair” (RECQL4, TREX1,PMS2, TP53, q-value = 8.0E−02; supplemental Table 3B). Several genes (PTPN11, TP53, POT1, and PMS2) are associated with known AD cancer–predisposition syndromes.
P/LP variants identified in patients with WM from 64 families. Oncoprint representation of 46 P/LP variants residing in 44 genes identified in 55 patients with WM. Variants are shown in rows, and patients are shown in columns. Clinical and demographic metadata are shown on the top of the table. Individuals from the same family are denoted by the same symbols (eg, circles, squares, triangles, etc) in the color-filled squares of the oncoprint. Color code for Count is the same as for LPD Diagnosis. Related B-cell disorder includes Hodgkin lymphoma, non-Hodgkin lymphoma and chronic lymphocytic leukemia. LPD, lymphoproliferative disease.
P/LP variants identified in patients with WM from 64 families. Oncoprint representation of 46 P/LP variants residing in 44 genes identified in 55 patients with WM. Variants are shown in rows, and patients are shown in columns. Clinical and demographic metadata are shown on the top of the table. Individuals from the same family are denoted by the same symbols (eg, circles, squares, triangles, etc) in the color-filled squares of the oncoprint. Color code for Count is the same as for LPD Diagnosis. Related B-cell disorder includes Hodgkin lymphoma, non-Hodgkin lymphoma and chronic lymphocytic leukemia. LPD, lymphoproliferative disease.
P and PL variants in WM families
| Patient ID . | Chr . | Position . | Variant ID . | Ref . | Var . | Gene symbol . | Mode of inheritance . | Variant pathogenicity . | ClinVariant ID . | AF_popmax . | MetaSVM . | REVEL . | CADD phred . | spliceAI . | Tier . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fam20_Pt4 | 17 | 7577568 | rs730882005 | C | A | TP53 | AD | P | 376574 | . | Damaging | 0.958 | 31 | Tolerated | 1 |
| Fam26_Pt2 | 2 | 25469529 | rs1234388246 | G | GC | DNMT3A | AD | P | 985294 | . | . | . | . | Tolerated | 1 |
| Fam41_Pt1 | 9 | 139412682 | rs1290954710 | C | T | NOTCH1 | AD | LP | 445806 | . | Damaging | 0.439 | 28.9 | Tolerated | 1 |
| Fam41_Pt2 | 9 | 139412682 | rs1290954710 | C | T | NOTCH1 | AD | LP | 445806 | . | Damaging | 0.439 | 28.9 | Tolerated | 1 |
| Fam54_Pt2 | 17 | 7577120 | rs28934576 | C | T | TP53 | AD | P | 12366 | . | Damaging | 0.868 | 23.9 | Tolerated | 1 |
| Fam55_Pt1 | 17 | 48703927 | rs767121010 | G | A | CACNA1G | AD | LP | 426883 | . | Tolerated | 0.075 | 23.5 | Tolerated | 1 |
| Fam60_Pt1 | 12 | 112888193 | rs397516801 | A | G | PTPN11 | AD | P | 44603 | . | Damaging | 0.668 | 25.9 | Tolerated | 1 |
| Fam37_Pt1 | 20 | 35555631 | rs515726146 | A | AC | SAMHD1 | AD, AR | P | 126413 | . | . | . | . | Tolerated | 1 |
| Fam37_Pt2 | 20 | 35555631 | rs515726146 | A | AC | SAMHD1 | AD, AR | P | 126413 | . | . | . | . | Tolerated | 1 |
| Fam37_Pt3 | 9 | 111662096 | rs111033171 | A | G | ELP1 | AD, AR | P | 6085 | 0.00006479 | . | . | . | Tolerated | 1 |
| Fam37_Pt4 | 9 | 111662096 | rs111033171 | A | G | ELP1 | AD, AR | P | 6085 | 0.00006479 | . | . | . | Tolerated | 1 |
| Fam2_Pt1 | 9 | 111662096 | rs111033171 | A | G | ELP1 | AD, AR | P | 6085 | 0.00006479 | . | . | . | Tolerated | 1 |
| Fam2_Pt2 | 9 | 111662096 | rs111033171 | A | G | ELP1 | AD, AR | P | 6085 | 0.00006479 | . | . | . | Tolerated | 1 |
| Fam2_Pt4 | 9 | 111662096 | rs111033171 | A | G | ELP1 | AD, AR | P | 6085 | 0.00006479 | . | . | . | Tolerated | 1 |
| Fam3_Pt1 | 3 | 48508395 | rs72556554 | G | A | TREX1 | AD, AR | P | . | 0.0003 | Damaging | 0.828 | 31 | Tolerated | 1 |
| Fam3_Pt2 | 3 | 48508395 | rs72556554 | G | A | TREX1 | AD, AR | P | 4179 | 0.0003 | Damaging | 0.828 | 31 | Tolerated | 1 |
