Evidence suggests that T lymphocyte–mediated inhibition of hematopoiesis in myelodysplastic syndrome (MDS) contributes to cytopenia in some patients and can be reversed by treatment with immunosuppression. We examined the T-cell repertoires of 12 patients with MDS before and after antithymocyte globulin (ATG)–based treatment by T-cell receptor Vβ (TCR-Vβ) spectratype analysis. The average number of TCR-Vβ families with skewed spectratypes, representative of clonal or oligoclonal T-cell populations, was 7.6 in MDS patients before treatment and 3.2 in healthy controls (P = .02). Four patients who recovered effective hematopoiesis after treatment lost prominent, skewed peaks on their spectratypes, suggesting loss or diminution of overrepresented clonal T-cell populations. In contrast, patients who did not recover effective hematopoiesis showed persistently skewed repertoires 3 to 6 months after treatment. In 3 patients with skewed repertoires, cDNA from the complementarity-determining region 3 (CDR3) of 4 TCR-Vβ families was cloned and repetitively sequenced, confirming clonal T-cell dominance in each family. In one nonresponder, 16 of 19 CDR3 sequences were identical, demonstrating that 9.3% of the total T-cell population was made up of a single clone. By 6 months after treatment, this clone persisted on both spectratype and DNA sequence complementarity and when analyzed by flow cytometry was shown to be CD8+/CD45RA+/HLA-DR−. T-cell clones were not anergic because they could be expanded 4-fold in vitro. Our results demonstrate that predominant clonal T cells that appear to be antigen-driven persist in patients with MDS unresponsive to immunosuppression, but predominant clones regress in responders to immunosuppression.
Introduction
Myelodysplastic syndrome (MDS) is a bone marrow disorder characterized by dysplasia, cytopenia, and a propensity for conversion to acute myeloid leukemia (AML). Although the etiology of the ineffective hematopoiesis and resultant cytopenia is not certain, evidence suggests that T cell–mediated suppression of hematopoietic precursors contributes to the cytopenia in some patients.1-4 Similarly, in aplastic anemia there is also evidence of T cell–mediated inhibition of hematopoietic precursors and prolonged recovery of hematopoiesis is observed in up to 70% of patients after immunosuppressive treatment with antithymocyte globulin (ATG).5-7 In 25 patients with MDS and significant cytopenia who were treated with ATG, we found that 35% of patients had recovery of effective hematopoiesis.8 We studied 5 of those patients for lymphocyte-mediated suppression and found that removal of CD3+ or CD8+ T cells from bone marrow progenitor cultures resulted in a significant increase in granulocyte-macrophage colony-forming units (CFU-GMs) in responders to ATG, but not in nonresponders. In 1 of 2 patients with refractory anemia, there also appeared to be increased T-cell polyclonality after clinical response to ATG treatment when T-cell receptor (TCR) diversity was crudely analyzed by single-stranded DNA conformational polymorphism (SSCP), suggesting that diversification of the T-cell repertoire might occur in patients who respond to immunosuppression.2
To investigate whether dominant T-cell clones are present in patients with MDS, we studied 12 patients with cytopenia who were enrolled in a randomized clinical trial of ATG, ATG plus fludarabine, or ATG plus cyclosporine. We hypothesized that dominant clonal T lymphocytes contribute to cytopenia by suppressing hematopoietic precursors and that these clones become less prominent in patients who respond to immunosuppression. TCR-Vβ spectratype analysis was used to study peripheral blood T-cell diversity before and 3 to 6 months after ATG-based treatment, and dominant clones were analyzed further by flow cytometry. In this study, we demonstrate that all 12 MDS patients have T-cell repertoires that show dominant clonal and oligoclonal T-cell expansions. Moreover, we demonstrate a significantly increased number of TCR-Vβ families with skewed complementarity-determining region (CDR3) length distributions in MDS patients compared with healthy controls. This increased amount of CDR3 skewing strongly suggests an increased number of predominant clonal and oligoclonal T-cell expansions in the patients with MDS compared with the healthy controls. In one patient, 28% of total CD8+ T lymphocytes, representing 9.3% of total T cells, was derived from a single clone. These T-lymphocyte clones persisted in patients who did not respond to immunosuppressive therapy, but there was loss of overrepresented T-cell expansions in all 4 patients with a hematologic response to immunosuppressive therapy. Together with our previous studies, these results suggest that predominant clonal T lymphocytes contribute to the cytopenia of MDS and that patients who respond to immunosuppression lose clonal dominance, permitting polyclonal T cells to re-emerge with reversal of hematopoietic suppression and recovery of effective hematopoiesis.
