Key Points
AML with high SLFN11 expression is more sensitive to cytarabine and has a better prognosis, suggesting that it may be a new biomarker for AML.
SLFN11 prevents overactivation of the ATR pathway promoting apoptosis in response to cytarabine.
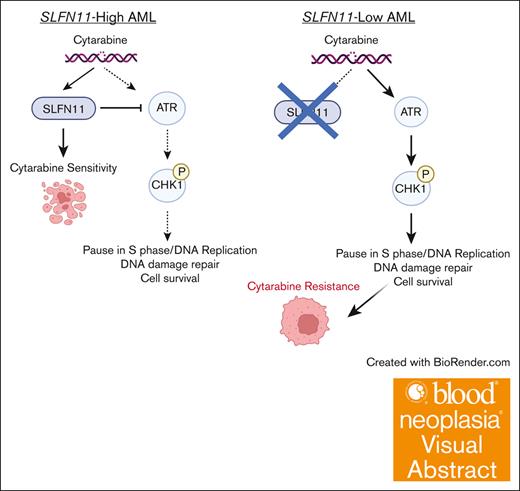
Visual Abstract
Chemoresistance represents an ongoing challenge in treating patients with acute myeloid leukemia (AML), and a better understanding of the resistance mechanisms can lead to the development of novel AML therapies. Here, we demonstrated that low expression of the DNA damage response gene Schlafen 11 (SLFN11) correlates with poor overall survival and worse prognosis in patients with AML. Moreover, we showed that SLFN11 plays an essential role in regulating chemotherapy sensitivity in AML. AML cells with suppressed levels of SLFN11 do not undergo apoptosis in response to cytarabine because of aberrant activation of the Ataxia telangiectasia and Rad3-related protein (ATR)/Checkpoint kinase 1 (Chk1) pathway, allowing for DNA damage repair, whereas sensitivity to cytarabine can be restored by inhibiting the ATR pathway. Importantly, SLFN11 knockout AML cells retain sensitivity to hypomethylating agents and the B-cell lymphoma 2 (BCL-2) inhibitor venetoclax. Altogether, these results reveal SLFN11 as an important regulator and predictor of chemotherapy sensitivity in AML and suggest that targeting pathways suppressed by SLFN11 may offer potential combination therapies to enhance and optimize chemotherapy responses in AML.
Introduction
Acute myeloid leukemia (AML) is a deadly disease, with >20 000 new cases in the United States each year and an average 5-year survival rate of >30%, with even lower survival rates in older patients.1 These poor survival rates are due to the aggressiveness of the disease, inherent chemotherapy resistance, high relapse rates, and severe side effects of treatment that limit the use of potentially curative therapies. Curative therapies consist of intensive cytarabine (AraC)–based combination chemotherapy regimens followed by allogeneic stem cell transplantation in certain groups of patients. Patients who cannot tolerate intensive treatment, or whose disease is not expected to respond to combination chemotherapy, are frequently treated with hypomethylating agents (HMAs) combined with the B-cell lymphoma 2 (BCL-2) inhibitor venetoclax (VEN).2 Unfortunately, this combination is not considered curative, and ultimately most patients succumb to their disease.
Schlafen 11 (SLFN11) is a member of the interferon-regulated Schlafen family of proteins.3 It localizes to sites of DNA damage (stalled replication forks) by binding to replication protein A (RPA1).4 Once recruited to DNA, SLFN11 causes irreversible replication arrest and ultimately apoptosis.5-7 In the absence of SLFN11, cells are less primed to undergo apoptosis in response to DNA damage. Accordingly, high SLFN11 expression is associated with sensitivity to multiple DNA-damaging agents (DDAs).8-11 Because of its demonstrated role as a predictive marker for chemotherapy response in preclinical models, SLFN11 is being used as a biomarker in clinical trials for lung (ClinicalTrials.gov identifier: NCT04939662)12 and uterine cancer (NCT03880019).13 SLFN11’s role in AML, however, has not yet been explored. Here, we demonstrate that SLFN11 is a key element in the pathophysiology of chemotherapy resistance in AML. Our findings raise the possibility that SLFN11 may be a predictive and prognostic marker for AML. SLFN11 sensitizes AML cells to AraC by counteracting Ataxia telangiectasia and Rad3-related protein (ATR)/Checkpoint 1 (CHK1) pathway activation; in cells that lack SLFN11, AraC causes sustained activation of the ATR/Chk1 pathway, allowing for DNA damage repair and cell survival.
Methods
Cell lines and primary cells
The cells were incubated in a 37°C humidified incubator with 5% CO2. The U937 (U-937, catalogue no. CRL-1593.2) and human erythroleukemia (HEL 92.1.7, catalogue no. TIB-180) cell lines were obtained from the American Type Culture Collection and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). All cell lines were tested for mycoplasma before use and authenticated every 6 months to 1 year using short tandem repeat analysis. Studies on patient-derived primary AML cells were approved by the institutional review board of Northwestern University. Peripheral blood and bone marrow (BM) samples were collected from patients with AML after obtaining written informed consent. Mononuclear cells were separated using Ficoll Hypaque and subjected to red blood cell lysis. Leukemic blasts were enriched through positive selection by incubation with CD34-conjugated (STEMCELL Technologies; EasySep Kit, catalog no. 17856) or CD117-conjugated (Miltenyi Biotec, catalog no. 130-091-332) magnetic microbeads and running through an EasySep magnetic column (STEMCELL Technologies catalog no. 18000). Primary leukemic blasts were used to perform quantitative real-time polymerase chain reaction (qRT-PCR) and colony formation assays.
Correlation between SLFN11 expression and survival in AML
Using the University of California Santa Cruz (UCSC) Xena browser https://xena.ucsc.edu/14 data were extracted from the The Cancer Genome Atlas (TCGA) AML data set. Data of patients with AML were split into 2 groups according to SLFN11 expression levels measured by RNA sequencing (Illumina HiSeq), with a cutoff of 11.18 (n = 81 in the SLFN11-low group and n = 80 in the SLFN11-high groups) and graphed according to the probability of survival. The Kaplan-Meier method was used to estimate overall survival (OS), and the groups were compared using the log-rank test.
Correlation between DNA methylation of SLFN11 and survival in AML
The survival probability of patients with AML based on methylation of transcriptional start sites (TSSs) on SLFN11 was determined using the web tool MethSurv https://biit.cs.ut.ee/methsurv/15, in which the DNA methylation status is represented as values from 0 to 1 based on probe intensities. TSSs within the UCSC cytosine and guanine (CpG) Island were analyzed by the “best” splitting option. For cg11309496, higher and lower levels of methylation were defined as above and <0.022, respectively. For cg26573518, higher and lower levels of methylation were defined as above and <0.092, respectively. Raw methylation data used in the MethSurv web tool were extracted from the TCGA AML data set publicly available through the Broad Institute website58 https://gdac.broadinstitute.org/. The Kaplan-Meier method was used to estimate OS, and the groups were compared using the log-rank test.
Comparison of SLFN11 expression in AML subgroups
Relative SLFN11 expression for different AML subgroups was obtained from the public Beat AML data set available through the data viewer Vizome59,http://www.vizome.org, and comparisons were made based on survival (Beat AML 2.0), risk category, relapse status, de novo vs secondary- or therapy-related AML, nucleophosmin 1 (NPM1) mutation status, and CEBPA mutation status. For stratification based on TP53 mutation status, data were obtained from the Oregon Health & Science University (OHSU) AML cohort57, available through cBioPortal53-55 https://www.cbioportal.org/. SLFN11 expression in different cytogenetic groups was obtained from the TCGA AML data set available through BloodSpot56 https://servers.binf.ku.dk/bloodspot/. Number of patients in each group is listed in the respective figure legend Fig. 2.
Correlation between drug activity and SLFN11 expression in cancer cell lines
Raw data on AraC and 5'-azacitidine (AZA) drug activity (based on 50% growth-inhibitory levels, determined by the National Institutes of Health Developmental Therapeutics Program using the sulforhodamine B assay), SLFN11 expression, and methylation of the National Cancer Insitute (NCI)-60 panel of cell lines were obtained from the CellMiner Cross-Database https://discover.nci.nih.gov/rsconnect/cellminercdb/35. Data were analyzed using GraphPad Prism version 9.5.1.