| Fam3_Pt3 | 3 | 48508395 | rs72556554 | G | A | TREX1 | AD, AR | P | 4179 | 0.0003 | Damaging | 0.828 | 31 | Tolerated | 1 |
| Fam24_Pt1 | 17 | 40693092 | rs104894592 | C | T | NAGLU | AD, AR | P | 1562 | 0.0001 | . | . | 35 | Tolerated | 1 |
| Fam32_Pt3 | 7 | 6029587 | rs587780064 | C | A | PMS2 | AD, AR | P | 127802 | . | . | . | 35 | Damaging | 1 |
| Fam44_Pt3 | 1 | 169500043 | rs118203907 | T | C | F5 | AD, AR | P | 649 | . | Damaging | 0.972 | 31 | Tolerated | 1 |
| Fam45_Pt1 | 8 | 55537568 | rs760689800 | C | T | RP1 | AD, AR | P | 1065648 | . | . | . | 25.2 | Tolerated | 1 |
| Fam45_Pt2 | 8 | 55537568 | rs760689800 | C | T | RP1 | AD, AR | P | 1065648 | . | . | . | 25.2 | Tolerated | 1 |
| Fam48_Pt1 | 7 | 124481134 | . | G | T | POT1 | AD, AR | P | 1042147 | . | . | . | 38 | Tolerated | 1 |
| Fam53_Pt1 | X | 153363118 | rs179363901 | G | A | MECP2 | XL | P | 11845 | . | Tolerated | 0.229 | 22.4 | Tolerated | 1 |
| Fam2_Pt1 | 12 | 49312533 | rs1272967209 | GTA | G | CCDC65 | AR | P | 88685 | . | . | . | . | Tolerated | 2 |
| Fam7_Pt2 | 1 | 63868019 | rs199682486 | G | A | ALG6 | AR | P | 95529 | 0.0008 | . | . | . | Damaging | 2 |
| Fam3_Pt3 | 2 | 31596738 | rs148412639 | C | G | XDH | AR | LP | 505602 | 0.0008 | . | . | 34 | Damaging | 2 |
| Fam4_Pt2 | 1 | 17313566 | rs765632065 | AG | A | ATP13A2 | AR | P | 465253 | 0.0012 | . | . | . | Tolerated | 2 |
| Fam6_Pt2 | 15 | 56736015 | rs185005213 | G | A | MNS1 | AR | P | 973691 | 0.0001 | . | . | 37 | Tolerated | 2 |
| Fam6_Pt3 | 15 | 56736015 | rs185005213 | G | A | MNS1 | AR | P | 973691 | 0.0001 | . | . | 37 | Tolerated | 2 |
| Fam10_Pt1 | 8 | 67986544 | rs766020802 | CAA | C | CSPP1 | AR | P | 575640 | 0.0001 | . | . | . | Tolerated | 2 |
| Fam10_Pt3 | 8 | 67986544 | rs766020802 | CAA | C | CSPP1 | AR | P | 575640 | 0.0001 | . | . | . | Tolerated | 2 |
| Fam11_Pt1 | 1 | 43221287 | rs118203996 | G | A | P3H1 | AR | P | 1258 | . | . | . | 36 | Tolerated | 2 |
| Fam11_Pt2 | 1 | 43221287 | rs118203996 | G | A | P3H1 | AR | P | 1258 | . | . | . | 36 | Tolerated | 2 |
| Fam14_Pt2 | 1 | 27121547 | rs139073416 | C | A | PIGV | AR | P | 1284 | 0.0001 | Damaging | 0.83 | 25.1 | Tolerated | 2 |
| Fam14_Pt3 | 6 | 135754219 | rs372659908 | G | A | AHI1 | AR | P | 217525 | 0.00006483 | . | . | 36 | Tolerated | 2 |
| Fam14_Pt6 | 11 | 88070745 | rs587777533 | A | C | CTSC | AR | P | 139655 | 0.0001 | . | . | 36 | Tolerated | 2 |
| Fam19_Pt1 | 2 | 26644264 | rs142371860 | C | T | DRC1 | AR | P | 55840 | 0.0006 | . | . | 36 | Tolerated | 2 |
| Fam19_Pt2 | 2 | 26644264 | rs142371860 | C | T | DRC1 | AR | P | 55840 | 0.0006 | . | . | 36 | Tolerated | 2 |
| Fam20_Pt1 | 11 | 68548130 | rs80356779 | G | A | CPT1A | AR | P | 65644 | 0.0012 | Damaging | 0.785 | 24.8 | Tolerated | 2 |
| Fam22_Pt1 | 10 | 104590667 | rs777638364 | C | T | CYP17A1 | AR | P | 1338524 | . | Damaging | 0.908 | 31 | Tolerated | 2 |
| Fam24_Pt1 | 19 | 44012925 | rs761661864 | A | C | ETHE1 | AR | LP | 577939 | . | . | . | 35 | Damaging | 2 |
| Fam24_Pt2 | 19 | 44012925 | rs761661864 | A | C | ETHE1 | AR | LP | 577939 | . | . | . | 35 | Damaging | 2 |
| Fam27_Pt1 | 8 | 68030481 | rs766633448 | A | C | CSPP1 | AR | LP | 1464866 | . | . | . | 32 | Damaging | 2 |
| Fam27_Pt1 | 6 | 74348214 | rs727504156 | TG | T | SLC17A5 | AR | P | 167693 | 0.0001 | . | . | . | Tolerated | 2 |
| Fam27_Pt2 | 17 | 18058028 | rs184435771 | G | A | MYO15A | AR | P | 228276 | 0.0002 | Damaging | 0.89 | 32 | Tolerated | 2 |
| Fam27_Pt2 | 6 | 74348214 | rs727504156 | TG | T | SLC17A5 | AR | P | 167693 | 0.0001 | . | . | . | Tolerated | 2 |
| Fam29_Pt1 | 16 | 88876475 | rs745594160 | T | TA | APRT | AR | P | 203396 | 0.0001 | . | . | . | Tolerated | 2 |
