Congenital afibrinogenemia is a rare inherited coagulopathy, characterized by very low or unmeasurable plasma levels of immunoreactive fibrinogen. So far, 25 mutations have been identified in afibrinogenemia, 17 in the Aα, 6 in the γ, and only 2 in the Bβ fibrinogen–chain genes. Here, 2 afibrinogenemic probands, showing undetectable levels of functional fibrinogen, were screened for causative mutations at the genomic level. Sequence analysis of the 3 fibrinogen genes disclosed 2 novel homozygous mutations in introns 6 and 7 of the Bβ-chain gene (IVS6 + 13C > T and IVS7 + 1G > T), representing the first Bβ-chain gene splicing mutations described in afibrinogenemia. The IVS6 + 13C > T mutation predicts the creation of a donor splice site in intron 6, whereas the IVS7 + 1G > T mutation causes the disappearance of the invariant GT dinucleotide of intron 7 donor splice site. To analyze the effect of these mutations, expression plasmids containing Bβ-chain minigene constructs, either wild-type or mutant, were transfected in HeLa cells. Assessed by semiquantitative analysis of reverse transcriptase–polymerase chain reaction products, the IVS7 + 1G > T mutation resulted in multiple aberrant splicings, while the IVS6 + 13C > T mutation resulted in activation of a new splice site 11 nucleotides downstream of the physiologic one. Both mutations are predicted to determine protein truncations, supporting the importance of the C-terminal domain of the Bβ chain for fibrinogen assembly and secretion.
Introduction
Fibrinogen is a hexameric, heavily disulfide-linked molecule, assembled from 3 pairs of subunits (Aα, Bβ, and γ chain), each subunit being the product of a single copy gene (for a review, see Blomback1). Fibrinogen Aα-, Bβ-, and γ-chain genes are clustered2 on human chromosome 4q28 and arranged in γ, Aα, and Bβ order, with the Bβ-chain gene in opposite transcriptional orientation.3 Circulating fibrinogen is expressed in hepatic parenchymal cells at wide-ranging rates (1.7 to 5 g/d) and is secreted as a fully assembled 340-kDa glycoprotein.4 Fibrinogen is the predominant acute-phase protein5 and plays a crucial role in the hemostasis process. Fibrin monomers, formed by proteolytic cleavage of fibrinogen molecules, polymerize to form a fibrin network and mediate platelet aggregation.6
Hereditary fibrinogen disorders include quantitative alterations (hypofibrinogenemia and afibrinogenemia) and qualitative alterations (dysfibrinogenemia). Among them, congenital afibrinogenemia (Mendelian Inheritance in Man no. #202400, found athttp://www.ncbi.nlm.nih.gov/omim/) is the less characterized from a molecular point of view. For this rare autosomal recessive trait, characterized by the concomitant absence of coagulant activity and immunoreactive protein and by hemorrhagic diathesis of variable severity,7,8 only 25 different genetic defects have been reported so far. In particular, 23 are mutations leading to protein truncations and are spread over Aα- and γ-chain genes (1 large deletion, 6 splicing mutations, 7 frameshift mutations, and 9 nonsense mutations); only 2 missense mutations, leading to an impairment of fibrinogen secretion, have been characterized so far and are located in the Bβ-chain gene. These represent the only afibrinogenemia causing mutations described in the Bβ-chain gene.9 10
In this study, 2 novel point mutations in the Bβ-chain gene, each identified in a different afibrinogenemic proband, are reported. Both mutations are located in intronic regions: a homozygous C > T transition at nucleotide position +13 of intron 6 (IVS6 + 13C > T) and a homozygous G > T transversion affecting the first nucleotide of intron 7 (IVS7 + 1G > T). Production of mutant transcripts in transfected HeLa cells demonstrated that both IVS6 + 13C > T and IVS7 + 1G > T mutations alter the pattern of mRNA processing.
Materials and methods
This study was approved by the institutional review board of the University of Milan. After acquiring informed consent, citrate-anticoagulated blood samples from all individuals were collected for biochemical and genetic analyses.
Coagulation tests
Activated partial thromboplastin time, prothrombin time, thrombin time, and reptilase time were obtained by using routine methods (reference intervals: 24-38 seconds, 10.5-12.2 seconds, 15-18 seconds, and 19-21 seconds, respectively). Plasma fibrinogen concentration was determined by a functional assay based on fibrin polymerization time11 and an enzyme-linked immunosorbent assay12 (normal range for both tests: 160-400 mg/dL). The sensitivities of the functional and of the immunologic assay were 5 mg/dL and 0.02 mg/dL, respectively.
Mutation analysis
Genomic DNA was purified from venous blood by using the PUREGENE kit (Gentra Systems, Minneapolis, MN) according to the manufacturer's protocol. Genomic DNA was polymerase chain reaction (PCR)–amplified by using sense and antisense primers designed on the basis of known sequences of the fibrinogen cluster (GenBank accession numbers M64982,M64983, M10014, U36478, and AF229198). PCR conditions and primer (Life Technologies, Inchinnan, Paisley, United Kingdom) sequences can be provided on request. Direct sequencing of PCR products was performed on both strands by the fluorescent dideoxy terminator method (BigDye Terminator Cycle Sequencing Kit, Applied Biosystems, Foster City, CA) and analyzed by using an automated multicapillary 3100 DNA sequencer (Applied Biosystems). Factura and Sequence Navigator software were used for mutation detection (Applied Biosystems).