CRISPR/CRISPR associated protein 9 (Cas9) gene editing
The Edit-R Lentiviral hEF1α-Blast-Cas9 nuclease plasmid DNA (Horizon, Dharmacon Reagents, catalog no. CAS10138) was used to generate U937 and HEL cells stably expressing Cas9 nuclease, and blasticidin treatment was used as the selection marker, as in our previous studies.16,17 U937-Cas9 and HEL-Cas9 expressing cells were then transduced with lentiviral particles expressing Edit-R human SLFN11 lentiviral single guide RNA (sgRNA) (Horizon, Dharmacon Reagents, catalog no. VSGH10142–246458929; target sequence: GGTCAGCAGGATCCGAGTTA) to generate U937-SLFN11 knockout (KO) and HEL-SLFN11 KO cells using puromycin treatment as the selection marker (pooled clones). U937-SLFN11 KO and U937-Cas9 single clones were generated by performing serial cell dilutions of the respective pooled clones and by growing single cells in a conditioned medium collected and filtered from U937 cell cultures. U937-Cas9 (pooled and single clones) and HEL-Cas9 (pooled clones) stably transduced cells were maintained in culture using RPMI 1640 medium supplemented with 10% FBS and blasticidin. U937-SLFN11 KO (pooled and single clones) and HEL-SLFN11 KO (pooled clones) cells were maintained in culture using RPMI 1640 medium supplemented with 10% FBS, blasticidin, and puromycin. SLFN11 protein expression was assessed by immunoblotting.
Cell viability assays
Cells were plated in quintuplicate into 96-well plates and treated with either respective vehicle control (VC) or increasing concentrations of AraC (TargetMol, catalog no. T1272), AZA (TargetMol, catalog no. T1339), decitabine (DAC; Chemietek, catalog no. CT-DECI), VEN (TargetMol, catalog no. T2119), and/or the ATR inhibitors (ATRis) VE822 (Cayman Chemicals, catalog no. CAY24198) and AZD6738 (Cayman Chemicals, catalog no. CAY21035). Twenty-four or 48 hours after treatment, the cell proliferation reagent, water-soluble tetrazolium 1 (WST-1) (Hoffman-La Roche Inc/Sigma, catalog no. 5015944001) was added to the wells, and cell viability was assessed and quantified according to the manufacturer’s instructions as in our previous study.18 For trypan blue exclusion assay, cells were plated in triplicate in 6-well plates and treated with either vehicle-control dimethyl sulfoxide or 0.3 μM VE822 (ATRi). After 24 hours, cells were diluted 1:1 with trypan blue (Bio-Rad, #1450021) and 10 μL of the cell mixture was loaded onto cell counting dual chambered slides (Bio-Rad, catalog no. 1450019) and the percentage of live cells was determined using a TC20 automated cell counter (Bio-Rad, catalog no. 1450102).
qRT-PCR analyses
Total RNA was isolated from leukemic blasts of patients with AML using the RNeasy Mini Kit (Qiagen) per the manufacturer’s instructions, as in our previous study.19 Total RNA was reverse-transcribed into complementary DNA using a high-capacity complementary DNA reverse transcription kit (Applied Biosystems, catalog no. 4368814) and qRT-PCR was performed using FAM-labeled TaqMan primer/probe sets for SLFN11 (Thermo Fisher Scientific catalog no. Hs00536981_m1) and glyceraldehyde-3-phosphate hydrogenase (GAPDH) (Thermo Fisher Scientific, catalog no. Hs02758991_g1). Relative SLFN11 expression was normalized to GAPDH expression using the formula 2−deltaCt.
Colony formation assays
Leukemic blasts from patients with AML were plated in MethoCult H4534 classic medium (STEMCELL Technologies, catalog no. 04534) and treated with either vehicle control (water) or with 3 different concentrations of AraC (0.08 ng/mL, 0.8 ng/mL, and 8 ng/mL) (TargetMol. Catalog no. T1272). Leukemic colony-forming units were counted after 10 to 14 days as in our previous study.20
AML xenograft in vivo studies
All animal studies were approved by the Northwestern University Institutional Animal Care and Use Committee and were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals. U937-Cas9 or U937-SLFN11 KO cells (single clones) (2.5 × 106) were implanted into the left flank of 6 to 10-week-old female natural cytotoxicity receptor 1-deficient (NCr) nude mice (CrTac:NCr-Foxn1nu, Taconic, NCRNU-F) by subcutaneous injection. Eight days after tumor cell implantation, tumors were measured using a caliper, and mice in both genotype-tumor groups were randomized into 2 treatment groups based on tumor volume and body weight. Mice were treated with either vehicle control (phosphate-buffered saline [PBS]) or AraC (Fresenius Kabi, pharmaceutical grade, catalog no. 102020) at 10 mg/kg of body weight by intraperitoneal injection once per day for a total of 10 days (days 9-18). Tumor size was measured using calipers 3 times per week, and the volume was calculated according to the formula (D × d2) × π/6, where D is the longest diameter and d is the shorter diameter. The mice were followed up for survival until day 42. The criteria used for euthanasia was weight loss ≥20% compared with the body weight before tumor cell implantation or tumor size ≥2 cm3. The in vivo study was performed twice, with 4 mice per group (16 mice total) in the first study and 8 to 9 mice per group (33 mice total) in the second study.
Cell cycle analysis
These assays were performed as previously described.21,22 Cells were plated and treated with either the vehicle control (water) or 500 ng/mL AraC for 6 hours. The cells were then collected, washed with PBS, and fixed in ice-cold 70% ethanol. The fixed cells were washed using PBS with 2% FBS and resuspended in PBS containing 8 μg/mL propidium iodide (Sigma, catalog no. R4642) and 18 μg/mL RNase (Calbiochem, catalog no. 537059). Cell cycle analysis was performed using a FACSymphony instrument (BD Biosciences), and the data were analyzed using FlowJo version v10.8.1.
Cell lysis and immunoblotting
Cells were lysed in Triton-X 100 lysis buffer (40 mM HEPES pH 7, 120 mM NaCl, 1 mM EDTA, 10 mM sodium pyrophosphate, 50 mM sodium fluoride, 10 mM B-glycerophosphate, and 0.5% Triton-X 100) with 1 mmol/L phenylmethylsulfonylfluoride, protease inhibitor cocktail set V (EMD Millipore, catalog no. 539137), and phosphatase inhibitor cocktail set I (EMD Millipore, catalog no. 524624). Total protein lysates were resolved by sodium dodecyl sulfate−polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Bio-Rad). Immunoblotting and detection of bands by chemiluminescence were performed as previously described.23 The following primary antibodies were used: anti-SLFN11 (Santa Cruz Biotechnology, catalog no. SC-374339), anti-GAPDH (Millipore, catalog no. MAB374), anti-PARP (Cell Signaling Technology, catalog no. 9542), anti–phospho-CHK1 (S345; Cell Signaling Technology, catalog no. 2348), anti-CHK1 (Cell Signaling Technology, catalog no. 2360), and anti–phospho-histone H2A.X (Ser139; γH2AX; Cell Signaling Technology, catalog no. 2577).
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 9.5.1. and R version 4.2.1. The statistical tests used are described in the figure legends. P values <.05 were considered statistically significant.
Results
Low SLFN11 expression correlates with poor OS in AML
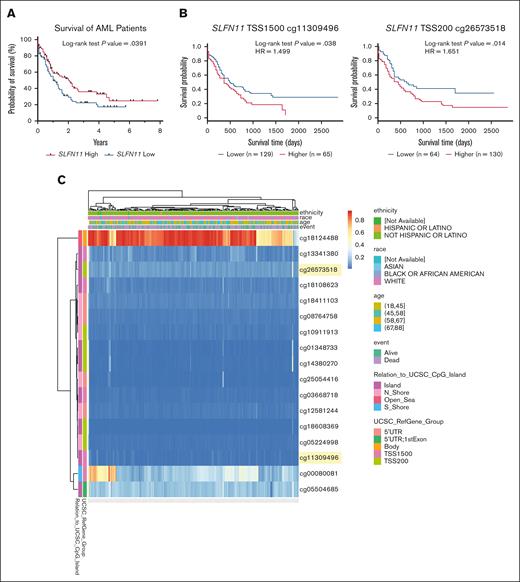
We first used the publicly available TCGA AML data set, available through the UCSC Xena browser, to explore the potential role of SLFN11 as a prognostic marker in AML.14 We observed a statistically significant difference in survival based on SLFN11 expression, in patients with AML who were SLFN11-high expressers demonstrating improved survival vs SLFN11-low expressers (Figure 1A). Because SLFN11 expression has been shown to be regulated in some contexts by promoter methylation,24,25 with less methylated promoter regions corresponding with higher expression of SLFN11, we stratified the same cohort of patients based on SLFN11 promoter methylation at 2 different locations, 1500 and 200 base pairs upstream of the TSS, using the MethSurv web tool.15 We observed that patients with AML with lower methylation levels in these SLFN11 promoter regions presented increased OS (Figure 1B). Additionally, by graphing the entire methylation pattern for SLFN11 using data from the TCGA AML data set and the MethSurv web tool, we noted that the gene body of SLFN11 (cg18124488) was highly methylated in most patients (Figure 1C), although the effect of gene body methylation in this context is unknown.26 In summary, our results support that low levels of SLFN11 expression, potentially as a result of increased methylation of its promoter region, correlate with lower OS in patients with AML.