| Fam34_Pt2 | 17 | 8076835 | rs201558321 | T | C | SNORD118 | AR | P | 929280 | . | . | . | . | Tolerated | 2 |
| Fam35_Pt1 | 3 | 52409343 | rs762545991 | T | TG | DNAH1 | AR | P | 478376 | . | . | . | . | Tolerated | 2 |
| Fam35_Pt1 | 1 | 12026314 | . | AAGG | A | PLOD1 | AR | LP | 14367 | . | . | . | . | Tolerated | 2 |
| Fam38_Pt1 | 8 | 145740366 | rs386833845 | CA | C | RECQL4 | AR | P | 6066 | 0.0012 | . | . | . | Tolerated | 2 |
| Fam38_Pt2 | 8 | 145740366 | rs386833845 | CA | C | RECQL4 | AR | P | 6066 | 0.0012 | . | . | . | Tolerated | 2 |
| Fam40_Pt2 | 11 | 66637890 | rs200030109 | C | A | PC | AR | LP | 203916 | 0.0012 | Damaging | 0.79 | 25.5 | Tolerated | 2 |
| Fam42_Pt1 | 2 | 69627594 | rs374514431 | C | A | NFU1 | AR | P | 30700 | 0.0012 | Damaging | 0.972 | 29.5 | Tolerated | 2 |
| Fam43_Pt1 | 9 | 133946976 | . | AG | A | LAMC3 | AR | LP | 504359 | . | . | . | . | Tolerated | 2 |
| Fam43_Pt2 | 7 | 50544323 | rs201951824 | C | T | DDC | AR | P | 202181 | 0.0012 | Tolerated | 0.381 | 31 | Tolerated | 2 |
| Fam43_Pt2 | 3 | 46747402 | rs375470385 | G | A | TMIE | AR | LP | 504683 | 0.00006479 | . | . | . | Damaging | 2 |
| Fam43_Pt3 | 3 | 46747402 | rs375470385 | G | A | TMIE | AR | LP | 504683 | 0.00006479 | . | . | . | Damaging | 2 |
| Fam43_Pt3 | 6 | 151751289 | rs144972972 | T | C | RMND1 | AR | P | 225255 | 0.0002 | Tolerated | 0.803 | 25.2 | Tolerated | 2 |
| Fam57_Pt2 | 2 | 211460259 | . | G | C | CPS1 | AR | LP | 449391 | . | Damaging | 0.959 | 27.4 | Tolerated | 2 |
| Fam60_Pt1 | 14 | 76201609 | rs541400148 | C | T | TTLL5 | AR | P | 1072044 | 0.0012 | . | . | 33 | Tolerated | 2 |
| Fam60_Pt2 | 14 | 76201609 | rs541400148 | C | T | TTLL5 | AR | P | 1072044 | 0.0012 | . | . | 33 | Tolerated | 2 |
| Fam61_Pt1 | 3 | 49137706 | rs751537797; rs149346696 | G | A | QARS1 | AR | P | 941594 | . | . | . | 34 | Tolerated | 2 |
| Patient ID . | Chr . | Position . | Variant ID . | Ref . | Var . | Gene symbol . | Mode of inheritance . | Variant pathogenicity . | ClinVariant ID . | AF_popmax . | MetaSVM . | REVEL . | CADD phred . | spliceAI . | Tier . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fam20_Pt4 | 17 | 7577568 | rs730882005 | C | A | TP53 | AD | P | 376574 | . | Damaging | 0.958 | 31 | Tolerated | 1 |
| Fam26_Pt2 | 2 | 25469529 | rs1234388246 | G | GC | DNMT3A | AD | P | 985294 | . | . | . | . | Tolerated | 1 |
| Fam41_Pt1 | 9 | 139412682 | rs1290954710 | C | T | NOTCH1 | AD | LP | 445806 | . | Damaging | 0.439 | 28.9 | Tolerated | 1 |
| Fam41_Pt2 | 9 | 139412682 | rs1290954710 | C | T | NOTCH1 | AD | LP | 445806 | . | Damaging | 0.439 | 28.9 | Tolerated | 1 |
| Fam54_Pt2 | 17 | 7577120 | rs28934576 | C | T | TP53 | AD | P | 12366 | . | Damaging | 0.868 | 23.9 | Tolerated | 1 |
| Fam55_Pt1 | 17 | 48703927 | rs767121010 | G | A | CACNA1G | AD | LP | 426883 | . | Tolerated | 0.075 | 23.5 | Tolerated | 1 |
| Fam60_Pt1 | 12 | 112888193 | rs397516801 | A | G | PTPN11 | AD | P | 44603 | . | Damaging | 0.668 | 25.9 | Tolerated | 1 |
| Fam37_Pt1 | 20 | 35555631 | rs515726146 | A | AC | SAMHD1 | AD, AR | P | 126413 | . | . | . | . | Tolerated | 1 |
| Fam37_Pt2 | 20 | 35555631 | rs515726146 | A | AC | SAMHD1 | AD, AR | P | 126413 | . | . | . | . | Tolerated | 1 |
| Fam37_Pt3 | 9 | 111662096 | rs111033171 | A | G | ELP1 | AD, AR | P | 6085 | 0.00006479 | . | . | . | Tolerated | 1 |
| Fam37_Pt4 | 9 | 111662096 | rs111033171 | A | G | ELP1 | AD, AR | P | 6085 | 0.00006479 | . | . | . | Tolerated | 1 |
| Fam2_Pt1 | 9 | 111662096 | rs111033171 | A | G | ELP1 | AD, AR | P | 6085 | 0.00006479 | . | . | . | Tolerated | 1 |
| Fam2_Pt2 | 9 | 111662096 | rs111033171 | A | G | ELP1 | AD, AR | P | 6085 | 0.00006479 | . | . | . | Tolerated | 1 |