Computer DNA analysis
Computer-assisted analysis for splice site prediction on DNA sequences was accomplished by using the Neural Network Promoter Prediction Tool program (http://www.fruitfly.org/seq_tools/splice.html)13 and the SpliceView program (http://l25.itba.mi.cnr.it/∼webgene/wwwspliceview.html).14
Minigene construction and mutagenesis
A minigene containing part of the human fibrinogen Bβ-chain gene (spanning from exon 6 to the beginning of the 3′ untranslated region [UTR] in exon 8) was PCR-amplified from genomic DNA of a healthy individual by using the primer couple FGB-In5-F (5′-GCTGTTGGTTAATATATGCTC-3′) and FGB-3′UTR-R (5′-TGTTGTCACATACAGAAGAGC-3′). PCR was performed on 100 ng genomic DNA in a standard 50 μL volume, containing 1× reaction buffer (200 mM Tris[tris(hydroxymethyl)aminomethane]-HCl, pH 8.4 and 500 mM KCl), 1.5 mM MgCl2, 0.4 μM of each primer, 200 μM deoxynucleoside triphosphates, and 2.5 U PlatinumTaq DNA polymerase (Life Technologies) in a PTC-100 (MJ Research, Watertown, MA) thermal cycler. Thermal conditions were 33 cycles of 95°C for 30 seconds, 53°C for 30 seconds, and 72°C for 3 minutes, preceded by 3 minutes at 95°C and followed by a final elongation step at 72°C for 10 minutes. The PCR product was A-tailed as previously described9 and cloned into the mammalian expression vector pTARGET (Promega, Milan, Italy). Plasmid DNA was isolated by the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany), and the pT-Bβ–wild-type (wt) recombinant plasmid was ascertained by sequencing. The 2 identified mutations were independently introduced in the pT-Bβ-wt by means of the QuickChange Site-Direct Mutagenesis Kit (Stratagene, La Jolla, CA), as instructed by the manufacturer. The 2 mutant plasmids (pT-Bβ-IVS6 + 13C > T and pT-Bβ-IVS7 + 1G > T) were checked by sequencing.
Cell cultures and transfections
Human cervix carcinoma HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 10% calf serum, 1% glutamine, and antibiotics (100 IU/mL penicillin and 100 μg/mL streptomycin). Cells were grown in a humidified atmosphere of 5% CO2 and 95% air at 37°C and cultured according to standard procedures. Transient transfections were performed in dishes 10 centimeters in diameter (2 × 106 cells/dish) by using the calcium phosphate technique, essentially as described.15 For each construct (pT-Bβ-wt, pT-Bβ-IVS6 + 13C > T, or pT-Bβ-IVS7 + 1G > T), 20 μg was independently transfected. After 16 hours of exposure to CaPO4-DNA precipitates, cells were washed twice with phosphate-buffered saline, and the medium was replaced with a fresh one. Cells were incubated for an additional 48 hours before RNA extraction.
RNA extraction and reverse transcriptase–polymerase chain reaction
Total RNA was isolated from transfected cells by using the RNAWIZ reagent (Ambion, Austin, TX). Random nonamers and Enhanced Avian RT-PCR Kit (Sigma, St Louis, MO) were used to perform first-strand complementary DNA (cDNA) synthesis starting from 1 μg of total RNA, according to the manufacturer's instructions. Of a total of 20 μL, 2.5 μL were used as template to amplify wild-type and mutant transcripts by using the exonic primer couple FGB-Ex6-F (5′-AGTGATTCAGAACCGTCAAG-3′) and FGB-Ex8-R (5′-TCCACCACCGTCTTCTTTAG-3′). Fluorescent hot-stop technique was performed on each PCR to quantify the Bβ transcripts, as described.9 In particular, each PCR product was subjected to a final cycle with the addition of 0.4 μM FGB-Ex8-R 6-Fam–labeled primer, and the labeled fragments were separated on a 3100 DNA sequencer (Applied Biosystems). Peak areas and molecular weights were evaluated by means of the GeneScan Analysis Software 3.1 (Applied Biosystems).
Subcloning of RT-PCR products
Prior to the addition of the 6-Fam–labeled FGB-Ex8-R primer, an aliquot (5 μL of a total of 50 μL) of each fluorescent hot-stop PCR was withdrawn, in order to subclone the PCR-amplified fibrinogen Bβ products. PCR products were blunt cloned into SmaI linearized pUC9 vector. Screening of recombinants was carried out by PCR on X-gal–selected white colonies. Cells were directly dissolved in a standard PCR mixture, and PCR was performed by using the 2 totally vector-derived primers UP and RP. PCR reactions were carried out under standard conditions, with the exception of an extended initial denaturation step (7 minutes) to allow the lysis of bacteria. All PCR products were subjected to direct sequencing, as described above.