Patients, materials, and methods
Patients
Twelve patients with refractory anemia (RA), refractory anemia with ringed sideroblasts (RARS), refractory anemia with excess blasts (RAEB), or refractory anemia with excess blasts in transformation (RAEBt) subtypes of MDS and transfusion-dependent cytopenia who were enrolled in a phase 2 ATG-based treatment protocol were studied. These patients were the first 12 patients for which both pretreatment and posttreatment samples were available. Informed consent was obtained prior to enrollment and all research was approved by the institutional review board of the M. D. Anderson Cancer Center. Heparinized blood and bone marrow samples were obtained just before and again 3 to 6 months after treatment. All patients studied were confirmed by morphologic examination to have MDS by hematopathologists not otherwise involved in this research. All of the patients were dependent on red blood cell or platelet transfusions, requiring at least 2 U red blood cells monthly. All patients were evaluated for T-cell large granular lymphocyte (T-LGL) leukemia by standard morphologic and immunophenotypic criteria.9 Patients with T-LGL leukemia were excluded from this study. Patients were also evaluated for paroxysmal nocturnal hemoglobinuria (PNH) by flow cytometry and were similarly excluded. The mean age of the MDS patients studied was 58 years. Patient characteristics are summarized in Table1. All patients reported here received treatment consisting of either ATG (Upjohn, Kalamazoo, MI), 40 mg/kg for 4 days, or the same schedule of ATG plus either fludarabine (Berlex, Richmond, CA), 20 mg/m2 daily for 4 days, or cyclosporine 6 mg/kg daily for 3 months. Hematologic recovery was defined previously and required at minimum a 50% increase in blood counts with transfusion independence for more than 2 months.8 Patients were all free of active infections at the time of treatment. The results of this ongoing clinical trial will be reported elsewhere.
In addition, peripheral blood was collected from 13 healthy individuals for use in normal control spectratype experiments. Spectratype analysis was performed on all 13 healthy control subjects and the results were compared with the results from the 12 patients with MDS. The mean age of the healthy controls was 52 years.
Mononuclear cell isolation
Both blood and bone marrow were collected in heparin-containing vials. As soon as possible after collection, the bone marrow mononuclear cells (BMCs) and peripheral blood mononuclear cells (PBMCs) were collected by centrifugation over a Histopaque 1077 (Sigma, St Louis, MO) density gradient and cryopreserved in liquid nitrogen.
Polymerase chain reaction and CDR3 size analysis
In all instances, 4 to 8 × 106 BMCs or PBMCs were thawed, washed once, and used for RNA extraction. For UPN 3, 11, and 12, BMCs were used for RNA extraction. In all other MDS patients and all healthy controls PBMCs were the source of RNA. RNA was extracted using the RNA Stat 60 kit (Tel Test, Friendswood, TX) according to the manufacturer's instructions. Then, 1 μg of the extracted RNA was treated with DNAse (Gibco BRL, Rockville, MD) to remove contaminating gDNA. All of the DNAse-treated RNA was used to synthesize cDNA by reverse transcription using the manufacturer's protocol with the Gene Amp RNA kit (Perkin Elmer, Foster City, CA).
Polymerase chain reaction (PCR) was then performed as previously described2 by combining 25 pmol of one Vβ primer for each of the different TCR-Vβ families with 25 pmol of a 6-FAM–labeled Cβ primer, which was used for each of the 25 reactions. The reactions were carried out in a 25-μL reaction volume with 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, 200 μM each of dNTP and 2 U Taq polymerase. One thirtieth of the total cDNA synthesized from 1 μg RNA was used in each reaction. The PCRs were cycled as follows: 95°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute for 30 cycles, with a final extension step of 72°C for 8 minutes. Thirty cycles of PCR allowed semiquantitative comparative analysis of the PCR products on an ethidium bromide gel by densitometry using the Gel Dock 1000 imager system (Biorad, Hercules, CA). The relative percentage of the total TCR-Vβ repertoire made up by each family was determined by dividing the intensity of the signal for each TCR-Vβ family by the sum of the intensities of the signals from all of the 25 TCR-Vβ families. A negative control in which cDNA was replaced by nuclease-free water was included in all PCR experiments.