| Fam2_Pt4 | 9 | 111662096 | rs111033171 | A | G | ELP1 | AD, AR | P | 6085 | 0.00006479 | . | . | . | Tolerated | 1 |
| Fam3_Pt1 | 3 | 48508395 | rs72556554 | G | A | TREX1 | AD, AR | P | . | 0.0003 | Damaging | 0.828 | 31 | Tolerated | 1 |
| Fam3_Pt2 | 3 | 48508395 | rs72556554 | G | A | TREX1 | AD, AR | P | 4179 | 0.0003 | Damaging | 0.828 | 31 | Tolerated | 1 |
| Fam3_Pt3 | 3 | 48508395 | rs72556554 | G | A | TREX1 | AD, AR | P | 4179 | 0.0003 | Damaging | 0.828 | 31 | Tolerated | 1 |
| Fam24_Pt1 | 17 | 40693092 | rs104894592 | C | T | NAGLU | AD, AR | P | 1562 | 0.0001 | . | . | 35 | Tolerated | 1 |
| Fam32_Pt3 | 7 | 6029587 | rs587780064 | C | A | PMS2 | AD, AR | P | 127802 | . | . | . | 35 | Damaging | 1 |
| Fam44_Pt3 | 1 | 169500043 | rs118203907 | T | C | F5 | AD, AR | P | 649 | . | Damaging | 0.972 | 31 | Tolerated | 1 |
| Fam45_Pt1 | 8 | 55537568 | rs760689800 | C | T | RP1 | AD, AR | P | 1065648 | . | . | . | 25.2 | Tolerated | 1 |
| Fam45_Pt2 | 8 | 55537568 | rs760689800 | C | T | RP1 | AD, AR | P | 1065648 | . | . | . | 25.2 | Tolerated | 1 |
| Fam48_Pt1 | 7 | 124481134 | . | G | T | POT1 | AD, AR | P | 1042147 | . | . | . | 38 | Tolerated | 1 |
| Fam53_Pt1 | X | 153363118 | rs179363901 | G | A | MECP2 | XL | P | 11845 | . | Tolerated | 0.229 | 22.4 | Tolerated | 1 |
| Fam2_Pt1 | 12 | 49312533 | rs1272967209 | GTA | G | CCDC65 | AR | P | 88685 | . | . | . | . | Tolerated | 2 |
| Fam7_Pt2 | 1 | 63868019 | rs199682486 | G | A | ALG6 | AR | P | 95529 | 0.0008 | . | . | . | Damaging | 2 |
| Fam3_Pt3 | 2 | 31596738 | rs148412639 | C | G | XDH | AR | LP | 505602 | 0.0008 | . | . | 34 | Damaging | 2 |
| Fam4_Pt2 | 1 | 17313566 | rs765632065 | AG | A | ATP13A2 | AR | P | 465253 | 0.0012 | . | . | . | Tolerated | 2 |
| Fam6_Pt2 | 15 | 56736015 | rs185005213 | G | A | MNS1 | AR | P | 973691 | 0.0001 | . | . | 37 | Tolerated | 2 |
| Fam6_Pt3 | 15 | 56736015 | rs185005213 | G | A | MNS1 | AR | P | 973691 | 0.0001 | . | . | 37 | Tolerated | 2 |
| Fam10_Pt1 | 8 | 67986544 | rs766020802 | CAA | C | CSPP1 | AR | P | 575640 | 0.0001 | . | . | . | Tolerated | 2 |
| Fam10_Pt3 | 8 | 67986544 | rs766020802 | CAA | C | CSPP1 | AR | P | 575640 | 0.0001 | . | . | . | Tolerated | 2 |
| Fam11_Pt1 | 1 | 43221287 | rs118203996 | G | A | P3H1 | AR | P | 1258 | . | . | . | 36 | Tolerated | 2 |
| Fam11_Pt2 | 1 | 43221287 | rs118203996 | G | A | P3H1 | AR | P | 1258 | . | . | . | 36 | Tolerated | 2 |
| Fam14_Pt2 | 1 | 27121547 | rs139073416 | C | A | PIGV | AR | P | 1284 | 0.0001 | Damaging | 0.83 | 25.1 | Tolerated | 2 |
| Fam14_Pt3 | 6 | 135754219 | rs372659908 | G | A | AHI1 | AR | P | 217525 | 0.00006483 | . | . | 36 | Tolerated | 2 |
| Fam14_Pt6 | 11 | 88070745 | rs587777533 | A | C | CTSC | AR | P | 139655 | 0.0001 | . | . | 36 | Tolerated | 2 |
| Fam19_Pt1 | 2 | 26644264 | rs142371860 | C | T | DRC1 | AR | P | 55840 | 0.0006 | . | . | 36 | Tolerated | 2 |
| Fam19_Pt2 | 2 | 26644264 | rs142371860 | C | T | DRC1 | AR | P | 55840 | 0.0006 | . | . | 36 | Tolerated | 2 |
| Fam20_Pt1 | 11 | 68548130 | rs80356779 | G | A | CPT1A | AR | P | 65644 | 0.0012 | Damaging | 0.785 | 24.8 | Tolerated | 2 |
| Fam22_Pt1 | 10 | 104590667 | rs777638364 | C | T | CYP17A1 | AR | P | 1338524 | . | Damaging | 0.908 | 31 | Tolerated | 2 |
| Fam24_Pt1 | 19 | 44012925 | rs761661864 | A | C | ETHE1 | AR | LP | 577939 | . | . | . | 35 | Damaging | 2 |
| Fam24_Pt2 | 19 | 44012925 | rs761661864 | A | C | ETHE1 | AR | LP | 577939 | . | . | . | 35 | Damaging | 2 |
| Fam27_Pt1 | 8 | 68030481 | rs766633448 | A | C | CSPP1 | AR | LP | 1464866 | . | . | . | 32 | Damaging | 2 |
| Fam27_Pt1 | 6 | 74348214 | rs727504156 | TG | T | SLC17A5 | AR | P | 167693 | 0.0001 | . | . | . | Tolerated | 2 |