Results
Case reports and laboratory data
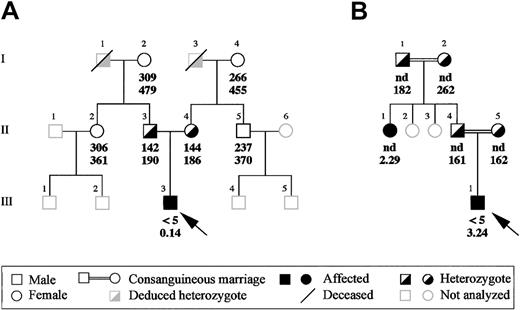
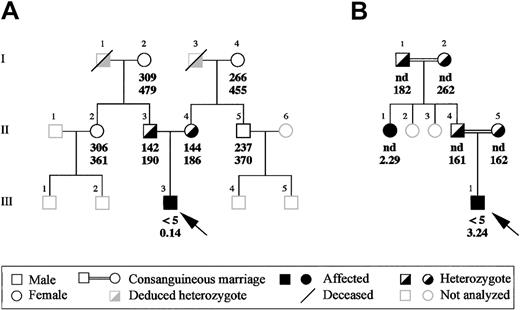
Two patients with congenital afibrinogenemia, an Italian and an Iranian, were analyzed. Proband A, a 2-year-old child born in Torino, Italy, originally was diagnosed at birth after routine medical examinations, which disclosed infinite activated partial thromboplastin time, prothrombin time, thrombin time, and reptilase time. Further investigations showed that all plasma-clotting factors were normal except fibrinogen, which was undetectable when measured by the Clauss method and extremely reduced (0.14 mg/dL) when assayed by the enzyme-linked immunosorbent assay. Nevertheless, no bleeding tendency has been observed so far in this proband, spontaneously or at the time of the fall of the umbilical cord. All of this proband's relatives had normal functional and immunologic fibrinogen levels except his parents, who had approximately half the normal values, with good concordance between functional and immunologic measurements (Figure1A). All family members so far have been asymptomatic.
Family pedigrees of the 2 afibrinogenemic probands.
(A) Pedigree of the Italian proband and (B) of the Iranian proband. Functional and immunoreactive plasma fibrinogen values (mg/dL) of the probands and of their available family members are indicated in this order below the corresponding symbols. Probands are indicated by an arrow. nd indicates not done.
Family pedigrees of the 2 afibrinogenemic probands.
(A) Pedigree of the Italian proband and (B) of the Iranian proband. Functional and immunoreactive plasma fibrinogen values (mg/dL) of the probands and of their available family members are indicated in this order below the corresponding symbols. Probands are indicated by an arrow. nd indicates not done.
Proband B is an 18-year-old Iranian boy born from a consanguineous marriage (Figure 1B). Consanguinity is a recurrent feature in the proband's family; besides his parents, who are first cousins, paternal grandparents also were consanguineous, even though the degree of relationship is unknown. The diagnosis of afibrinogenemia was made at birth after prolonged bleeding from the umbilical cord. During childhood and puberty, the proband bled from the nose and mouth, developed hemarthrosis, and repeatedly suffered from hematomas and excessive bleeding after dental extractions. Proband's plasma fibrinogen levels were unclottable by the functional assay (< 5 mg/dL), whereas immunoreactive fibrinogen was 3.24 mg/dL. Immunoreactive fibrinogen was assayed in all available family members (Figure 1B): in individuals I-1, I-2, I-4, and I-5, all asymptomatic, plasma fibrinogen levels were normal or in the lower part of the normal range, whereas individual II-1 had abnormally low concentration of immunoreactive fibrinogen (2.29 mg/dL). A subsequent ascertainment of individual II-1's clinical history revealed bleeding symptoms similar to those of proband B.
Mutation analysis
The whole coding regions, all intron-exon boundaries, and about 500 base pair (bp) of the promoter region of the fibrinogen Aα-, Bβ-, and γ-chain genes were PCR-amplified from probands' genomic DNA. Direct sequencing of all the amplified fragments from Aα- and γ-chain genes did not detect any mutation, whereas sequence analysis of the Bβ-chain gene enabled the identification of 2 novel point mutations (1 in proband A and 1 in proband B), each present in the homozygous state.
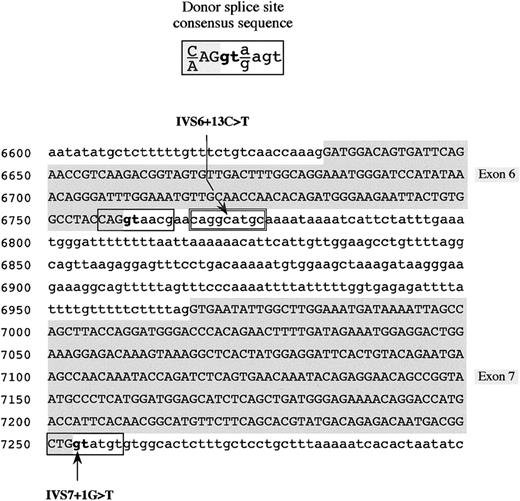
In proband A, a G-to-T transversion was found at the first nucleotide (position 7253, numbering according to GenBank accession number M64983) of intron 7 (IVS7 + 1G > T; Figure2). This mutation, which involves the extremely conserved GT dinucleotide of the donor splice site consensus sequence, was detected, in the heterozygous state, in both parents of proband A. The remaining analyzed family members (individuals I-2, I-4, II-2, and II-5; Figure 1A) did not carry this molecular change.