High SLFN11 expression correlates with increased OS in patients with AML. (A) Kaplan-Meier survival curves for patients with AML expressing high (above the median, n = 80) vs low (below the median, n = 81) SLFN11 expression. Data were extracted from the TCGA AML data set using the UCSC Xena browser. (B) Kaplan-Meier survival curves for patients with AML presenting (left) high (n = 65) vs low (n = 129) methylation levels at TSS 1500 of SLFN11 and (right) high (n = 130) vs low (n = 64) methylation levels at TSS 200 of SLFN11. Raw methylation data used in the MethSurv web tool were extracted from the TCGA AML data set available on the Broad Institute website. (C) Heat map showing the methylation levels of various CpG sites on SLFN11 in patients with AML. Data were extracted from the TCGA AML data set available on the Broad Institute website and graphed using the MethSurv web tool. CpG sites shown in panel B are highlighted in yellow. (A-B) Statistical analyses were performed using the log-rank test, and P values are shown. HR, hazard ratio; UTR, untranslated region.
High SLFN11 expression correlates with increased OS in patients with AML. (A) Kaplan-Meier survival curves for patients with AML expressing high (above the median, n = 80) vs low (below the median, n = 81) SLFN11 expression. Data were extracted from the TCGA AML data set using the UCSC Xena browser. (B) Kaplan-Meier survival curves for patients with AML presenting (left) high (n = 65) vs low (n = 129) methylation levels at TSS 1500 of SLFN11 and (right) high (n = 130) vs low (n = 64) methylation levels at TSS 200 of SLFN11. Raw methylation data used in the MethSurv web tool were extracted from the TCGA AML data set available on the Broad Institute website. (C) Heat map showing the methylation levels of various CpG sites on SLFN11 in patients with AML. Data were extracted from the TCGA AML data set available on the Broad Institute website and graphed using the MethSurv web tool. CpG sites shown in panel B are highlighted in yellow. (A-B) Statistical analyses were performed using the log-rank test, and P values are shown. HR, hazard ratio; UTR, untranslated region.
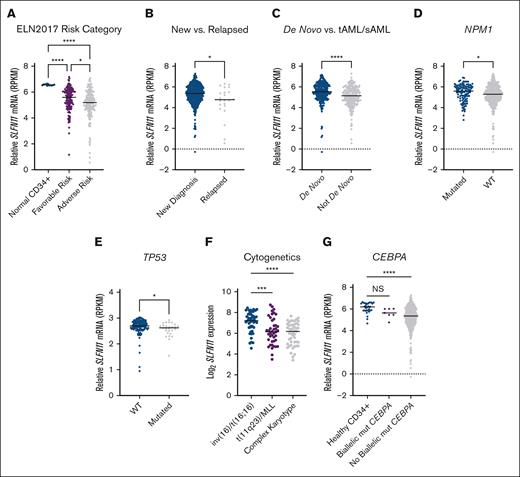
Next, using the Beat AML 2.0 data set,27 we assessed the association between OS of patients with AML and SLFN11 expression using 3 groups based on quartiles of SLFN11 levels (Q1, Q2 + Q3, and Q4). We found that OS was worse in patients expressing lower SLFN11 (P = .04), and we also found a statistically significant (P = .01) difference between patients expressing high (top quartile) vs low (bottom quartile) levels of SLFN11 (supplemental Figure 1). We then stratified patients with AML based on the ELN2017 risk category, a commonly adapted prognostic model in AML28,29 and plotted these risk categories relative to SLFN11 expression in patients with favorable-risk AML, adverse risk AML, and normal CD34+ cells from healthy donors (Figure 2A). Compared with normal CD34+ cells, patients with either adverse- or favorable-risk AML showed significantly lower SLFN11 expression. Patients in the adverse risk group presented even lower levels of SLFN11 expression than patients in the favorable-risk group (Figure 2A). Using data from the same data set, we then observed that patients with AML with relapsed disease present lower SLFN11 expression than patients with newly diagnosed disease (Figure 2B). Patients with AML with secondary or therapy-related disease, which are generally more resistant to chemotherapy,30 also showed lower SLFN11 expression than patients with de novo AML (Figure 2C).
High SLFN11 expression is associated with better prognostic groups in AML. (A) Relative SLFN11 messenger RNA expression in normal CD34+ cells isolated from healthy donors (n = 12) and in patients with favorable-risk (n = 117) or adverse risk (n = 164) AML based on the ELN 2017 risk criteria. Data were extracted from the Beat AML data set, available through Vizome. (B-D) Relative SLFN11 mRNA expression in patients with (B) newly diagnosed (n = 428) and relapsed (n = 23) AML, (C) de novo (n = 223) and therapy-related AML (tAML) or secondary AML (sAML) (n = 228), and (D) NPM1-mutated (mut) (n = 108) or NPM1 wild-type (WT; n = 340) AML. Data were extracted from the Beat AML data set, and accessed through Vizome. (E) Relative SLFN11 mRNA expression in TP53-WT (n = 117) and TP53-mutated (n = 27) AML. Data were extracted from the OSHU AML cohort using the cBioPortal. (F) Log2 of SLFN11 mRNA expression in patients with AML with inv(16)/t(16;16) (n = 47), t(11q23)/mixed lineage leukemia (MLL) (n = 42), or a complex karyotype (n = 49). Data were extracted from the TCGA AML data set available through BloodSpot. (G) Relative SLFN11 mRNA expression in normal CD34+ cells (n = 32) and patients with (n = 7) or without (n = 444) biallelic CEBPA mutations. Data were extracted from the Beat AML data set available through Vizome. (A,F,G) Statistical analysis was performed using the Kruskal-Wallis test followed by Dunn multiple comparison adjustment. (B-E) Statistical analysis was performed using the 2-tailed Mann-Whitney test. (A-G) The median is represented by the black line for each AML subgroup. ∗P < .05; ∗∗∗P < .001; ∗∗∗∗P < .0001. NS, not significant; RPKM, reads per kilobase million.
High SLFN11 expression is associated with better prognostic groups in AML. (A) Relative SLFN11 messenger RNA expression in normal CD34+ cells isolated from healthy donors (n = 12) and in patients with favorable-risk (n = 117) or adverse risk (n = 164) AML based on the ELN 2017 risk criteria. Data were extracted from the Beat AML data set, available through Vizome. (B-D) Relative SLFN11 mRNA expression in patients with (B) newly diagnosed (n = 428) and relapsed (n = 23) AML, (C) de novo (n = 223) and therapy-related AML (tAML) or secondary AML (sAML) (n = 228), and (D) NPM1-mutated (mut) (n = 108) or NPM1 wild-type (WT; n = 340) AML. Data were extracted from the Beat AML data set, and accessed through Vizome. (E) Relative SLFN11 mRNA expression in TP53-WT (n = 117) and TP53-mutated (n = 27) AML. Data were extracted from the OSHU AML cohort using the cBioPortal. (F) Log2 of SLFN11 mRNA expression in patients with AML with inv(16)/t(16;16) (n = 47), t(11q23)/mixed lineage leukemia (MLL) (n = 42), or a complex karyotype (n = 49). Data were extracted from the TCGA AML data set available through BloodSpot. (G) Relative SLFN11 mRNA expression in normal CD34+ cells (n = 32) and patients with (n = 7) or without (n = 444) biallelic CEBPA mutations. Data were extracted from the Beat AML data set available through Vizome. (A,F,G) Statistical analysis was performed using the Kruskal-Wallis test followed by Dunn multiple comparison adjustment. (B-E) Statistical analysis was performed using the 2-tailed Mann-Whitney test. (A-G) The median is represented by the black line for each AML subgroup. ∗P < .05; ∗∗∗P < .001; ∗∗∗∗P < .0001. NS, not significant; RPKM, reads per kilobase million.
To test whether SLFN11 levels correlate with other prognostic markers of AML, we examined patients with AML and mutated NPM1, a more favorable prognostic indicator.29,31 We observed that these patients have higher SLFN11 expression than patients with wild-type NPM1 (Figure 2D). Using data from the OSHU AML data set, we also observed that SLFN11 expression was lower in patients with mutated TP53, an adverse risk indicator,32 than in patients with wild-type TP53 (Figure 2E). Next, we assessed the expression of SLFN11 in patients with AML with specific cytogenetic abnormalities using data from the TCGA AML data set.33 Patients with AML with inversion 16 or t,16 a favorable prognostic indicator, have higher SLFN11 expression than patients with t(11q23)/Mixed Lineage Leukemia (MLL) rearrangement or complex karyotype AML, which have an unfavorable prognosis29 (Figure 2F). Moreover, using the Beat AML data set, we found that patients with biallelic CEBPA mutations, a favorable prognostic indicator in AML,34 present higher SLFN11 expression than those without biallelic CEBPA mutations (Figure 2G). Together, these data suggest that patients with AML with poorer prognoses express lower levels of SLFN11.