| Fam27_Pt2 | 17 | 18058028 | rs184435771 | G | A | MYO15A | AR | P | 228276 | 0.0002 | Damaging | 0.89 | 32 | Tolerated | 2 |
| Fam27_Pt2 | 6 | 74348214 | rs727504156 | TG | T | SLC17A5 | AR | P | 167693 | 0.0001 | . | . | . | Tolerated | 2 |
| Fam29_Pt1 | 16 | 88876475 | rs745594160 | T | TA | APRT | AR | P | 203396 | 0.0001 | . | . | . | Tolerated | 2 |
| Fam34_Pt2 | 17 | 8076835 | rs201558321 | T | C | SNORD118 | AR | P | 929280 | . | . | . | . | Tolerated | 2 |
| Fam35_Pt1 | 3 | 52409343 | rs762545991 | T | TG | DNAH1 | AR | P | 478376 | . | . | . | . | Tolerated | 2 |
| Fam35_Pt1 | 1 | 12026314 | . | AAGG | A | PLOD1 | AR | LP | 14367 | . | . | . | . | Tolerated | 2 |
| Fam38_Pt1 | 8 | 145740366 | rs386833845 | CA | C | RECQL4 | AR | P | 6066 | 0.0012 | . | . | . | Tolerated | 2 |
| Fam38_Pt2 | 8 | 145740366 | rs386833845 | CA | C | RECQL4 | AR | P | 6066 | 0.0012 | . | . | . | Tolerated | 2 |
| Fam40_Pt2 | 11 | 66637890 | rs200030109 | C | A | PC | AR | LP | 203916 | 0.0012 | Damaging | 0.79 | 25.5 | Tolerated | 2 |
| Fam42_Pt1 | 2 | 69627594 | rs374514431 | C | A | NFU1 | AR | P | 30700 | 0.0012 | Damaging | 0.972 | 29.5 | Tolerated | 2 |
| Fam43_Pt1 | 9 | 133946976 | . | AG | A | LAMC3 | AR | LP | 504359 | . | . | . | . | Tolerated | 2 |
| Fam43_Pt2 | 7 | 50544323 | rs201951824 | C | T | DDC | AR | P | 202181 | 0.0012 | Tolerated | 0.381 | 31 | Tolerated | 2 |
| Fam43_Pt2 | 3 | 46747402 | rs375470385 | G | A | TMIE | AR | LP | 504683 | 0.00006479 | . | . | . | Damaging | 2 |
| Fam43_Pt3 | 3 | 46747402 | rs375470385 | G | A | TMIE | AR | LP | 504683 | 0.00006479 | . | . | . | Damaging | 2 |
| Fam43_Pt3 | 6 | 151751289 | rs144972972 | T | C | RMND1 | AR | P | 225255 | 0.0002 | Tolerated | 0.803 | 25.2 | Tolerated | 2 |
| Fam57_Pt2 | 2 | 211460259 | . | G | C | CPS1 | AR | LP | 449391 | . | Damaging | 0.959 | 27.4 | Tolerated | 2 |
| Fam60_Pt1 | 14 | 76201609 | rs541400148 | C | T | TTLL5 | AR | P | 1072044 | 0.0012 | . | . | 33 | Tolerated | 2 |
| Fam60_Pt2 | 14 | 76201609 | rs541400148 | C | T | TTLL5 | AR | P | 1072044 | 0.0012 | . | . | 33 | Tolerated | 2 |
| Fam61_Pt1 | 3 | 49137706 | rs751537797; rs149346696 | G | A | QARS1 | AR | P | 941594 | . | . | . | 34 | Tolerated | 2 |
Variants are shown in GRCh37 genomic coordinates. AF_popmax - highest population allele frequency in gnomAD version 3. Tier 1 variants include variants in the genes associated with either AD or AD, AR, or XL mode of inheritance (highlighted in bold face); tier 2 variants include variants found in AR genes.
Chr, chromosome; Fam, family; ID, identification; Pt, patient; Ref, reference.
Segregation pattern of prioritized variants in 64 WM pedigrees
Next, we investigated whether any variants and genes were shared by all affected members in each of 64 WM families. The number of affected members per family available for analysis ranged from 1 to 8. Thirty pedigrees included 2 affected members, 5 pedigrees included a single affected individual, 1 pedigree had 8 individuals, and the rest of the families included 3 to 5 individuals with WM or other B-cell neoplasms (supplemental Figure 3). In aggregation, there were 30 040 shared variants in 12 506 genes across all 64 families. To further prioritize segregating variants, we selected P/LP ClinVar variants, putative loss-of-function, missense variants exceeding threshold in silico scores, noncanonical splicing variants with spliceAI delta_score of ≥0.5, nonframeshifting deletions/insertions, stop-loss variants, and all remaining VUSs for subsequent analysis. This prioritization resulted in 1288 variants in 1148 genes across 59 families (in 5 families there were no shared variants after prioritization; supplemental Table 4).