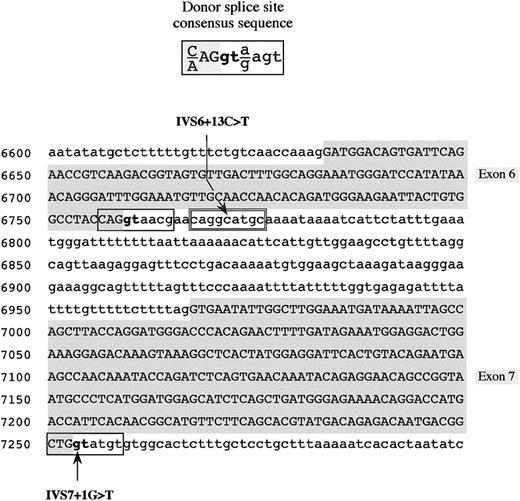
Mutations in the fibrinogen Bβ-chain gene identified in the 2 afibrinogenemic patients.
Donor splice site consensus sequence16 (top) and partial fibrinogen Bβ-chain gene sequence showing the position of the identified mutations (IVS6 + 13C > T and IVS7 + 1G > T) (bottom) are shown. The genomic Bβ-chain sequence spans from nucleotide positions 6600 to 7299. Identified mutations are indicated by an arrow. Exonic sequences are in uppercase letters and shaded in gray, whereas intronic sequences are in lowercase letters. Physiologic donor splice sites are boxed; the cryptic donor splice site in intron 6 is double-boxed. The invariant GT dinucleotide of the donor splice site is in boldface.
Mutations in the fibrinogen Bβ-chain gene identified in the 2 afibrinogenemic patients.
Donor splice site consensus sequence16 (top) and partial fibrinogen Bβ-chain gene sequence showing the position of the identified mutations (IVS6 + 13C > T and IVS7 + 1G > T) (bottom) are shown. The genomic Bβ-chain sequence spans from nucleotide positions 6600 to 7299. Identified mutations are indicated by an arrow. Exonic sequences are in uppercase letters and shaded in gray, whereas intronic sequences are in lowercase letters. Physiologic donor splice sites are boxed; the cryptic donor splice site in intron 6 is double-boxed. The invariant GT dinucleotide of the donor splice site is in boldface.
In proband B, a C-to-T transition located at nucleotide 13 (position 6771, numbering according to GenBank accession number M64983) of intron 6 (IVS6 + 13C > T) was detected (Figure 2). This variation involves a possible cryptic donor splice site, converting a GC dinucleotide into a canonical GT dinucleotide consensus. In the inbred family of proband B, the IVS6 + 13C > T mutation was recurrent: it was found in the heterozygous state in both parents and in both paternal grandparents (individuals II-4, II-5, I-1, and I-2; Figure 1B) and in the homozygous state in the afibrinogenemic aunt (individual II-1; Figure 1B).
The absence of IVS7 + 1G > T and IVS6 + 13C > T mutations in control Italian and Iranian populations was verified by sequencing 100 haploid genomes for each population.
To predict the possible effects of both mutations on the donor splice site pattern of the Bβ-chain gene, a computer-assisted analysis of the Bβ-gene sequence, spanning from exon 6 (nucleotide position 6633) to exon 8 (nucleotide position 7919) was performed by the Neural Network Promoter Prediction Tool and the SpliceView programs. As simulated by both software, the presence of the IVS7 + 1G > T mutation is sufficient to cause the disappearance of the physiologic donor splice site of intron 7. As far as the IVS6 + 13C > T mutation is concerned, its presence creates, in intron 6, an additional donor splice site, located 11 nucleotides downstream of the physiologic one (Figure 2). The scores predicted by using the Neural Network Promoter Prediction Tool and the SpliceView programs were 0.98 and 82, respectively (functional splice sites of the analyzed region of the Bβ-chain gene have scores ranging from 0.55 to 0.94 and from 74 to 85 for the 2 programs, respectively).