Higher levels of SLFN11 correlate with higher sensitivity to AraC
High expression of SLFN11 has been previously associated with increased sensitivity to DDAs, such as topoisomerase inhibitors, in multiple cancer cell lines.10 To determine whether there was a correlation between the efficacy of the DDA commonly used in AML treatment, AraC and SLFN11 promoter methylation, and gene expression, we next used the publicly available CellMiner database.35 Using the NCI-60 cell line panel as a reference, which included 6 leukemia cell lines, we observed a negative correlation between AraC activity and methylation levels of the SLFN11 promoter region (Figure 3A). In addition, we observed a positive correlation between AraC activity and SLFN11 expression levels (Figure 3B). We next treated U937 AML cells with the HMA AZA and observed an increase in SLFN11 levels after treatment by immunoblotting analysis (Figure 3C), supporting that SLFN11 expression is controlled by methylation in AML cells. Moreover, we next investigated whether SLFN11 expression in leukemic blasts of patients with AML could also correlate with sensitivity to AraC in vitro. For this, peripheral blood or BM aspirate samples were obtained from patients with AML, and leukemic blasts were isolated. Primary AML blasts with SLFN11 expression below and above the median (0.007), as assessed by qRT-PCR analyses, were categorized as SLFN11-low and SLFN11-high (Figure 3D, right panel), respectively, and plated for colony formation assays in the presence of either vehicle control or increasing concentrations of AraC. SLFN11-high AML blasts showed a trend toward decreased colony formation with increasing doses of AraC, whereas AraC treatment showed significantly lower inhibitory effects for SLFN11-low AML blasts (Figure 3D, left panel; P = .043). Taken together, these data indicate that low levels of SLFN11 correlate with lower sensitivity to AraC treatment in several tumor cell lines and primary AML blasts.
Low SLFN11 expression correlates with lower sensitivity to AraC. (A) Correlation analysis between AraC activity and SLFN11 gene methylation in cancer cell lines from the NCI-60 panel. Data were extracted from CellMiner Cross-Database (CDB) (n = 60). (B) Correlation analysis between AraC activity and Log2 of SLFN11 mRNA expression in cancer cell lines from the NCI-60 panel. Data were extracted from CellMiner CDB (n = 59). (A-B) Statistical analysis was performed using simple linear regression, and P values are shown for deviation of the line slope from 0. (C) U937 cells were treated with the vehicle control (dimethyl sulfoxide [DMSO]) or 1 μM AZA for 24 hours. Protein lysates from U937 cells were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblotting analysis with the indicated antibodies. GAPDH is shown as a loading control. (D) Clonogenic capability of primary leukemic blasts isolated from patients with AML expressing either high levels of SLFN11 (SLFN11-high, n = 3 for 8 ng/ml AraC and n = 4 for 0.08 and 0.8 ng/ml AraC) or low levels of SLFN11 (SLFN11-low, n = 3) treated with either vehicle control (VC; water) or increasing concentrations of AraC, as indicated (left panel). Data are expressed as the percentage of colony formation over VC-treated cells for each patient (Ctrl) and are shown as mean ± standard error of the mean (SEM). Statistical analyses were performed using Mixed-effects analysis followed by Tukey multiple comparisons test and P values are shown for the highest AraC dose for each group. There was a significant interaction (P = .043), that is, the relationship between concentration and percentage of colony formation was significantly different between patients with AML that were SLFN11-low vs SLFN11-high. (D) Relative SLFN11 mRNA expression, normalized to GAPDH, was assessed by qRT-PCR analysis and is shown for the primary leukemic blasts isolated from patients with AML and used in the clonogenic assays (SLFN11-low, n = 3 and SLFN11-high, n = 4; right panel).
Low SLFN11 expression correlates with lower sensitivity to AraC. (A) Correlation analysis between AraC activity and SLFN11 gene methylation in cancer cell lines from the NCI-60 panel. Data were extracted from CellMiner Cross-Database (CDB) (n = 60). (B) Correlation analysis between AraC activity and Log2 of SLFN11 mRNA expression in cancer cell lines from the NCI-60 panel. Data were extracted from CellMiner CDB (n = 59). (A-B) Statistical analysis was performed using simple linear regression, and P values are shown for deviation of the line slope from 0. (C) U937 cells were treated with the vehicle control (dimethyl sulfoxide [DMSO]) or 1 μM AZA for 24 hours. Protein lysates from U937 cells were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblotting analysis with the indicated antibodies. GAPDH is shown as a loading control. (D) Clonogenic capability of primary leukemic blasts isolated from patients with AML expressing either high levels of SLFN11 (SLFN11-high, n = 3 for 8 ng/ml AraC and n = 4 for 0.08 and 0.8 ng/ml AraC) or low levels of SLFN11 (SLFN11-low, n = 3) treated with either vehicle control (VC; water) or increasing concentrations of AraC, as indicated (left panel). Data are expressed as the percentage of colony formation over VC-treated cells for each patient (Ctrl) and are shown as mean ± standard error of the mean (SEM). Statistical analyses were performed using Mixed-effects analysis followed by Tukey multiple comparisons test and P values are shown for the highest AraC dose for each group. There was a significant interaction (P = .043), that is, the relationship between concentration and percentage of colony formation was significantly different between patients with AML that were SLFN11-low vs SLFN11-high. (D) Relative SLFN11 mRNA expression, normalized to GAPDH, was assessed by qRT-PCR analysis and is shown for the primary leukemic blasts isolated from patients with AML and used in the clonogenic assays (SLFN11-low, n = 3 and SLFN11-high, n = 4; right panel).
Targeting SLFN11 in AML cells promotes resistance to AraC, but does not affect responses to HMAs and VEN
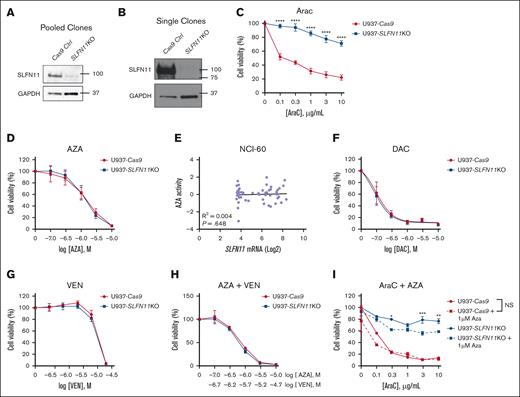
To further study the functional role of SLFN11 in AML, we next generated U937 cells, a human monocytic leukemia cell line that harbors a TP53 mutation expressing Cas9 (control). These cells were then transfected with guide RNA specific to SLFN11 to generate SLFN11 KO cells, pooled and single clones, and used for further studies. Knock out of SLFN11 was confirmed by immunoblotting analyses for the edited cells (Figure 4A- B). To determine the effects of knocking out SLFN11 on the efficacy of AraC treatment, U937-Cas9, and U937-SLFN11 KO (pooled clones) cells were treated with either the vehicle control or escalating doses of AraC, and cell viability was measured 24 hours later using WST-1 reagent (Figure 4C). We observed that knocking out SLFN11 in U937 cells promoted resistance to AraC treatment (Figure 4C). Similar results were obtained using U937-SLFN11 KO single clones compared with U937-Cas9 single clones, suggesting that these results were not due to clonal effects (supplemental Figure 2).
SLFN11 KO AML cells are resistant to AraC but not to HMAs and VEN. (A-B) Protein lysates from U937-Cas9 and U937-SLFN11 KO cells (pooled and single clones, as indicated) were resolved by SDS-PAGE, followed by immunoblotting with the indicated antibodies. GAPDH is shown as a loading control. (C) U937-Cas9 and U937-SLFN11 KO cells (pooled clones) were seeded in 96-well plates and treated with either VC (water) or increasing concentrations of AraC for 24 hours. (D) U937-Cas9 and U937-SLFN11 KO cells (single clones) were seeded in 96-well plates and treated with either VC (DMSO) or increasing concentrations of AZA for 24 hours. (E) Correlation analysis between AZA activity and Log2 of SLFN11 mRNA expression in cancer cell lines from the NCI-60 panel. Data extracted from CellMiner CDB (n = 59). Statistical analysis was performed using simple linear regression, and the P value is shown for the deviation of the line slope from 0. (F) U937-Cas9 and U937-SLFN11 KO cells (pooled clones) were seeded in 96-well plates and treated with either VC (DMSO) or increasing concentrations of DAC for 48 hours. (G) U937-Cas9 and U937-SLFN11 KO cells (single clones) were seeded in 96-well plates and treated with either VC (DMSO) or increasing concentrations of VEN for 24 hours. (H) U937-Cas9 and U937-SLFN11 KO cells (single clones) were seeded in 96-well plates and treated with either VC (DMSO) or increasing concentrations of AZA and VEN for 24 hours. (I) U937-Cas9 control cells and U937-SLFN11 KO cells (single clones) were seeded in 96-well plates and treated with either VC (DMSO) or increasing concentrations of AraC and/or AZA for 24 hours. (C,D,F-I) Cell viability was assessed using the WST-1 reagent. Data are expressed as a percentage of cell viability of VC-treated cells. Means ± SEM of 4 independent experiments in panel C or 3 independent experiments in panels D,F-I are shown. Statistical analyses were performed using a 2-way analysis of variance (ANOVA) followed by the Sidak multiple comparison test. In panel C, there is a significant interaction between cell type and concentration (P < .0001) in AraC-treated cells (the effect of AraC concentration on cell viability was significantly different between U937-Cas9 and U937-SLFN11 KO cells). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. NS, not significant.