Next, we investigated whether there were genes harboring prioritized variants shared by multiple pedigrees (supplemental Table 5A). We found 130 such genes present in ≥2 families. Variants in ZC3H18 were shared by 4 families; variants in GLMN, CP, and VWA2 were shared by 3 families, and the rest were shared by 2 families. Pathway analysis of these genes identified highly statistically significant enrichment for collagen fibril organization (q-value = 1.1E−04). We also observed significant association with telomere maintenance (q-value = 2.9E−02) and DNA Repair genes (q-value = 3.4E−02; supplemental Table 5B).
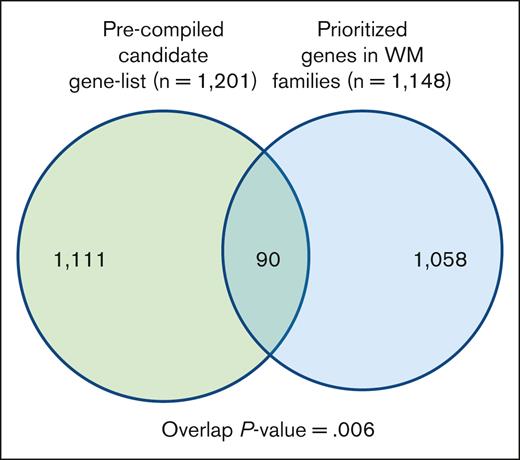
To gain additional insight into biological relevance of the prioritized variants/genes segregating in the families, we compiled a list of genes known for their involvement in WM, other lymphoproliferative diseases (LPDs), lymphocyte biology, and tumor predisposition disorders (supplemental Table 2). We then performed a hypergeometric test for the significance of the overlap between the 2 gene sets (Figure 3; supplemental Table 6). There were 90 genes in the overlap, which was statistically significant (P value = .006). Of 90 genes, 16 genes including BLK, MLH1, HERC2, and IKBKG were shared by multiple families.
Overlap between a precompiled list of genes related to WM biology. The list of segregating prioritized genes in families with WM.
Overlap between a precompiled list of genes related to WM biology. The list of segregating prioritized genes in families with WM.
In 59 of 64 pedigrees that shared prioritized variants, we observed 21.8 variants per family on average (median, 18; range, 1-88). Most of the pedigrees (51 of 59, 86.4%) shared at least 1 variant or a variant-carrying gene with at least 1 other pedigree.
Association analysis in 166 WM cases vs 681 controls, rare variants (population frequency ≤ 0.01)
Next, we analyzed the association of single variants with the malignancy using the SAIGE statistical package. SAIGE methodology considers relatedness between samples, which facilitates inclusion of multiple related individuals from the same family, thus increasing the analyses’ power. Before the analysis, 7 samples that did not cluster closely with the rest of the samples on principal component analysis were filtered out, thus leaving 166 WM cases, which were compared with 681 unrelated controls. There were no variants significant after multiple testing correction (supplemental Table 7A). Among the variants with nominal P values <.001, we identified 5 that resided in biologically plausible genes including PTPRK, ITGA1, PDGFB, FLT3LG, and TOPBP1. On the gene-level SAIGE analysis (supplemental Table 7B; supplemental Figure 2), there were no significant associations after multiple testing correction. Thirty-nine genes were associated with WM at a nominal P value <.01. As in the variant-level analysis, PDGFB was also found among the genes that were nominally associated with WM (supplemental Table 7B). Other biologically plausible genes included EXO1 (DNA repair), IGLL5 (B-cell receptor signaling), and RBPJ (ERBB signaling).
Association analyses in 166 WM cases vs 681 controls, common variants (0.01 < population frequency ≤ 0.05)
Association analysis of common variants with WM resulted in no significant associations after multiple testing correction (supplemental Table 7C). A single SNV (rs9838238) in DCBLD2 (discoidin, CUB and LCCL domain containing 2) was nearly significant after Bonferroni correction. The function of this gene is incompletely understood, and the consequences of this substitution appear to be modest as reflected by in silico scores, such as Combined Annotation Dependent Depletion (CADD) (22.3), Rare Exome Variant Ensemble Learner (REVEL) (0.177), and MetaSVM (tolerated; supplemental Table 7C).
Discussion
In this study, we exome-sequenced 64 WM pedigrees, most of which had at least 3 affected members. To our knowledge, this is the largest set of WM families reported to date. We analyzed the sequenced data to identify genetic risk factors for WM. We examined the frequency of P/LP variants among affected members across all families, performed variant- and gene-level association analyses in 166 WM cases vs 681 unaffected controls, and investigated the segregation pattern of deleterious variants in each pedigree.