Analysis of fibrinogen Bβ-chain transcripts
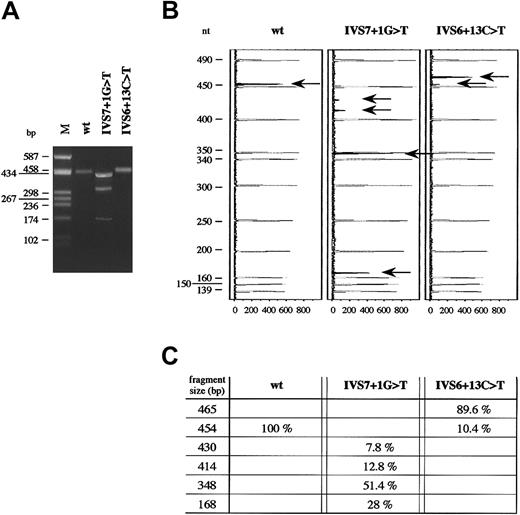
To examine the effects of IVS7 + 1G > T and IVS6 + 13C > T mutations on messenger RNA (mRNA), mutant Bβ-chain mRNAs were transiently produced in HeLa cells. For this purpose, a minigene construct composed of a portion of exon 6 (119 nucleotides), intron 6 (208 nucleotides), exon 7 (286 nucleotides), intron 7 (618 nucleotides), and a portion of exon 8 (273 nucleotides, comprising the first 41 nucleotides of the 3′UTR) was generated, as described in “Materials and methods.” The minigene construct, cloned into the mammalian expression vector pTARGET, was used as template to obtain, by site-directed mutagenesis, 2 recombinant vectors, each containing either IVS7 + 1G > T or IVS6 + 13C > T mutation. Plasmids expressing wild-type and each mutant fibrinogen Bβ-chain mRNA were independently transfected in HeLa cells (not expressing fibrinogen). Total RNA was extracted from transfected cells and analyzed by RT-PCR. To increase the sensitivity of the assay and to enable quantitation and sizing of mRNA species, the RT-PCRs were performed by using the fluorescent hot-stop technique and the exonic oligonucleotides FGB-Ex6-F (located in exon 6, nucleotide position 6640-6659) and FGB-Ex8-R (located in exon 8, nucleotide position 7919-7903). RT-PCR products were separated both by agarose-gel electrophoresis and by an automated DNA sequencer (Figure 3A-B).
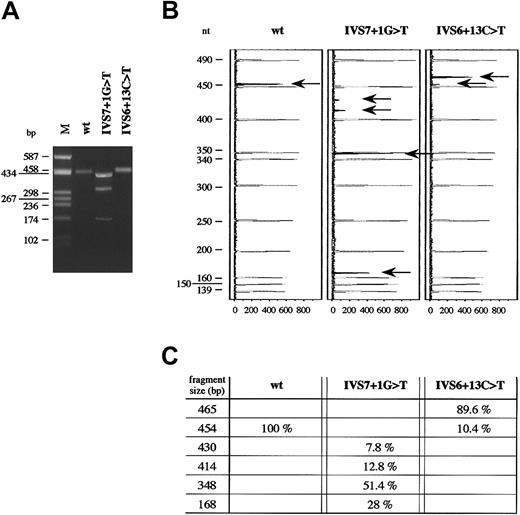
RT-PCR performed on total RNA of HeLa cells transfected with the pT-Bβ-IVS7 + 1G > T construct gave rise to 3 products, all shorter than the wild-type and with approximate lengths between 430 and 170 bp (Figure 3A). Conversely, only 1 band was visible after agarose-gel electrophoresis for both the pT-Bβ-wt and the pT-Bβ-IVS6 + 13C > T constructs. However, the molecular weight of the band obtained from the mutant construct was slightly higher than that expected for the correctly spliced wild-type fragment (454 bp) (Figure 3A). Analysis of fluorochrome-labeled RT-PCR products confirmed the presence of a unique splicing product only for the pT-Bβ-wt minigene (Figure 3B). In the case of the pT-Bβ-IVS7 + 1G > T minigene, a fourth splicing product, besides those evidenced by agarose-gel electrophoresis, was detected. In particular, the most retarded band, as visualized by agarose-gel electrophoresis, was composed of 2 different products (430 bp and 414 bp, respectively) (Figure 3B-C). In the case of IVS6 + 13C > T mutation, 2 peaks were detected: a larger one, corresponding to an aberrantly spliced product 11 bp longer than the wild-type one (465 bp vs 454 bp), and a very small one, corresponding to the wild-type transcript (Figure 3B-C). Quantitation of both wild-type and mutant splicing products, performed by measuring fluorescent peak areas, is shown in Figure 3C.
RT-PCR analysis of wild-type and mutant fibrinogen Bβ-chain mRNAs.
(A) Agarose (2%)-gel electrophoresis of RT-PCR products, amplified from total RNA extracted from HeLa cells after transfection either with IVS6 + 13C > T or IVS7 + 1G > T mutant minigenes. M indicates molecular weight marker (pUC8HaeIII). (B) Analysis of the same RT-PCR products as in panel A, labeled by fluorescent hot-stop technique, separated on an automated DNA sequencer, and analyzed by GeneScan software. In order, from left to right, are shown GeneScan Analysis windows of the capillary-electrophoretic runs of RT-PCR products for the wild-type, IVS7 + 1G > T, and IVS6 + 13C > T minigenes, respectively. Molecular weight standard peaks are in gray; peaks corresponding to the labeled fragments, indicated by arrows, are in black. GeneScan fluorescence units (x-axis) and length of molecular weight fragments (y-axis) are reported. (C) Fragment size and relative amounts of fluorochrome-labeled RT-PCR products, indicated by arrows in panel B, are shown. Molecular weights and peak areas were evaluated by means of GeneScan software. The percentage values correspond to the peak areas, setting the sum of RT-PCR black areas of each experiment equal to 100%.