The PCR products were next diluted in nuclease-free water so that 1.5 ng of the PCR product from each TCR-Vβ family was subjected to capillary electrophoresis using a 310 Genetic Analyzer single capillary electrophoresis machine (Perkin Elmer), and then analyzed using Genescan software (Perkin Elmer). Because the positions of the 5′ Vβ and the 3′ Cβ primers are fixed, variation in length of the PCR fragments within any TCR Vβ family is due to differences in length of the CDR3 regions. Genescan data are presented as fluorescence intensity versus DNA fragment length. The TCR-Vβ 10 and 19 families are pseudogenes and were therefore excluded from analysis.
Objective scoring of TCR-Vβ family skewing
Genescan software provides TCR-Vβ spectratypes in a graph format and also gives quantitative fluorescence intensity for each peak in a CDR3 length distribution. The number of peaks within each of the 23 TCR-Vβ family CDR3 length distributions were counted in all 12 patients with MDS and all 13 healthy subjects. Using a previously established method,10 a peak was defined as a fluorescence signal with at least 15% of the intensity of the strongest signal within its TCR-Vβ family CDR3 length distribution. To be counted as a peak, a fluorescence signal also had to be separated from the nearest neighboring signal by at least 2 nucleotides. A TCR-Vβ family was scored as normal if its CDR3 length distribution contained at least 6 peaks. TCR-Vβ families with fewer than 6 peaks were scored as abnormal.
Cloning and sequencing of PCR-amplified cDNA
The PCR products were visualized on an agarose gel, then excised and purified using the Qiaquick gel extraction kit (Qiagen, Valencia, CA). Next, the purified PCR product was cloned into the PCR 2.1 vector and expressed in Escherichia coli using the Original TA Cloning Kit (Invitrogen, Carlsbad, CA). Colonies were randomly selected and DNA was obtained by miniprep using the Qiaprep miniprep kit (Qiagen) according to the manufacturer's instructions. DNA from 20 randomly selected colonies containing inserts of the appropriate size was sequenced using M13 primers and an ABI 377 DNA Sequencer (Perkin Elmer). DNA sequences were compared and amino acid sequences deduced using MacVector software (Genetics Computer Group, Madison, WI).
Flow cytometry
The phenotype of T cells from patients shown to have clonal T-cell expansions was determined by flow cytometry using TCR-Vβ family-specific antibodies combined with antibodies for activation and memory phenotype. PBMCs were thawed and washed twice and 500 000 cells were labeled with combinations of the following monoclonal antibodies: Vβ 6.7 (Immunotech, Marseille, France), CD8 (Caltag, Burlingame, CA), CD45RO (Immunotech), CD4 (Caltag), CD45 RA (Caltag), HLA-DR (Pharmingen, San Jose, CA), CD 25 (Caltag), Vβ 1 (Immunotech), CD45 RA (Immunotech), HLA-DR (Caltag), CD8 (Immunotech), and Vβ 8 (Pharmingen). Cells were incubated 30 minutes at 4°C with antibodies and washed once and 6-color analysis was performed using the MoFlo flow cytometer (Cytomation, Ft Collins, CO).
Detection of specific CDR3 sequences
A detailed method for detection of specific CDR3 sequences from reverse-transcribed PCR products has been previously described.11 Target cDNA products, which were reverse-transcribed from the PCR products of a TCR-Vβ family shown to contain a clonal T-cell expansion with a known CDR3 sequence, were loaded onto a 1.5% agarose gel and electrophoresed at 70 V for 1 hour. The cDNA was transferred to a positively charged nylon membrane using the Gel Blot 758 device (Biorad). Next, an oligonucleotide probe (probe sequence: TCCGACAGGGGGGCTCCTTG) corresponding to the CDR3 sequence of interest was labeled using the DIG oligonucleotide tailing kit (Roche, Indianapolis, IN) following the manufacturer's instructions. Prehybridization was carried out using 20 mL buffer made up of 5 × standard sodium citrate (SSC), 2 mg poly A, 1% blocking solution (Roche), 0.1% N-lauroylsakosine, and 0.02% sodium dodecyl sulfate (SDS). Next, the prehybridization buffer was discarded and hybridization buffer was added, along with 0.3 pmol/mL labeled oligonucleotide probe. Hybridization was carried out for 6 hours at an incubation temperature specific for each probe. The membrane was washed 2 times for 5 minutes at room temperature in 2 × SSC, 0.1% SDS, and then 2 times at room temperature for 15 minutes in 0.1% SDS. Finally, the probe was detected as instructed by the manufacturer using the Dig Luminesence detection kit (Roche).