SLFN11 KO AML cells are resistant to AraC but not to HMAs and VEN. (A-B) Protein lysates from U937-Cas9 and U937-SLFN11 KO cells (pooled and single clones, as indicated) were resolved by SDS-PAGE, followed by immunoblotting with the indicated antibodies. GAPDH is shown as a loading control. (C) U937-Cas9 and U937-SLFN11 KO cells (pooled clones) were seeded in 96-well plates and treated with either VC (water) or increasing concentrations of AraC for 24 hours. (D) U937-Cas9 and U937-SLFN11 KO cells (single clones) were seeded in 96-well plates and treated with either VC (DMSO) or increasing concentrations of AZA for 24 hours. (E) Correlation analysis between AZA activity and Log2 of SLFN11 mRNA expression in cancer cell lines from the NCI-60 panel. Data extracted from CellMiner CDB (n = 59). Statistical analysis was performed using simple linear regression, and the P value is shown for the deviation of the line slope from 0. (F) U937-Cas9 and U937-SLFN11 KO cells (pooled clones) were seeded in 96-well plates and treated with either VC (DMSO) or increasing concentrations of DAC for 48 hours. (G) U937-Cas9 and U937-SLFN11 KO cells (single clones) were seeded in 96-well plates and treated with either VC (DMSO) or increasing concentrations of VEN for 24 hours. (H) U937-Cas9 and U937-SLFN11 KO cells (single clones) were seeded in 96-well plates and treated with either VC (DMSO) or increasing concentrations of AZA and VEN for 24 hours. (I) U937-Cas9 control cells and U937-SLFN11 KO cells (single clones) were seeded in 96-well plates and treated with either VC (DMSO) or increasing concentrations of AraC and/or AZA for 24 hours. (C,D,F-I) Cell viability was assessed using the WST-1 reagent. Data are expressed as a percentage of cell viability of VC-treated cells. Means ± SEM of 4 independent experiments in panel C or 3 independent experiments in panels D,F-I are shown. Statistical analyses were performed using a 2-way analysis of variance (ANOVA) followed by the Sidak multiple comparison test. In panel C, there is a significant interaction between cell type and concentration (P < .0001) in AraC-treated cells (the effect of AraC concentration on cell viability was significantly different between U937-Cas9 and U937-SLFN11 KO cells). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. NS, not significant.
Next, to determine whether SLFN11 is also required for the potent antileukemic effects of other drugs used for the treatment of AML, we tested the effects of knocking out SLFN11 on the efficacy of HMAs and VEN, a combination used frequently in AML treatment.36 U937-Cas9 and U937-SLFN11 KO cells (single clones) were treated with either vehicle control or increasing concentrations of AZA and 24 hours later, cell viability was measured by WST-1 assay (Figure 4D). In contrast to the results seen with AraC, U937-SLFN11 KO cells showed no notable differences in response to AZA treatment compared with U937-Cas9 cells (Figure 4D). Consistent with these results, we observed no correlation between SLFN11 expression and AZA activity in the NCI-60 cell line panel35 (Figure 4E). We then performed cellular viability assays using U937-Cas9 and U937-SLFN11 KO cells (pooled clones) treated with either the vehicle control or increasing doses of DAC, another HMA commonly used in the treatment of AML and Myelodysplastic syndromes.36,37 Similar to the treatment with AZA, sensitivity to DAC was independent of SLFN11 expression (Figure 4F). Furthermore, U937-Cas9 and U937-SLFN11 KO cells (single clones) were found to be equally sensitive, in cell viability assays, to the B-cell lymphoma 2 (BCL-2) inhibitor VEN alone (Figure 4G) and in combination with AZA (Figure 4H), a treatment frequently used in elderly patients with AML and patients with adverse risk AML.36 Because AZA treatment was found to increase SLFN11 expression in U937 cells (Figure 3C), we next evaluated the combination of AZA with AraC in U937-Cas9 and U937-SLFN11 KO cells (single clones) compared with AraC treatment alone in WST-1 assays. Our results showed that the combination of AraC with 1 μM of AZA decreased U937-Cas9 cell viability compared with AraC treatment alone only at the lowest dose of AraC used (Figure 4I), suggesting that the basal levels of SLFN11 in these cells are sufficient to sustain sensitivity to AraC at higher doses. Interestingly, the combination of AZA and AraC decreased U937-SLFN11 KO cell viability compared with AraC treatment alone at some doses (Figure 4I), suggesting that methylation of genes with antileukemic effects could be increased in the absence of SLFN11 in U937 cells, but this needs to be further explored. Together, these results suggest that SLFN11 plays a key role in promoting AraC-mediated antileukemic responses but is not required for the antileukemic effects of HMAs and VEN in AML.
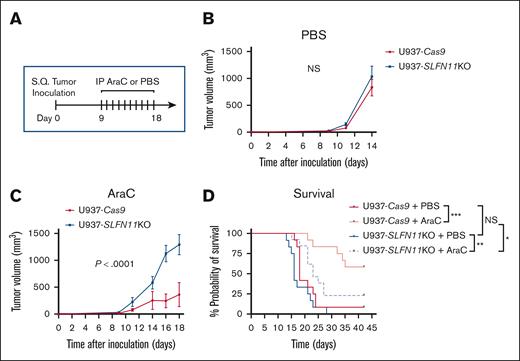
Absence of SLFN11 expression renders AML cells resistant to AraC treatment in vivo
Given our in vitro results, we next used a xenograft mouse model of AML to test the effects of SLFN11 absence in response to AraC treatment in vivo using U937-Cas9 (control; Ctrl) and U937-SLFN11 KO cells (single clones). U937-Cas9 or U937-SLFN11 KO cells were implanted subcutaneously into NCr nude mice. Eight days later, the mice bearing tumors of either genotype were randomized by tumor volume and body weight into 2 treatment groups: (1) vehicle control (PBS) and (2) 10 mg/kg AraC. Mice were treated with intraperitoneal injection daily for 10 days, and tumor volumes were measured 3 times per week until the study end point (Figure 5A). Although no significant differences were observed between tumor growth in PBS-treated mice bearing either U937-Cas9 or U937-SLFN11 KO tumors (Figure 5B), tumor growth was significantly decreased in AraC-treated mice bearing U937-Cas9 tumors compared with that in AraC-treated mice bearing U937-SLFN11 KO tumors (Figure 5C). Individual tumor growth patterns and body weights of PBS-treated– and AraC-treated mice are shown in supplemental Figure 3. Moreover, although there was no difference in survival between vehicle-treated mice groups, there was a statistically significant increase in the survival of mice bearing U937-Cas9 tumors compared with that of mice bearing U937-SLFN11 KO tumors in response to AraC treatment (Figure 5D). Additionally, the percentage of mice initially implanted with U937-Cas9 tumors that achieved a complete response to AraC (defined as tumor regressing to an unpalpable size) was greater than that of mice implanted with U937-SLFN11 KO tumors: 7 of 12 (58%) vs 3 of 13 (23%), respectively (Fisher exact test, P = .11).
Effects of knocking out SLFN11 in AML cells and in response to AraC treatment in vivo. (A) Schematic illustration of the AML xenograft mouse model and therapeutic regimen. Briefly, mice were injected SQ with U937-Cas9 or U937-SLFN11 KO cells (single clones). On day 8, mice from each tumor genotypic group were randomized by tumor volume and body weight into 2 treatment groups: VC (PBS) or AraC and daily treatments by intraperitoneal (IP) injection were given on days 9 to 18. Tumor volumes were measured 3 times per week until the study end point. Two independent in vivo studies were performed. (B-C) Tumor volumes (mean ± SEM) are shown for (B) PBS-treated or (C) AraC-treated mice implanted with either U937-Cas9 or U937-SLFN11 KO cells until the first tumor for either cohort reached 2000 mm3 (n = 8 for the PBS-treated groups and U937-Cas9 AraC-treated group and n = 9 for the U937-SLFN11 KO AraC-treated group). Mixed effects regression models were used to compare tumor growth between the groups. Tumor volume was the outcome variable, and log(volume + 1) transformation was used to stabilize the variance and satisfy the normality assumption. Day, group, and their interaction were included as fixed effects were fitted, and the within animal correlation between repeated tumor measurements over time was accounted for using a first-order autoregressive covariance structure (AR(1)). P value from the day × group interaction is reported. (D) Kaplan-Meier survival curves for the 4 treatment/genotypic groups (data compiled from the 2 in vivo studies, n = 12 for the PBS-treated and U937-Cas9 AraC-treated groups and n = 13 for the U937-SLFN11 KO AraC-treated group). Survival curves were compared using the log-rank test, and P values were adjusted for 4 pairwise comparisons (denoted in the figure) using the method of Holm-Sidak. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. NS, not significant; S.Q, subcutaneous.