RT-PCR analysis of wild-type and mutant fibrinogen Bβ-chain mRNAs.
(A) Agarose (2%)-gel electrophoresis of RT-PCR products, amplified from total RNA extracted from HeLa cells after transfection either with IVS6 + 13C > T or IVS7 + 1G > T mutant minigenes. M indicates molecular weight marker (pUC8HaeIII). (B) Analysis of the same RT-PCR products as in panel A, labeled by fluorescent hot-stop technique, separated on an automated DNA sequencer, and analyzed by GeneScan software. In order, from left to right, are shown GeneScan Analysis windows of the capillary-electrophoretic runs of RT-PCR products for the wild-type, IVS7 + 1G > T, and IVS6 + 13C > T minigenes, respectively. Molecular weight standard peaks are in gray; peaks corresponding to the labeled fragments, indicated by arrows, are in black. GeneScan fluorescence units (x-axis) and length of molecular weight fragments (y-axis) are reported. (C) Fragment size and relative amounts of fluorochrome-labeled RT-PCR products, indicated by arrows in panel B, are shown. Molecular weights and peak areas were evaluated by means of GeneScan software. The percentage values correspond to the peak areas, setting the sum of RT-PCR black areas of each experiment equal to 100%.
To further characterize spliced mRNA forms, RT-PCR products from wild-type and mutant transfected cells were subcloned into a plasmid vector, and the inserts of at least 60 recombinant plasmids for each subcloning experiment were analyzed by sequencing. Subclone frequencies, splicing events, and their predicted effects at the protein level are summarized in Figure4. Analysis of the cDNA fragments derived from wild-type minigene transfected cells showed that a correct splicing product was present in 96.6% of clones, whereas 3.4% of clones were characterized by aberrantly processed inserts (2 misspliced forms) not detected by fluorescent hot-stop PCR. As far as the splicing products caused by the IVS7 + 1G > T mutation are concerned, several (9) aberrant splicing events, which account for 96.6% of clones, were detected. Among them, the 414-, 348-, and 168-bp-long fragments are predominant forms and, together with the 430-bp-long fragment, correspond to the products detected by fluorescent hot-stop PCR. The remaining alternatively spliced products (aberrant products 5-9; Figure 4B) were found in single recombinant clones, while 3.4% of clones showed normal splicing. RT-PCR products derived from the pT-Bβ-IVS6 + 13C > T construct gave rise to 4 subtypes of clones. In the majority (86.7%), an aberrant splicing product of 465 bp, which retained into the mature mRNA the first 11 nucleotides of intron 6, was present. Of the transcripts, 10.3% were correctly spliced, whereas the remaining 3% of clones were constituted by 2 incorrectly spliced forms, not detected by fluorescent hot-stop PCR.
Effects of IVS6 + 13C > T and IVS7 + 1G > T mutations on splicing of mRNA molecules and on predicted proteins.
(A) Left: schematic representation of the fibrinogen Bβ-chain gene showing the position of the 2 identified mutations. Exons (numbered) and introns are indicated by boxes and lines, respectively, and are drawn to scale. The white part of the box representing exon 8 corresponds to the 3′UTR. Right: schematic representation of the fibrinogen Bβ chain. Numbers refer to amino acid positions at exon/exon boundaries. The last number (461) corresponds to the C-terminal amino acid. Numbering omits the signal peptide. (B) Left: schematic representation of the splicing events produced in HeLa cells transfected with the pT-Bβ-wt, pT-Bβ-IVS7 + 1G > T, and pT-Bβ-IVS6 + 13C > T constructs. RT-PCR products, amplified from total RNA of transfected cells, were subcloned and sequenced. A region of the Bβ-chain gene spanning from exon 6 (nucleotide position 6633) to exon 8 (nucleotide position 8102) is represented for each splicing event. Exons and introns are indicated by boxes and lines, respectively, and are drawn to scale. For each splicing event, used donor and acceptor splice sites are shown; nucleotides retained in each mature mRNA are indicated in uppercase letters. Clone frequencies (indicated as percentage value; left) and RT-PCR product size (right) are shown beside each splicing event scheme; boldface type values correspond to RT-PCR products also detected by fluorescent hot-stop RT-PCR. Right: schematic representation of the predicted Bβ-chain polypeptides, corresponding to wild-type and mutant transcripts. Only the C-terminal regions, starting from residue 248, are shown. Rectangles hatched in dark gray represent normal amino acid sequences; rectangles hatched in light gray, aberrant amino acid sequences originating from frameshifts.
Effects of IVS6 + 13C > T and IVS7 + 1G > T mutations on splicing of mRNA molecules and on predicted proteins.