Statistical analysis
The 2-tailed, unpaired Student t test was used to compare the number of TCR-Vβ families with abnormal CDR3 length distributions in patients with MDS and healthy control subjects.
Results
Restricted T-cell repertoires in MDS patients are characterized by increased skewing of CDR3 lengths compared with healthy individuals
We have previously shown that T lymphocyte–mediated suppression of hematopoiesis occurs in a subset of MDS patients, which is reversed by ATG treatment in clinical responders. In this study, we have examined the T-cell repertoire of 12 MDS patients before and after treatment with ATG-based immunosuppressive regimens using TCR-Vβ spectratyping of CDR3 lengths. Ordinarily, the CDR3 length distributions of each TCR-Vβ family in a healthy individual contain at least 6 peaks in a gaussian distribution, indicating a polyclonal T-cell population. A markedly nongaussian CDR3 length distribution, referred to as skewing, suggests a clonal or oligoclonal T-cell population. For comparison, we also performed TCR-Vβ spectratype analysis on a panel of 13 healthy individuals. An objective scoring method10 was used to determine the number of TCR-Vβ families with normal (gaussian, diverse) and abnormal (skewed, restricted) CDR3 length distributions in each MDS patient prior to treatment and in each of 13 healthy control subjects.
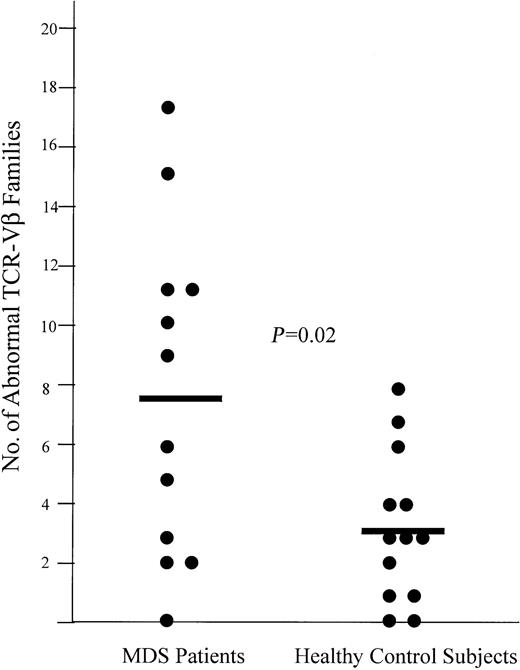
All MDS patients showed skewing of TCR-Vβ spectratypes (Table 1). The MDS patients had a mean of 7.6 (± 5.4) TCR-Vβ families with skewed spectratypes, whereas the healthy control subjects had a mean of 3.2 (± 2.6) TCR-Vβ families with abnormal CDR3 length distributions (P = .02; Figure 1). Representative examples of the complete TCR-Vβ spectratypes of 3 healthy control subjects are shown in Figure2. The complete spectratypes of each subject include the individual CDR3 length distributions of each of the 23 TCR-Vβ families.
Representative T-cell repertoire in MDS patients compared with healthy control subjects.
Comparison of the number of TCR-Vβ families with skewed spectratypes in MDS patients with healthy control subjects (P = .02). The mean number of TCR-Vβ families with skewed spectratypes is denoted by the horizontal bars.
Representative T-cell repertoire in MDS patients compared with healthy control subjects.
Comparison of the number of TCR-Vβ families with skewed spectratypes in MDS patients with healthy control subjects (P = .02). The mean number of TCR-Vβ families with skewed spectratypes is denoted by the horizontal bars.
T-cell repertoire diversity in healthy subjects.
Three representative example of spectratype analyses performed on 13 healthy control subjects show that most TCR-Vβ families exhibit a diverse, gaussian distribution of CDR3 lengths. However, some TCR-Vβ families do exhibit a skewed or restricted distribution of CDR3 lengths. The ages of the control subjects are (A) 31, (B) 41, and (C) 61 years.
T-cell repertoire diversity in healthy subjects.
Three representative example of spectratype analyses performed on 13 healthy control subjects show that most TCR-Vβ families exhibit a diverse, gaussian distribution of CDR3 lengths. However, some TCR-Vβ families do exhibit a skewed or restricted distribution of CDR3 lengths. The ages of the control subjects are (A) 31, (B) 41, and (C) 61 years.