Effects of knocking out SLFN11 in AML cells and in response to AraC treatment in vivo. (A) Schematic illustration of the AML xenograft mouse model and therapeutic regimen. Briefly, mice were injected SQ with U937-Cas9 or U937-SLFN11 KO cells (single clones). On day 8, mice from each tumor genotypic group were randomized by tumor volume and body weight into 2 treatment groups: VC (PBS) or AraC and daily treatments by intraperitoneal (IP) injection were given on days 9 to 18. Tumor volumes were measured 3 times per week until the study end point. Two independent in vivo studies were performed. (B-C) Tumor volumes (mean ± SEM) are shown for (B) PBS-treated or (C) AraC-treated mice implanted with either U937-Cas9 or U937-SLFN11 KO cells until the first tumor for either cohort reached 2000 mm3 (n = 8 for the PBS-treated groups and U937-Cas9 AraC-treated group and n = 9 for the U937-SLFN11 KO AraC-treated group). Mixed effects regression models were used to compare tumor growth between the groups. Tumor volume was the outcome variable, and log(volume + 1) transformation was used to stabilize the variance and satisfy the normality assumption. Day, group, and their interaction were included as fixed effects were fitted, and the within animal correlation between repeated tumor measurements over time was accounted for using a first-order autoregressive covariance structure (AR(1)). P value from the day × group interaction is reported. (D) Kaplan-Meier survival curves for the 4 treatment/genotypic groups (data compiled from the 2 in vivo studies, n = 12 for the PBS-treated and U937-Cas9 AraC-treated groups and n = 13 for the U937-SLFN11 KO AraC-treated group). Survival curves were compared using the log-rank test, and P values were adjusted for 4 pairwise comparisons (denoted in the figure) using the method of Holm-Sidak. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. NS, not significant; S.Q, subcutaneous.
SLFN11 KO AML cells demonstrate prolonged activation of ATR/Chk1 pathway in response to AraC treatment
Next, to identify the potential mechanisms driving the resistance to AraC treatment observed in SLFN11 KO U937 cells, we determined whether there were any differences in cell cycle progression between U937-Cas9 and U937-SLFN11 KO cells. For this, U937-Cas9 and U937-SLFN11 KO cells were treated with either vehicle control (water) or 500 ng/mL AraC for 6 hours, and cell cycle progression was assessed by flow cytometry analyses using propidium iodine staining. The cell cycle profile was similar between vehicle-treated U937-Cas9 and U937-SLFN11 KO cells (Figure 6A-B), suggesting that the difference in sensitivity to AraC, a DDA that acts primarily in cells in the S phase,38,39 is not due to differences in cell cycle progression at baseline between the 2 cell types. In the presence of AraC, U937-Cas9 cells showed a significant decrease in the percentage of cells in the S phase and a significant increase in the percentage of cells in the sub-G1 phase compared with vehicle-treated U937-Cas9 cells. These results suggest that cells in the S phase undergo apoptosis, consistent with AraC’s known mechanism of action.38,39 AraC-treated U937-SLFN11 KO cells, on the other hand, maintained a similar percentage of cells in the S phase and no increase in cells in the sub-G1 phase compared with vehicle-treated U937-SLFN11 KO cells, suggesting that the absence of SLFN11 blocks AraC-induced cell death in S phase.
SLFN11 KO AML cells present enhanced activation of the ATR/CHK1 pathway and decreased cell death in response to AraC treatment. (A-B) U937-Cas9 and U937-SLFN11 KO cells (single clones) were treated with VC (water) or AraC (500 ng/mL) for 6 hours and stained with propidium iodide (PI), followed by flow cytometry analysis. (A) Representative flow cytometry histograms showing U937-Cas9 and U937-SLFN11 KO cell populations in the sub-G1, G1, S, and G2-M phases of the cell cycle before (untreated [UT]) and after AraC treatment. (B) The bar graph of flow cytometry data shows the cell cycle distribution for each cell type and treatment condition. The mean ± standard deviation of 3 independent experiments are shown. For each phase of the cell cycle (sub-G1, G1, S, and G2/M), statistical analysis was performed using 2-way ANOVA followed by Sidak multiple comparisons test (∗∗∗P < .001; ∗∗∗∗P < .0001). (C) U937-Cas9 and U937-SLFN11 KO cells (pooled clones) were treated with either VC (water) or AraC (500 ng/mL) for the indicated lengths of time (hours). The cells were then lysed, and protein lysates were resolved by SDS-PAGE, followed by immunoblotting analyses using the indicated antibodies. GAPDH is shown as a loading control. Immunoblots are representative of 3 independent experiments. (D) U937-Cas9 and U937-SLFN11 KO cells (pooled clones) were seeded into 96-well plates and treated with either VC (DMSO) or increasing concentrations of the ATRi VE822 for 24 hours. Cell viability was assessed using the WST-1 assay. Data are expressed as a percentage of cell viability of VC-treated cells. Means ± SEM of 3 independent experiments are shown. (E) U937-Cas9 (left panel) and U937-SLFN11 KO cells (right panel; pooled clones) were seeded in 96-well plates and treated with either VC (DMSO), 0.3 μM VE822, or increasing concentrations of AraC alone or in combination with 0.3 μM VE822 for 24 hours. Cell viability was assessed by WST-1 assay. Data are expressed as a percentage of cell viability of VC-treated cells (for AraC alone) or 0.3 μM VE822-treated cells (for AraC + 0.3 μM VE822). Means ± SEM of 3 independent experiments are shown. Statistical analysis was performed using 2-way ANOVA followed by the Sidak multiple comparison test. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. (F) U937-SLFN11 KO cells (pooled clones) were treated with VC (DMSO), AraC (500 ng/mL), and/or VE822 (0.3 μM) for 1, 4, or 8 hours, as indicated. Cells were lysed and protein lysates were resolved by SDS-PAGE, followed by immunoblotting analyses using the indicated antibodies. GAPDH is shown as a loading control. Immunoblots are representative of 3 independent experiments. ns, not significant.
SLFN11 KO AML cells present enhanced activation of the ATR/CHK1 pathway and decreased cell death in response to AraC treatment. (A-B) U937-Cas9 and U937-SLFN11 KO cells (single clones) were treated with VC (water) or AraC (500 ng/mL) for 6 hours and stained with propidium iodide (PI), followed by flow cytometry analysis. (A) Representative flow cytometry histograms showing U937-Cas9 and U937-SLFN11 KO cell populations in the sub-G1, G1, S, and G2-M phases of the cell cycle before (untreated [UT]) and after AraC treatment. (B) The bar graph of flow cytometry data shows the cell cycle distribution for each cell type and treatment condition. The mean ± standard deviation of 3 independent experiments are shown. For each phase of the cell cycle (sub-G1, G1, S, and G2/M), statistical analysis was performed using 2-way ANOVA followed by Sidak multiple comparisons test (∗∗∗P < .001; ∗∗∗∗P < .0001). (C) U937-Cas9 and U937-SLFN11 KO cells (pooled clones) were treated with either VC (water) or AraC (500 ng/mL) for the indicated lengths of time (hours). The cells were then lysed, and protein lysates were resolved by SDS-PAGE, followed by immunoblotting analyses using the indicated antibodies. GAPDH is shown as a loading control. Immunoblots are representative of 3 independent experiments. (D) U937-Cas9 and U937-SLFN11 KO cells (pooled clones) were seeded into 96-well plates and treated with either VC (DMSO) or increasing concentrations of the ATRi VE822 for 24 hours. Cell viability was assessed using the WST-1 assay. Data are expressed as a percentage of cell viability of VC-treated cells. Means ± SEM of 3 independent experiments are shown. (E) U937-Cas9 (left panel) and U937-SLFN11 KO cells (right panel; pooled clones) were seeded in 96-well plates and treated with either VC (DMSO), 0.3 μM VE822, or increasing concentrations of AraC alone or in combination with 0.3 μM VE822 for 24 hours. Cell viability was assessed by WST-1 assay. Data are expressed as a percentage of cell viability of VC-treated cells (for AraC alone) or 0.3 μM VE822-treated cells (for AraC + 0.3 μM VE822). Means ± SEM of 3 independent experiments are shown. Statistical analysis was performed using 2-way ANOVA followed by the Sidak multiple comparison test. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. (F) U937-SLFN11 KO cells (pooled clones) were treated with VC (DMSO), AraC (500 ng/mL), and/or VE822 (0.3 μM) for 1, 4, or 8 hours, as indicated. Cells were lysed and protein lysates were resolved by SDS-PAGE, followed by immunoblotting analyses using the indicated antibodies. GAPDH is shown as a loading control. Immunoblots are representative of 3 independent experiments. ns, not significant.