(A) Left: schematic representation of the fibrinogen Bβ-chain gene showing the position of the 2 identified mutations. Exons (numbered) and introns are indicated by boxes and lines, respectively, and are drawn to scale. The white part of the box representing exon 8 corresponds to the 3′UTR. Right: schematic representation of the fibrinogen Bβ chain. Numbers refer to amino acid positions at exon/exon boundaries. The last number (461) corresponds to the C-terminal amino acid. Numbering omits the signal peptide. (B) Left: schematic representation of the splicing events produced in HeLa cells transfected with the pT-Bβ-wt, pT-Bβ-IVS7 + 1G > T, and pT-Bβ-IVS6 + 13C > T constructs. RT-PCR products, amplified from total RNA of transfected cells, were subcloned and sequenced. A region of the Bβ-chain gene spanning from exon 6 (nucleotide position 6633) to exon 8 (nucleotide position 8102) is represented for each splicing event. Exons and introns are indicated by boxes and lines, respectively, and are drawn to scale. For each splicing event, used donor and acceptor splice sites are shown; nucleotides retained in each mature mRNA are indicated in uppercase letters. Clone frequencies (indicated as percentage value; left) and RT-PCR product size (right) are shown beside each splicing event scheme; boldface type values correspond to RT-PCR products also detected by fluorescent hot-stop RT-PCR. Right: schematic representation of the predicted Bβ-chain polypeptides, corresponding to wild-type and mutant transcripts. Only the C-terminal regions, starting from residue 248, are shown. Rectangles hatched in dark gray represent normal amino acid sequences; rectangles hatched in light gray, aberrant amino acid sequences originating from frameshifts.
Discussion
In this study, 2 afibrinogenemic patients, 1 Italian and 1 Iranian, had unmeasurable functional levels of plasma fibrinogen, whereas the immunologic fibrinogen level was 20-fold lower in the Italian proband (0.14 mg/dL) compared with the Iranian one (3.24 mg/dL). Severity of bleeding did not directly correlate with fibrinogen levels. In fact, the Iranian proband (18 years old) suffered from repeated, even though generally mild, bleeding symptoms, whereas the Italian proband (2 years old) has been asymptomatic up to now. A possible explanation for the different symptomatology between the 2 patients could be the young age of the Italian proband, who has been less exposed to hemorrhagic challenges.
Direct sequence analysis of the 3 fibrinogen genes in both patients revealed 2 novel homozygous point mutations (IVS6 + 13C > T and IVS7 + 1G > T), both located in intronic regions of fibrinogen Bβ-chain gene. In each pedigree the corresponding nucleotide substitution cosegregated with the clinical phenotype, that is, afibrinogenemia (homozygous individuals) or mild hypofibrinogenemia (heterozygous individuals) (Figure 1). Both mutations were predicted by computer-assisted analysis to perturb the correct splicing of the Bβ-chain gene pre-mRNA, IVS6 + 13C > T mutation by creating a new perfect donor splice site 11 nucleotides downstream of the physiologic one, IVS7 + 1G > T mutation by altering the invariant GT dinucleotide at the donor splice site of intron 7.
To evaluate whether the identified mutations were responsible for afibrinogenemia by causing an aberrant splicing of Bβ-chain mRNA, minigene constructs containing a genomic region spanning from exon 6 to exon 8 of the fibrinogen Bβ-chain gene were prepared, mutagenized, and transiently transfected in HeLa cells. This ex vivo approach was adopted due to the liver-confined fibrinogen expression17 and to ethical constraints, which deny access to liver biopsies in afibrinogenemic and hypofibrinogenemic patients.
Sequencing of cloned RT-PCR products, obtained from RNA of cells expressing the IVS6 + 13C > T minigene transcripts, demonstrated that this mutation causes the activation of a cryptic donor splice site. Aberrant splicing determines the retention of the first 11 nucleotides of intron 6 into the mature transcript and causes the synthesis of a predicted mutant protein, containing the first normal 290 amino acids followed by 24 aberrant residues and 2 consecutive stop codons (Figure 4B). The activation of the cryptic donor splice site does not completely abolish the physiologic site, as demonstrated by the results of both fluorescent hot-stop RT-PCR and subcloning experiments, which revealed that about 10% of transcripts were the product of a normal splicing of intron 6 (Figure 3B-C). IVS6 + 13C > T is the first afibrinogenemia-causing mutation that creates a new donor splice site, since all previously reported splicing mutations in the fibrinogen genes lead to the inactivation of a physiologic splice site.18-22 The presence of about 10% normal splicing can be justified by taking into consideration that the wild-type consensus sequence of intron 6 donor splice site is not affected by the mutation.