Loss of T-cell clonal dominance on CDR3 spectratype analysis occurs in responders to ATG-based treatment
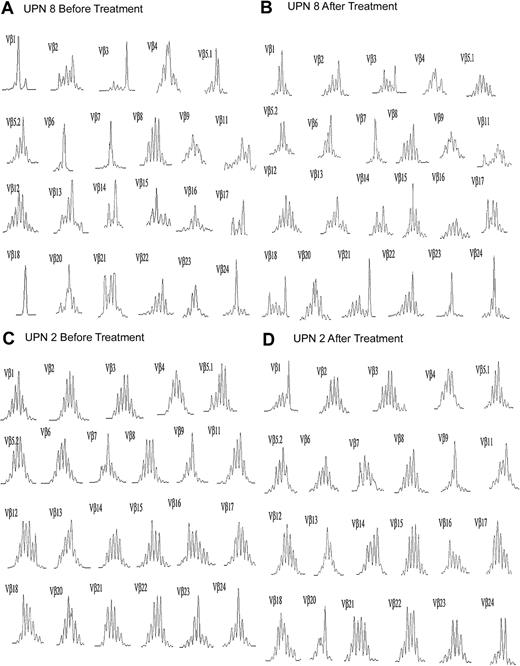
All of the MDS patients showed skewing on pretreatment spectratypes, and complete spectratypes prior to ATG-based treatment and again 3 to 6 months after treatment are shown for comparison in Figure 3 for one responder and one nonresponder. This skewing persisted for at least 3 to 6 months after treatment in all nonresponders. In some cases, in addition to the same predominant peak, new peaks appeared in the spectratypes of previously skewed TCR-Vβ families, as represented in the TCR-Vβ 18 family from UPN 8 (Figure 3). Four patients responded to therapy with ATG-based therapies with recovery of effective hematopoiesis (UPN 1-3, and UPN 5). In all 4 patients, complete loss or marked diminution of a dominant skewed peak on spectratype analysis occurred in at least one TCR-Vβ family. Figure 4 shows the persistence of skewed spectratypes in the nonresponders (Figure 4A), and the loss of skewing in the responders (Figure 4B).
Representative TCR-Vβ spectratype before and after ATG-based treatment.
Complete spectratype for (A) UPN 8, a clinical nonresponder, and (B) UPN 2, a responder.
Representative TCR-Vβ spectratype before and after ATG-based treatment.
Complete spectratype for (A) UPN 8, a clinical nonresponder, and (B) UPN 2, a responder.
Representative TCR-Vβ spectratypes from nonresponders and responders before and after ATG-based treatment.
Overrepresented CDR3 lengths on spectratype analysis persist in nonresponders to immunosuppression, but become less dominant in responders. The length in base pairs of the dominant peak is indicated on the x-axis. The y-axis represents relative fluorescence intensity. The percentages with each spectratype give the percentage of the total T-cell repertoire made up by the indicated TCR Vβ family. (A) Two representative nonresponders, and (B) the 4 responders.
Representative TCR-Vβ spectratypes from nonresponders and responders before and after ATG-based treatment.
Overrepresented CDR3 lengths on spectratype analysis persist in nonresponders to immunosuppression, but become less dominant in responders. The length in base pairs of the dominant peak is indicated on the x-axis. The y-axis represents relative fluorescence intensity. The percentages with each spectratype give the percentage of the total T-cell repertoire made up by the indicated TCR Vβ family. (A) Two representative nonresponders, and (B) the 4 responders.
Presence of skewing on spectratype analysis suggests, but does not conclusively demonstrate, a clonal T-cell expansion. Therefore, we further analyzed several TCR-Vβ families with extensive skewing on spectratype analysis by cloning cDNA from these families followed by direct repetitive sequencing of multiple clones. The presence of identical CDR3 sequences from plasmid clones recovered from randomly selected separate E coli colonies demonstrates a clonal expansion.