The P/LP analysis in the families identified several biologically plausible genes. We observed a P/LP variant in TREX1 in 3 patients with WM from a single pedigree, and a P/LP variant in SAMHD1 in 2 patients with WM from a different family. These 2 genes are associated with Aicardi-Goutières syndrome (AGS, Online Mendelian Inheritance in Man [OMIM] nos. 225750, 612952), a rare disorder affecting the brain, immune system, and the skin.30 TREX1 is a 3′-to-5′ exonuclease that degrades both single- and double-stranded free cytoplasmic DNA fragments, thus regulating the amount of interferon-stimulatory DNA present in the cell and suppressing the senescence-associated secretory phenotype, a process known to turn a senescent cell into a proinflammatory cell with a potential for tumor initiation.31 Another AGS-associated gene, SAMHD1, encodes a protein that functions at the interface between inflammation and DNA repair: it is involved in innate immune response to viruses via regulation of deoxyribonucleotide triphosphare (dNTP) pools, as well as in repairing DNA via stimulation of the exonuclease activity of MRE11 at the sites of stalled replication forks.31 Both proteins function in cytosolic DNA-sensing pathway (https://www.kegg.jp/pathway/hsa04623), which intersects with the larger NF-κB signal transduction network including MYD88, the most frequently somatically mutated gene in WM.20 The P/LP variants in SAMHD1 (rs515726146) and TREX1 (rs72556554) identified in this study were heterozygous, whereas in patients with AGS these SNVs were observed in homozygous or compound heterozygous states.32-34 It is therefore unlikely that heterozygous rs515726146 or rs72556554 would cause symptoms of AGS, a predominantly AR disease.35 However, the rs515726146 in TREX1 was reported in a heterozygous state as a P/LP variant associated with systemic lupus erythematosus,36,37 a clinically related disorder of an aberrant immune response and a well-known risk factor for WM. It is noteworthy that 2 rare variants in the genes functioning in the same cellular process and associated with the same rare genetic disease were identified in multiple patients from multiple WM pedigrees. These findings warrant further investigation of possible roles of TREX1 and SAMHD1 in the etiology of WM.
In addition, we identified P/LP variants in the genes associated with telomere maintenance and DNA repair, including POT1 (protection of telomeres 1) and RECQL4 (RecQ-like helicase 4), in patients with WM from 2 pedigrees. POT1 tumor predisposition disorder is AD and is associated with a lifetime increased risk of several solid malignancies as well as chronic lymphocytic leukemia.38 Biallelic inactivating variants in RECQL4 are associated with several AR disorders (Rothmund-Thomson syndrome, Baller-Gerold syndrome, and RAPADILINO [radial ray malformations, patella and palate abnormalities, diarrhea and dislocated joints, limb abnormalities and little size, and slender nose and normal intelligence] syndrome, predisposing affected individuals to multiple malignancies, including lymphomas [https://www.ncbi.nlm.nih.gov/books/NBK1237/]).
PTPN11 and TP53 are well-known genetic disorder-causing genes involved in the etiology of AD Noonan (OMIM no. 163950) and Li-Fraumeni (OMIM no. 151623) syndromes, respectively. Cancer risk may be elevated in Noonan syndrome compared with the general population and is enriched for hematological malignancies.39 Li-Fraumeni is a tumor-predisposing disorder and is associated with multiple solid tumors as well as blood malignancies, including acute lymphoblastic leukemi40; however, TP53 is one of the most frequently mutated genes that are associated with clonal hematopoiesis of indeterminate potential41 and, therefore, the possibility of the somatic origin of 2 TP53 P/LP variants in 2 patients with WM cannot be excluded. Moreover, an association of P/LP somatic changes in TP53 with progression and unfavorable prognosis in WM has been reported.42 Of note, before the analyses, all variants were filtered by their VAF (≥0.35 and ≤0.70) and the variants in PTPN11 (rs39751680, VAF = 0.43) and TP53 (rs730882005, VAF = 0.62; and rs28934576, VAF = 0.69) were within this range.
Pathogenic variants in PMS2 are associated with AD Lynch syndrome (OMIM no. 614337) and AR constitutional mismatch repair deficiency syndrome (CMMRDS, OMIM no. 619101). Lynch syndrome is typically associated with solid malignancies (colorectal, endometrium, ovary, stomach, small bowel, and few others), whereas hematological neoplasms have not been traditionally associated with this disorder.43 In contrast, leukemia and lymphomas are frequent in patients with CMMRDS44; however, a P/LP PMS2 variant identified in 1 of the patients with WM was heterozygous, and no other P/LP variants in this gene were detected in that patient.
The P/LP variants in TREX1, SAMHD1, RECQL4, PTPN11, TP53 (rs28934576), and PMS2 are listed in both ClinVar and The Human Gene Mutation Database, and the p.(Ser421∗) nonsense substitution in POT1 and p.(Cys238Phe) missense variant in TP53 (rs730882005) are listed in ClinVar only. In the literature, all of these variants were reported in patients with associated disorders. We also examined available clinical records for this study’s patients with WM but did not identify specific symptoms suggestive of AGS, Noonan, Li-Fraumeni, Lynch, CMMRDS, Rothmund-Thomson, Baller-Gerold, or RAPADILINO syndromes. However, it should be noted that in a substantial subset, the available clinical information was limited to blood malignancies and clinical follow-up for all patients was not feasible. In addition, a literature search did not demonstrate association of WM/LPL with these disorders.
In the segregation patterning analyses of the WM pedigrees, a comparison of the list of genes carrying prioritized segregating variants with the list of biologically plausible genes (supplemental Table 2) showed an overlap of 90 genes, 16 of which were found in multiple families. Most of these 16 genes’ functions are related to B-cell biology and immunoregulation. For instance, BLK (B-lymphoid tyrosine kinase proto-oncogene) and PRF1 (perforin 1) are highly specifically expressed in the bone marrow, spleen, and lymph nodes, and play important roles in B-cell receptor signaling, B-cell development, and in lymphoid malignancies. Finally, a pathway analysis of the set of 130 genes harboring prioritized variants that were observed in ≥2 pedigrees demonstrated a statistically significant overlap (q-value ≤ 0.05) with telomere maintenance (RTEL1 and SLX4) and DNA repair genes (HERC2, MC1R, SLX4, IGHMBP2, ERCC5, ACTR8, MLH1, and FAN1).