To further investigate the mechanism underlying the low AraC sensitivity observed in SLFN11-deficient U937 cells, we delved into the mechanics of the DNA damage response. SLFN11 is known to be recruited to stalled replication forks created by DDAs via binding to RPA1.4,5 Current models suggest that under situations of replication stress, SLFN11 causes RPA1 disassembly from single-stranded DNA, irreversible cell cycle arrest in the S phase, and ultimately cell death.6,40 ATR is also recruited to stalled replication forks, after which it phosphorylates CHK1. In the presence of replication stress, CHK1 phosphorylation leads to a pause of the cell cycle in the S phase, giving cells time to repair DNA damage,41 supporting cell survival. Therefore, we compared the kinetics of ATR/CHK1 pathway activation in U937-Cas9 and U937-SLFN11 KO cells in response to AraC treatment. Both cell lines showed robust activation of ATR, as indicated by an increase in the phosphorylation of CHK1, within 1 hour of treatment with AraC (Figure 6C). However, although U937-Cas9 cells showed a gradual reduction in phospho-CHK1 by 4 and 8 hours after treatment with AraC, phospho-CHK1 levels remained elevated in U937-SLFN11 KO cells (Figure 6C). Additionally, U937-Cas9 cells showed a large increase in γH2AX, a marker of DNA damage,42 in response to AraC treatment, whereas the U937-SLFN11 KO cells did not (Figure 6C). Moreover, U937-Cas9 cells showed increased levels of cleaved poly (ADP-ribose) polymerase (PARP) compared with U937-SLFN11 KO cells in response to AraC, indicating that AraC-induced apoptosis increased in the presence of SLFN11 in AML cells (Figure 6C). Together, these results suggest that SLFN11 decreases AraC-induced activation of the ATR-dependent DNA damage response, increasing the accumulation of DNA damage, and ultimately, apoptosis. When SLFN11 is absent, AraC-treated AML cells have a prolonged activation of ATR, potentially allowing the repair of DNA damage and evading apoptosis.
Combination of ATR inhibition with AraC treatment induces apoptosis and reduces viability of SLFN11 KO AML cell lines
Previous studies have shown that the loss of SLFN11 sensitizes some solid tumor cell lines to ATR inhibition alone,5 and that chemotherapy resistance in some SLFN11 KO cell lines, such as human prostate cancer cells, can be reversed by the combination of an ATRi and chemotherapy drugs, such as camptothecin43,44. The effects of ATR inhibition in combination with AraC in AML, however, have not yet been studied. Taken together with our findings, this raised the possibility that inhibition of the ATR/CHK1 pathway might also sensitize SLFN11 KO AML cells to AraC treatment. To test this hypothesis, we first treated both U937-Cas9 and U937-SLFN11 KO cells with increasing concentrations of the ATRi VE822.45 No significant difference in cell viability was observed with increasing doses of the inhibitor between both cell lines (Figure 6D). We then used the highest dose of VE822 (0.3 μM) that did not decrease cell viability using the WST-1 assay (Figure 6D) and confirmed that at this concentration, VE822 caused no significant effects on cell proliferation using the trypan blue exclusion assay (supplemental Figure 4A) but still decreased AraC-induced phosphorylation of CHK1 (supplemental Figure 4B). We then treated U937-Cas9 and U937-SLFN11 KO cells with increasing doses of AraC alone or in combination with 0.3 μM VE822, and measured cell viability using the WST-1 assay. We found that the ATRi VE822 sensitized U937-Cas9 cells to AraC treatment, but this drug combination completely overcame the resistance to AraC in U937-SLFN11 KO cells (Figure 6E). Moreover, we observed an increase in cleaved PARP levels at 4 and 8 hours after cotreatment of U937-SLFN11 KO cells with AraC and VE822 compared with either treatment alone (Figure 6F). Using the CRISPR/Cas9 system and the human erythroleukemia cell line HEL, we next generated HEL-Cas9 and HEL-SLFN11 KO cells (supplemental Figure 4C) and, similar to U937 cells, control HEL cells showed increased levels of H2AX phosphorylation and lower levels of Chk1 phosphorylation after 8 hours of AraC treatment compared with SLFN11 KO HEL cells (supplemental Figure 4D). We then treated these cells with AraC alone or in combination with VE822 and observed that knocking out SLFN11 in HEL cells promoted resistance to AraC treatment, but that this resistance could be reversed by cotreatment with VE822, whereas the combination of VE822 and AraC treatment in control HEL cells did not further reduce cell viability compared with AraC treatment alone (supplemental Figure 4E). Additionally, as observed for VE822, we observed no differences in the reduction of cell viability between U937-Cas9 and U937-SLFN11 KO cells in response to increasing concentrations of AZD6738, another ATRi46 (supplemental Figure 4F). When U937-SLFN11 KO cells were cotreated with AZD6738 and AraC, however, they showed a significant reduction in cell viability compared with either treatment alone (supplemental Figure 4G). Together, our data support that ATR inhibition could restore responses to AraC treatment in AML cells expressing low levels or no SLFN11.
Discussion
In this study, we have identified SLFN11 as a potentially novel prognostic and predictive marker for AML, which could have important clinical implications in patients with AML. Currently, there are several prognostic markers used to stratify patients with AML into different risk groups in clinical practice, including gene mutations and cytogenetic abnormalities,32 but there remains a need for identifying additional markers that can further refine risk stratification and improve patient outcomes. Our study demonstrates that high expression of SLFN11 is significantly correlated with longer OS in AML, indicating its potential as a favorable prognostic marker. Higher SLFN11 expression also correlates with increased sensitivity to AraC, the main chemotherapeutic agent used in AML treatment.36 Consistent with this observation, we also show that AML cells lacking SLFN11 are more resistant to AraC treatment using both in vitro and in vivo models. In a previous study, SLFN11 was shown to preferentially suppress the translation of ATR and ataxia telangiectasia mutated (ATM), due to being encoded by genes with high leucine (thymine-thymine-adenine; TTA) content, upon treatment with DDAs, suggesting an SLFN11-dependent mechanism promoting responses to DDAs.47 Thus, the authors suggested that SLFN11-low/absent tumors could possibly be resensitized to DDAs by targeting ATR.47 Moreover, in another study, targeting ATR was found to enhance the antileukemic effects of AraC in AML.48 Consistent with these studies, we showed that the AraC resistance seen in SLFN11 KO AML cells is, at least in part, related to hyperactivation of the ATR/CHK1 pathway. Notably, our results indicate that ATR is an effective target, whose inhibition resensitizes cells lacking SLFN11 to AraC therapy. Based on these findings, incorporating SLFN11 expression analysis into treatment decisions for initial therapy in newly diagnosed AML could significantly improve the outcomes of patients with AML. For example, the combination of ATRis with AraC is a promising treatment strategy for patients with AML with low SLFN11 expression. Notably, a previous study using an ATM/ATRi (CGK733) has shown that inhibition of ATM/ATR signaling alone has no effects on cell viability and apoptosis in human CD34+ hematopoietic progenitor cells from cord blood.49 In another study, Di Tullio et al50 showed that drug-targeted inhibition of CHK1 in combination with AraC treatment reduces AML disease burden and delays leukemic relapse in vivo, but does not affect normal human hematopoietic stem and progenitor cells viability (Lin−CD34+CD38−) in vivo. Together, these studies suggest that there is a potential therapeutic window to treat SLFN11 low AML tumors with an ATRi or CHK1 inhibitor in combination with AraC, without affecting normal CD34+ progenitor cells. However, future studies are required to fully address this. In a recent study, Lasry et al51 performed single-cell RNA sequencing on 10 BM samples from healthy donors, 20 BM aspirates from newly diagnosed adult patients with AML, and 22 BM aspirates from newly diagnosed pediatric patients with AML. In this study, differential expression analysis comparing malignant hematopoietic stem progenitor cell (HSPC)–like cells with healthy donor HSPCs and malignant myeloid-like cells compared with healthy donor myeloid cells identified significant differences in the expression of a number of genes, but not in SLFN11 expression. However, this could be a result of the need of a higher number of samples per group to reach significance and/or the need to include BM aspirates from patients with relapsed AML, as these patients present lower levels of SLFN11 than newly diagnosed patients with AML. Thus, future studies will be important to also compare the levels of SLFN11 further in newly diagnosed and relapse patient-derived AML HSPCs and malignant myeloid cells with healthy donor HSPCs and myeloid cells.
Notably, we also showed that SLFN11-low cells remained sensitive to AZA plus VEN, suggesting that this regimen may be preferable to intensive chemotherapy in patients with SLFN11-low AML. These experiments, however, were performed in vitro and because HMAs have previously been shown to have effects on the immune system,52 it is possible that SLFN11 might also play a noncell intrinsic role in AML cell death in response to HMAs. Future experiments should include humanized mouse models to test the effects of AZA on SLFN11 KO leukemia cells in the presence of a functioning immune system.