Concerning the IVS7 + 1G > T mutation, a much more complex pattern of RT-PCR products was observed. The inactivation of the donor splice site caused by the G > T transversion, besides causing exon 7 skipping in about 17% of cases (which should cause the synthesis of a 290-residue polypeptide chain) (Figure 4B), also determines the activation of multiple cryptic splice sites within exon 7. In particular, in the more abundant mature mRNA species, a donor splice site located 106 nucleotides upstream of the physiologic one was activated. This transcript, which accounts for about 57% of clones, determined the translation of a mature Bβ chain truncated at residue 350. Two other cryptic splice sites, located 40 and 24 nucleotides upstream of the 3′-end of exon 7, also were activated. These give rise to a truncation of the Bβ chain at amino acid 372 and to a frameshift causing the production of a 453-residue-long Bβ chain, whose last 76 amino acids are unrelated to the wild-type protein. Relative quantitation, performed by semiquantitative fluorescent hot-stop PCR, of the different splicing products caused by IVS7 + 1G > T mutation is concordant with the observed clone frequencies. The only slight exception is represented by the 430-bp-long fragment that amounted to 7.8% by RT-PCR but accounted for only 2.2% of clones. This discrepancy, which could reflect a lower cloning efficiency of this fragment, stresses the need to quantify the relative abundance of aberrant splicings by different approaches, such as the evaluation of clone frequencies and direct quantitation by PCR. As described for the IVS6 + 13C > T mutation, residual normal splicing also was observed for the IVS7 + 1G > T mutation (3.4%). The corresponding fragment was undetectable by fluorescent hot-stop PCR, probably because its abundance was below the detection limit of the assay. The lower level of normal transcript, detected for the IVS7 + 1G > T mutation compared to the IVS6 + 13C > T mutation (3.4% vs 10.3%; Figure 4B), positively correlates with plasma fibrinogen levels measured in the corresponding proband (0.14 vs 3.24 mg/dL; Figure 1). Possible alternative explanations for the observed differences in plasma fibrinogen levels are differences in mRNA stability, efficiency of translation, as well as assembly and secretion of the mutant proteins.
A number of single clones containing inserts resulting from additional aberrant splicing events also were identified. Only 2 aberrant transcripts—aberrant transcript 3 for the IVS6 + 13C > T mutation and aberrant transcript 9 for the IVS7 + 1G > T mutation (Figure 4B)—originated from the use of the same splice sites. The rarity of these clones, their undetectability by RT-PCR, and the presence of 2 single aberrant clones also for the wild-type construct suggest that they could represent the result of a background aberrant splicing activity and probably do not have a pathophysiologic meaning.
All predominant aberrant splicing events are predicted to cause the synthesis of truncated Bβ-chain polypeptides, ranging from 290 to 453 residues in length. Truncations causing hypofibrinogenemia or afibrinogenemia have been reported for both fibrinogen Aα and γ chains. In particular, in the case of Aα-chain truncations, a correlation between the degree of the truncation and the severity of the fibrinogen deficiency could be established,18 the more C-terminal premature stop codons up to now identified in afibrinogenemic patients corresponding to residue 149.9,10,23 As far as the γ chain is concerned, recent data by Okumura et al24 demonstrated that the C-terminal region of γ chain is essential for fibrinogen assembly and secretion. Expression of a series of fibrinogen γ-chain mutants, truncated between residue 379 and the C-terminus (residue 411), established at amino acid 387 the minimum γ-chain length necessary for assembly and secretion of a stable fibrinogen molecule. For the Bβ chain, on the basis of expression experiments with deletion mutants,25the C-terminal domain (amino acids 208-461) was considered unnecessary for fibrinogen assembly and secretion. By contrast, our results support the importance of the C-terminal region of fibrinogen Bβ chain, since all predicted truncations caused by IVS6 + 13C > T and IVS7 + 1G > T mutations are located downstream of residue 208. A clear-cut evaluation of the importance of Bβ-chain C-terminal truncations per se is somewhat hampered by the fact that for both mutations some predicted mutant polypeptides contain a gibberish amino acid sequence at their C-terminus that could perturb the correct folding of the C-terminal globular domain. Nevertheless, in the case of IVS7 + 1G > T mutation, the majority (92.2%) of aberrant transcripts are predicted to cause only the deletion of the last 111, 171, or 89 residues (accounting for 51.4%, 28%, and 12.8% of mutant transcripts, respectively), making it possible to hypothesize that such Bβ-chain C-terminal truncations can prevent assembly and/or secretion of hexameric fibrinogen molecules, resulting in afibrinogenemia.
We thank all family members for their participation in this study. We wish to thank Dr Elena Santagostino (Angelo Bianchi Bonomi Hemophilia and Thrombosis Center) for the clinical identification of the probands, family history, blood sample collection, and helpful discussion. We are indebted to Prof Pier Mannuccio Mannucci (Angelo Bianchi Bonomi Hemophilia and Thrombosis Center and Fondazione Luigi Villa, Department of Internal Medicine, University of Milan) for critically reading this manuscript.
Prepublished online as Blood First Edition Paper, August 1, 2002; DOI 10.1182/blood-2002-06-1647.
Supported by the Ministero dell'Università e della Ricerca Scientifica e Tecnologica (MURST 60%) and by IRCCS Maggiore Hospital, Milan, Italy. This work also was founded in part by a grant from Fondazione Italo Monzino (F.P.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Maria Luisa Tenchini, Department of Biology and Genetics for Medical Sciences, via Viotti, 3/5-20133 Milano, Italy; e-mail: marialuisa.tenchini@unimi.it.