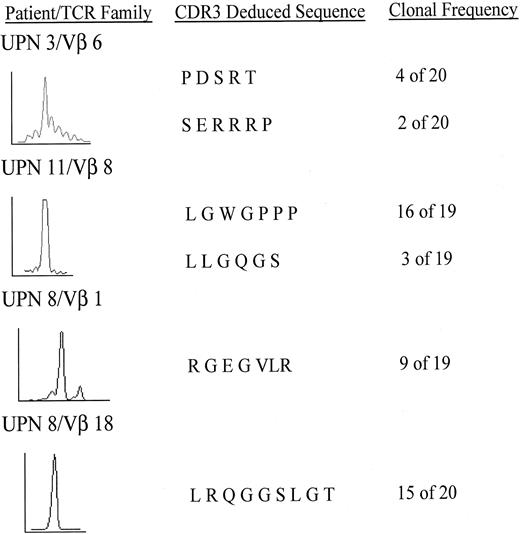
In UPN 11, the TCR-Vβ 8 family exhibited a single peak on spectratype analysis. After cloning and sequencing, 16 of 19 clones, or 84% of the T lymphocytes in the TCR-Vβ 8 family shared an identical sequence, as shown in Figure 5. Because the TCR-Vβ 8 family made up 11.3% of the total bone marrow TCR repertoire by semiquantitative analysis of all TCR-Vβ PCR products (data not shown), the clone made up 9.3% of the total T lymphocytes. In a control experiment, the cDNA from a healthy donor TCR-Vβ family was cloned, and 12 of 12 colonies exhibited unique CDR3 sequences.
Sequencing of CDR3 region in skewed TCR-Vβ families.
Cloned CDR3 PCR products were subjected to repetitive sequencing. Spectratype (left), deduced amino acid sequence (middle), and the number of clones with the identical sequence as a clonal frequency (right) are shown.
Sequencing of CDR3 region in skewed TCR-Vβ families.
Cloned CDR3 PCR products were subjected to repetitive sequencing. Spectratype (left), deduced amino acid sequence (middle), and the number of clones with the identical sequence as a clonal frequency (right) are shown.
In UPN 8, 2 clonal expansions were demonstrated, 1 in the TCR Vβ 18 family in which 15 of 20 clones shared an identical CDR3 sequence, and 1 in the TCR-Vβ 1 family in which 9 of 19 clones had the same CDR3 sequence. After determining the relative expression of all TCR-Vβ families, the clones corresponded to 3.4% and 2.5%, respectively, of the total T-lymphocyte population in this patient.
In UPN 3, a transient responder to immunosuppression, 4 of 20 clones from the TCR-Vβ 6 family exhibited identical CDR3 sequences before treatment. Because the TCR-Vβ 6 family made up 11.6% of the total T-lymphocyte repertoire, as determined by the ratio of the TCR-Vβ 6 PCR product to the total TCR-Vβ PCR products, the clone made up 2.3% of the total T lymphocyte population. The length of the clonal CDR3 sequence was identical to the length of the dominant peak seen on the pretreatment spectratype analysis (Figures 4B and 5). This patient had a clinical relapse characterized by decreasing blood counts on day 151 after treatment with ATG. CDR3 cloning and sequencing of the TCR-Vβ 6 family, performed on day 179 after ATG treatment, showed 2 of 20 clones to be identical to the prominent clone detected before treatment, although the corresponding peak on the CDR3 spectratype had decreased substantially after treatment (Figure 4B). The sequencing and spectratype data taken together suggest that although still present, the clone is no longer dominant in this relapsing patient. In light of the clinical relapse, this may represent the re-emergence of the clone.
Persistent clonal dominance in TCR-Vβ spectratype corresponds to a single clonal T cell
The TCR-Vβ 18 family of UPN 8 was shown to contain a dominant clonal expansion by spectratype and sequence analysis (Figure 5). To confirm that the predominant clonal T lymphocyte was still present after treatment with immunosuppression, a DNA probe specific for the dominant CDR3 sequence was designed. As shown by Southern blot in Figure 6, this dominant CDR3 sequence persisted 6 months after immunosuppression in this patient. This demonstrates the persistence of a dominant clonal T-cell expansion in a patient who did not have hematologic improvement after immunosuppressive therapy.
Detection of CDR3 sequences before and after ATG-based treatment.
The TCR-Vβ 18 family of UPN 8 contained a clonal T-cell expansion that persisted 6 months after ATG treatment. The detection of the target DNA sequence was performed on TCR-Vβ 18 PCR product using an oligonucleotide probe with the shared CDR3 sequence of the clonal T-cell expansion. Lane 1 was loaded with 1 ng and lane 2 with 0.1 ng target DNA product from pretreatment PBMCs of UPN 8. Lane 3 was loaded with 1 ng and lane 4 with 0.1 ng target DNA product from posttreatment PBMCs of UPN 8. Lane 5 is a negative control in which 2 ng of the reverse-transcribed PCR product of the TCR-Vβ 18 family from a healthy donor was probed.