Nevertheless, our study supports further validation of the clinical utility of SLFN11 expression as a prognostic and predictive marker in AML, using large patient cohorts. Further work will be important to determine how SLFN11 expression factors are involved in multiple gene mutations and cytogenetic changes. Building a biomarker hierarchy would help determine whether SLFN11 can be successfully used as part of a revised list of risk criteria. The integration of SLFN11 as a biomarker in AML could have significant implications in that it would help clinicians determine which patients with AML are more likely to respond to intensive chemotherapy and which patients are likely to require alternative treatments, such as HMA plus VEN or other combination treatment strategies. It also remains to be explored whether testing SLFN11 expression in AML cells at the single-cell level is required for proper risk stratification, or whether determining the average SLFN11 expression in multiple leukemia blasts from a patient is sufficient. As the average SLFN11 expression in relapsed AML is lower than that in newly diagnosed AML, it is possible that the cells with the lowest levels of SLFN11 expression are the cells that survive treatment with chemotherapy and thus lead to relapse. A more granular analysis, such as using single-cell RNA-sequencing methodologies, would elucidate the variability of SLFN11 expression within an individual and allow further tailoring of AML treatment. In summary, our study strongly supports the potential of using SLFN11 expression in patients with AML for better decision-making when selecting therapeutic regimens and to better define disease outcomes and prognosis.
Acknowledgments
Flow cytometry work was performed at the Northwestern University Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Facility.
This study was supported by National Institutes of Health (grants CA77816 and CA12192) and Veterans Affairs (VA) Merit Review (grant I01CX000916). S.H.S. is supported by the American Society of Hematology Research Training Award.
Authorship
Contribution: S.H.S. conceived the project, designed and performed experiments, analyzed data, and wrote the manuscript; R.E.P. performed experiments and analyzed data; E.M.B. performed experiments, analyzed data, and edited the manuscript; A.H.B. performed experiments; S.D.W. performed experiments and analyzed data; M.F. and M.S. contributed to the performance of experiments; M.K. analyzed the data; D.S. analyzed the data and edited the manuscript; L.C.P. conceived and supervised the project and edited the manuscript; and all authors critically reviewed and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Leonidas C. Platanias, Robert H. Lurie Comprehensive Cancer Center, 303 E. Superior St, Lurie 3-125, Chicago, IL 60611; email: l-platanias@northwestern.edu; and Sara H. Small, Fels Cancer Institute for Personalized Medicine, Lewis Katz School of Medicine, Temple University, 3307 N. Broad St, PAHB Room 204, Philadelphia, PA 19140; email: sara.small@fccc.edu.
References
Author notes
Original data are available on request from the corresponding authors, Sara H. Small (sara.small@fccc.edu) or Leonidas C. Platanias (l-platanias@northwestern.edu).
The full-text version of this article contains a data supplement.

![Low SLFN11 expression correlates with lower sensitivity to AraC. (A) Correlation analysis between AraC activity and SLFN11 gene methylation in cancer cell lines from the NCI-60 panel. Data were extracted from CellMiner Cross-Database (CDB) (n = 60). (B) Correlation analysis between AraC activity and Log2 of SLFN11 mRNA expression in cancer cell lines from the NCI-60 panel. Data were extracted from CellMiner CDB (n = 59). (A-B) Statistical analysis was performed using simple linear regression, and P values are shown for deviation of the line slope from 0. (C) U937 cells were treated with the vehicle control (dimethyl sulfoxide [DMSO]) or 1 μM AZA for 24 hours. Protein lysates from U937 cells were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblotting analysis with the indicated antibodies. GAPDH is shown as a loading control. (D) Clonogenic capability of primary leukemic blasts isolated from patients with AML expressing either high levels of SLFN11 (SLFN11-high, n = 3 for 8 ng/ml AraC and n = 4 for 0.08 and 0.8 ng/ml AraC) or low levels of SLFN11 (SLFN11-low, n = 3) treated with either vehicle control (VC; water) or increasing concentrations of AraC, as indicated (left panel). Data are expressed as the percentage of colony formation over VC-treated cells for each patient (Ctrl) and are shown as mean ± standard error of the mean (SEM). Statistical analyses were performed using Mixed-effects analysis followed by Tukey multiple comparisons test and P values are shown for the highest AraC dose for each group. There was a significant interaction (P = .043), that is, the relationship between concentration and percentage of colony formation was significantly different between patients with AML that were SLFN11-low vs SLFN11-high. (D) Relative SLFN11 mRNA expression, normalized to GAPDH, was assessed by qRT-PCR analysis and is shown for the primary leukemic blasts isolated from patients with AML and used in the clonogenic assays (SLFN11-low, n = 3 and SLFN11-high, n = 4; right panel).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodneoplasia/1/4/10.1016_j.bneo.2024.100037/2/m_bneo_neo-2024-000191-gr3.jpeg?Expires=1764598215&Signature=0HwN9PWhmncBpeyknJT-6RZgQRoTNjOLqdxWt9MbIn~BARg152iTUv33wroxVYwiKirHKgNxyHmaYJYLtNyjX48bb8RWTWFOnFaSLLtXX5s-aOAsds9qAUjSaLoBZfIhDK-F5ocqMZ7OHCzkkzVbypf415OizrQwGb9Ej95o5Hc3ngmvEh23Rufknjm5QHk9Gz9BnT2ipGpnZz5maBcjwljCO7qIVd2FCQfqSrocv5QL~IA94jpFTnyaNR2JwkUOvu71Uoz182NNHmtnSfzghabznSWFadBlhxcmCRCSfYNVpyVCiq5uuw31T~OvAz746xF5~9JHLgwZV~Xm1dvyLg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![SLFN11 KO AML cells present enhanced activation of the ATR/CHK1 pathway and decreased cell death in response to AraC treatment. (A-B) U937-Cas9 and U937-SLFN11 KO cells (single clones) were treated with VC (water) or AraC (500 ng/mL) for 6 hours and stained with propidium iodide (PI), followed by flow cytometry analysis. (A) Representative flow cytometry histograms showing U937-Cas9 and U937-SLFN11 KO cell populations in the sub-G1, G1, S, and G2-M phases of the cell cycle before (untreated [UT]) and after AraC treatment. (B) The bar graph of flow cytometry data shows the cell cycle distribution for each cell type and treatment condition. The mean ± standard deviation of 3 independent experiments are shown. For each phase of the cell cycle (sub-G1, G1, S, and G2/M), statistical analysis was performed using 2-way ANOVA followed by Sidak multiple comparisons test (∗∗∗P < .001; ∗∗∗∗P < .0001). (C) U937-Cas9 and U937-SLFN11 KO cells (pooled clones) were treated with either VC (water) or AraC (500 ng/mL) for the indicated lengths of time (hours). The cells were then lysed, and protein lysates were resolved by SDS-PAGE, followed by immunoblotting analyses using the indicated antibodies. GAPDH is shown as a loading control. Immunoblots are representative of 3 independent experiments. (D) U937-Cas9 and U937-SLFN11 KO cells (pooled clones) were seeded into 96-well plates and treated with either VC (DMSO) or increasing concentrations of the ATRi VE822 for 24 hours. Cell viability was assessed using the WST-1 assay. Data are expressed as a percentage of cell viability of VC-treated cells. Means ± SEM of 3 independent experiments are shown. (E) U937-Cas9 (left panel) and U937-SLFN11 KO cells (right panel; pooled clones) were seeded in 96-well plates and treated with either VC (DMSO), 0.3 μM VE822, or increasing concentrations of AraC alone or in combination with 0.3 μM VE822 for 24 hours. Cell viability was assessed by WST-1 assay. Data are expressed as a percentage of cell viability of VC-treated cells (for AraC alone) or 0.3 μM VE822-treated cells (for AraC + 0.3 μM VE822). Means ± SEM of 3 independent experiments are shown. Statistical analysis was performed using 2-way ANOVA followed by the Sidak multiple comparison test. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. (F) U937-SLFN11 KO cells (pooled clones) were treated with VC (DMSO), AraC (500 ng/mL), and/or VE822 (0.3 μM) for 1, 4, or 8 hours, as indicated. Cells were lysed and protein lysates were resolved by SDS-PAGE, followed by immunoblotting analyses using the indicated antibodies. GAPDH is shown as a loading control. Immunoblots are representative of 3 independent experiments. ns, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodneoplasia/1/4/10.1016_j.bneo.2024.100037/2/m_bneo_neo-2024-000191-gr6.jpeg?Expires=1764598215&Signature=RsbA6gXFApFLmvWrzp4VgmaOf~lDdvBb2LfSjqnpcOjbl~LGLwP0tbic5u2HvfylSyuWtjhG7qIqlPqJTOGpHzzSF0bv3pe6zb88cAhvr~3gKWuAWpLsNO7AoMw~4YF2wfpSsaQU4DN-F5kVi~ylSN8OFtrf6UgEbGlp1xd8tEq0jFJYybY86MpVlkw4ikcJhULRgqTEXFQVGTDTZXKcTllLYl5DGgF7MdW8wk77hiPc~DLdVJRSl8A92zWcOHyXKCjwfcrj5RaP3ODh2Aw1j3I8NqT9DqhFpGd2k0SmDG8nVuVVLlQ~JpX5noOXbnoi~q340fhkaRMIV2O~9EK5oA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)