The etiology of WM initiation and progression is largely unknown. Most WM cases are associated with a single somatic point mutation in MYD88 and with substantial chromosomal instability.20-22 In this study, we did not observe the p.(Leu265Pro) MYD88 P/LP variant in any of the WM cases, thus indirectly confirming its somatic origin; however, in pathway analysis, we identified significant enrichment in genes involved in telomere maintenance and DNA repair processes. A possible mechanism of WM development may be dependent on an increased rate of accumulation of somatic hits or chromosomal rearrangements in hematopoietic cells due to the presence of constitutive defects in genes that control proper DNA repair or chromosome maintenance. A modulating mutation or copy-number variant (CNV) in a driver gene (eg, MYD88, ARID1A, and CXCR4) may confer a proliferative advantage to a single cell thus initiating a clonal tumor progression.
SAIGE association analysis identified a variant in PDGFB (rs143980537, P value = 1.44 × 10−5), which was observed 5 times in 166 cases (5 heterozygous patients with WM in 3 pedigrees) but was not found in any of the 681 controls. This is a rare variant, it is classified as a VUS in ClinVar (VarID 2264365), has a highly conserved Genomic Evolutionary Rate Profiling (GERP++) score, and resides in the PDGF/VEGF domain; however, it has a mixture of high and low of in silico prediction scores (eg, CADD = 25.2, but REVEL = 0.122). PDGFB encodes platelet-derived growth factor B and is moderately expressed in the spleen and lymph nodes. It is a known oncogene associated with tumorigenesis in multiple tissues and organs, but its rate of somatic mutations in hematopoietic and lymphoid tissues is relatively low (0.22%) compared with other tissues (https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=PDGFB#tissue). The gene-level SAIGE analysis also identified PDGFB association with WM at the nominal P value < .01; however, after P value correction for multiple testing, neither the gene- nor the variant-level association was significant.
Our study has several limitations. Although we exome-sequenced and analyzed 1 of the largest sets of familial WM samples, including many multiplex families, the total number of cases (n = 166) was small for rare-variant association analyses. Although the cases and controls were sequenced in the same facility and on the same platform, the sequencing of the samples was done over a period of several years and on different Illumina instruments, thus possibly introducing a batch effect into the data. In the association analyses, we included samples from all affected family members and then took advantage of the SAIGE method, which accounts for the relatedness of individuals in the sample set. To mitigate a possible sequencing batch effect, all samples were bioinformatically jointly processed from FASTQ files to a VCF file as a single set. Another limitation of this study is that only the coding portion of the genome, which constitutes <2% of the genetic material of the cell, was analyzed. Understanding the function of regulatory regions of the genome and noncoding genes and their interaction with protein-coding loci will be necessary to master the complete knowledge of how genomes operate; however, this task remains immensely complex, labor-intensive, and costly, and was outside the scope of this study.
In summary, we identified multiple deleterious rare variants and plausible candidate genes in patients with WM. In 2 pedigrees, we identified multiple patients with WM with P/LP variants in TREX1 and SAMHD1, the genes that function at the interface between innate immune response, genotoxic surveillance, and DNA repair, and that are associated with the Aicardi-Goutières syndrome. There were additional P/LP variants residing in genes associated with well-known cancer-predisposing disorders, for example, POT1, RECQL4, PTPN11, and PMS2. On a pathway level, we observed statistically significant involvement of genes that play a role in telomere maintenance, DNA repair, and regulation of innate immune response. Affected members of each pedigree shared multiple deleterious variants (median, n = 18), but the overlap between the pedigrees was modest. In association analyses, we observed several VUSs including rs143980537 in PDGFB at the nominal P value <.001. This gene was also found to be associated with WM in SAIGE_SKAT-O analysis at the nominal P value <.01. We conclude that multiple genes are likely involved in the etiology of WM, each pedigree is largely unique in terms of its genetic risk architecture, and highly penetrant P/LP variants account for only a small proportion of deleterious variant load in families with WM. Larger studies are needed to identify a full catalog of genes associated with elevated risk for WM. Main challenges include the rarity of this disease and a likely oligogenic/polygenic nature of the genetic risk factors predisposing to WM. As a future research effort, methodological studies that examine the incorporation of variants derived from WM GWAS (only 1 was published to date) into polygenic risk score models may become feasible.
Acknowledgments
This work used resources of the National Institutes of Health High Performance Computing Biowulf cluster.
This work was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics of the National Cancer Institute, Bethesda, MD.
Authorship
Contribution: A.P. designed the research, analyzed data, and wrote and prepared the manuscript for publication; J.K. designed the research and analyzed data; W.L designed the research and analyzed data; J.L. oversaw the bioinformatics analyses; C.G. designed the research and analyzed data; K.J. oversaw patient sample processing and sequencing; D.D. oversaw sample collection and provided patients’ samples; N.D.F. oversaw sample collection and provided control samples; C.D. oversaw sample collection and provided patients’ samples; B.Z. designed the research, oversaw the data analyses, and cowrote the manuscript; M.L.M. designed the research, oversaw sample collection, provided patients’ samples, and cowrote the manuscript; and D.R.S. designed the research, oversaw the project, and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Douglas R. Stewart, Division of Cancer Epidemiology and Genetics, Clinical Genetics Branch, National Cancer Institute, 9609 Medical Center Dr, Room 6E450, Rockville, MD 20850; email: drstewart@mail.nih.gov.
References
Author notes
Genomic data are available through controlled access in dbGaP per the NIH genomic data sharing policy for the following studies: WM genotyping (phs001284.v1.p1); and chronic lymphocytic leukemia, Hodgkin, non-Hodgkin, and WM exome data (phs001219.v1.p1).
The full-text version of this article contains a data supplement.