Detection of CDR3 sequences before and after ATG-based treatment.
The TCR-Vβ 18 family of UPN 8 contained a clonal T-cell expansion that persisted 6 months after ATG treatment. The detection of the target DNA sequence was performed on TCR-Vβ 18 PCR product using an oligonucleotide probe with the shared CDR3 sequence of the clonal T-cell expansion. Lane 1 was loaded with 1 ng and lane 2 with 0.1 ng target DNA product from pretreatment PBMCs of UPN 8. Lane 3 was loaded with 1 ng and lane 4 with 0.1 ng target DNA product from posttreatment PBMCs of UPN 8. Lane 5 is a negative control in which 2 ng of the reverse-transcribed PCR product of the TCR-Vβ 18 family from a healthy donor was probed.
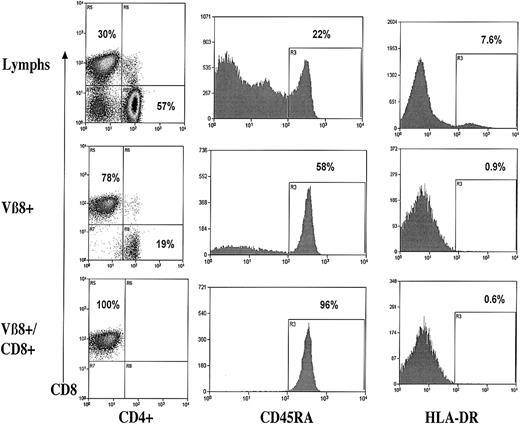
The prominent clonal T-cell expansion found in UPN 11 has a naive CD8+ phenotype
The TCR-Vβ 8 family of UPN 11 was shown to contain a large clonal population with 16 of 19 clones sharing an identical CDR3 sequence, as shown in Figure 5. We further analyzed this TCR-Vβ family by flow cytometry, as shown in Figure7. The TCR-Vβ 8 family in UPN 11 was 78% CD8+CD4−. The CD8+subpopulation of this TCR-Vβ family was a naive lymphocyte population with 96% CD45RA+ exhibiting low HLA-DR expression of 0.6%.
Flow cytometric analysis of total lymphocytes and the TCR-Vβ 8+ T cells from UPN 11.
CD4/CD8, CD45 RA, and HLA-DR expression on unfractionated lymphocytes (top row), TCR-Vβ 8+ lymphocytes (middle row), and CD8+ TCR-Vβ8+ lymphocytes (bottom row).
Flow cytometric analysis of total lymphocytes and the TCR-Vβ 8+ T cells from UPN 11.
CD4/CD8, CD45 RA, and HLA-DR expression on unfractionated lymphocytes (top row), TCR-Vβ 8+ lymphocytes (middle row), and CD8+ TCR-Vβ8+ lymphocytes (bottom row).
The PBMCs from 3 patients, UPN 3, UPN 8, and UPN 11, were expanded 3- to 5-fold after 7 to 10 days of coincubation with anti-CD3/anti-CD28 beads and analyzed again by flow cytometry. The percentage of T cells that expressed the skewed TCR-Vβ receptor remained constant, as evidenced by identical surface expression (data not shown). This implies that the dominant clonal T cells were not anergic because they persisted and underwent proportional expansion.
Discussion
In this study, we evaluated the total T-cell repertoire of 12 MDS patients using TCR spectratype analysis before and after treatment with ATG-based therapy to look for evidence of T-lymphocyte clonality. This analysis showed that all 12 patients exhibited extensive skewing of their pretreatment TCR spectratypes, suggesting clonal or oligoclonal T-cell expansions. Furthermore, we demonstrated a statistically significant increase in the number of TCR-Vβ families with abnormal CDR3 length distributions in MDS patients compared with normal controls. To directly demonstrate clonal expansions, we applied a strategy of cDNA cloning and sequencing to several TCR-Vβ families that showed skewing on spectratype analysis. We demonstrated 4 dominant clonal T-cell expansions, 3 in patients not responding to immunosuppression and 1 in a patient who responded hematologically to ATG, corresponding to dominant peaks on spectratype analysis. Phenotypic analysis in one nonresponder showed that the clonal T cells were CD8+, naive, and nonactivated. The clones from 3 patients were not anergic because they could be expanded in vitro. Finally, the dominant T-cell clones were replaced by a normal polyclonal T-cell distribution in the patients whose blood counts recovered after immunosuppressive treatment.
Dominant T-cell clones have also been found in other closely related bone marrow failure syndromes with cytopenia that are thought to be immune-mediated in part, such as aplastic anemia (AA)12,13and PNH.14 Like some patients with MDS, patients with these disorders frequently respond to treatment with ATG and other immunosuppressive drugs with restored effective hematopoiesis. This suggests a common immune pathogenesis for the development of cytopenia in these different disorders. In cases of PNH, overrepresented T-cell expansions within TCR-Vβ 4 have been noted to be present at increased frequency.14 Interestingly, we observed skewing in the pretreatment CDR3 length distributions of the TCR-Vβ 4 family in 7 of 12 (58%) MDS patients studied. Similar to other studies that found up to 40% of patients with AA and PNH with skewing in the TCR-Vβ 7 family,12 14 we found that 5 of 12 (42%) of our patients showed such skewing.
Myelodysplastic syndrome occurs more commonly among older individuals and increasing T-cell clonality has been demonstrated to occur in normal healthy individuals with increasing age.15-17 The T-cell clones found in aged healthy individuals are most commonly long-lived memory CD8+ T lymphocytes that are anergic.18 In contrast, the CD8+ clonal T cells in one MDS patient showed a naive phenotype and were not anergic. Our findings are similar to other reports demonstrating autoreactive clonal T-cell expansions in the blood and inflamed joint spaces of patients with rheumatoid arthritis.19 Importantly, the loss or reduction of T-cell clonality was associated with response to immunosuppressive treatment and recovery from cytopenia.
Because most of the patients in this study received multiple transfusions of red blood cells and platelets, it is possible that T-cell clones might have emerged as a consequence of alloimmunization. However, Karadimitris et al investigated this possibility in their report of increased T-cell clonality among patients with PNH and they found no TCR-Vβ skewing in a control group of patients who were heavily transfused and alloimmunized.14
Although hematopoiesis in MDS patients is also clonal in the majority of patients, it is unlikely that the clonal T-cell expansions are part of the MDS clone. In previous studies where T cells have been evaluated by studying the pattern of inactivation of X chromosomes in female patients, the majority of T lymphocytes were found to be polyclonal and therefore not derived from the clonal dysplastic hematopoietic precursors.20,21 Moreover, the clonal T-cell expansions evaluated in this study appear to have undergone normal development. The sequences of the CDR3 regions from these clonal populations contained nucleotide changes consistent with the normal junctional diversity that occurs during thymic development.22
Surprisingly, the dominant clonal T lymphocytes in MDS patients may constitute up to 9.3% of the total T-cell population, which suggests a dominant antigen-driven expansion of these cells. It is likely that the dominant clonal T cells retain effector function because they can be expanded in vitro and thus are not anergic. We have previously shown that T lymphocytes contribute to cytopenia in MDS patients by directly inhibiting CFU-GM hematopoietic precursors and that restored effective hematopoiesis after ATG treatment is associated with loss of T cell–mediated inhibition.2,8 Taken together with the results from the current study, we hypothesize that clonal T lymphocytes expand in response to a hematopoietic antigen-driven stimulus, which results in T cell–mediated inhibition of hematopoietic progenitors contributing to cytopenia. This may be a common mechanism of the cytopenia associated with other bone marrow failure disorders such as AA,7,12 PNH,14 T-LGL lymphoproliferative disorder,23 pure red cell aplasia,24 and perhaps hairy cell leukemia.25Formal proof of this hypothesis depends on demonstrating suppression of hematopoietic progenitors by the dominant clonal T lymphocytes, work that is currently underway in our laboratory.
Prepublished online as Blood First Edition Paper, July 5, 2002; DOI 10.1182/blood-2002-01-0155.
Supported by United States Public Health Service grant CA85843 to J.J.M. and by National Cancer Institute grant CA16672 to the DNA Core Facility of the M. D. Anderson Cancer Center. J.N.K. is supported by National Institutes of Health training grant TA-3209666.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jeffrey J. Molldrem, Section of Transplantation Immunology, Department of Blood and Marrow Transplantation, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Box 448, Houston, TX 77030; e-mail:jmolldre@notes.mdacc.tmc.edu.