Abstract
The cloning of the PIG-A gene has facilitated the unraveling of the complex pathophysiology of paroxysmal nocturnal hemoglobinuria (PNH). Of current major concern is the mechanism by which a PNH clone expands. Many reports have suggested that an immune mechanism operates to cause bone marrow failure in some patients with PNH, aplastic anemia, and myelodysplastic syndromes. Because blood cells of PNH phenotype are often found in patients with these marrow diseases, one hypothesis is that the PNH clone escapes immune attack, producing a survival advantage by immunoselection. To test this hypothesis, we examined the sensitivity of blood cells, with or withoutPIG-A mutations, to killing by natural killer (NK) cells, using 51Cr-release assay in vitro. To both peripheral blood and cultured NK cells, PIG-A mutant cells prepared from myeloid and lymphoid leukemic cell lines were less susceptible than their control counterparts (reverted from the mutant cells by transfection with a PIG-A cDNA). NK activity was completely abolished with concanamycin A and by calcium chelation, indicating that killing was perforin-dependent. There were no differences in major histocompatibility (MHC) class I expression or sensitivity to either purified perforin or to interleukin-2–activated NK cells betweenPIG-A mutant and control cells. From these results, we infer that PIG-A mutant cells lack molecules needed for NK activation or to trigger perforin-mediated killing. Our experiments suggest that PIG-A mutations confer a relative survival advantage to a PNH clone, contributing to selective expansion of these cells in the setting of marrow injury by cytotoxic lymphocytes.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is an acquired, clonal stem cell disorder that manifests clinically as intravascular hemolysis, venous thrombosis, frequent episodes of infection, and rare leukemic conversion.1,2 In addition, PNH is often a bone marrow (BM) failure syndrome, strongly related to aplastic anemia and the myelodysplastic syndromes (MDSs).3,4 All 3 processes may occur in a single patient.5,6 Among the clinical manifestations, the molecular events leading to hemolysis have been elucidated7-9 and the responsible gene PIG-Ahas been cloned.10 PNH cells with PIG-Amutation do not produce glycosylphosphatidylinositol (GPI) and lack cell surface expression of a large family of proteins that utilize GPI to localize in the plasma membrane.10 Therefore, PNH cells, deficient in complement regulatory GPI-linked proteins such as decay-accelerating factor (DAF) and CD59, undergo complement-mediated hemolysis characteristic of PNH.11 A mechanism of infection-associated precipitation of hemolysis also has been shown.12 Proclivity to thrombosis is also attributable toPIG-A mutations, but a molecular mechanism has not been found.13 14
Despite rapid progress in understanding PNH pathophysiology, the etiology and the mechanism by which a PNH clone expands are still unknown.15 Murine pig-a knockout models have shown that clonal expansion of PNH cells does not follow on the presence of the gene mutation alone.16-18 Furthermore, small numbers of PIG-A mutant cells are present in healthy individuals.19,20 Alternative theories have been proposed.15,21,22 An intrinsic alteration might confer a proliferative advantage to PNH cells over normal cells, according to one growth advantage hypothesis. Leukemic conversion can occur in PNH patients, albeit infrequently.6,23 A predominant proliferating clone is often detected among multiple PNH clones within the same patient.24-26 Transplantation experiments in mice have suggested an intrinsic growth alteration in PNH stem cells.22 However, the gene responsible for such behavior is not known.
Alternatively, the lack of GPI-linked membrane proteins might confer a relative survival advantage to PNH clones.15,21 There is evidence for15,27 and against such a survival advantage.28 For example, it is widely accepted that an immune-mediated BM injury occurs in patients with PNH,29-32 aplastic anemia,33,34 and MDS,35 and blood cells with PIG-A mutation (or PNH phenotype) are detected more abundantly in patients with BM failure syndromes than in healthy individuals.4,36-38 A PNH clone could escape immune-mediated BM attack and survive preferentially in such a pathologic environment. Lymphocytes of PNH phenotype appeared in patients with non-Hodgkin lymphoma after therapy with antibodies to the GPI-linked lymphocyte antigen CD52, an example of iatrogenic immunoselection of PIG-A mutant cells.39,40 To date, the incitement and molecular events in the immune attack to BM cells by cytotoxic lymphocytes are unclear. As candidates for the pathogenic effector cells, T cells34 and natural killer (NK) cells41,42 have been most studied. Involvement of NK cells in BM impairment is suggested by clinical observation43 and experimental data.44 Recent evidence has indicated a close relationship between NK cells (serving innate immunity) and cytotoxic T lymphocytes (acquired immunity).45-47 In this context, and in order to develop evidence relating to the survival advantage theory, we prepared leukemic cells with PIG-A mutations and their control counterparts by transfection with a PIG-A cDNA, and then compared their sensitivity to killing by NK cells in vitro.
Materials and methods
Preparation of target cells with and without PIG-Amutations
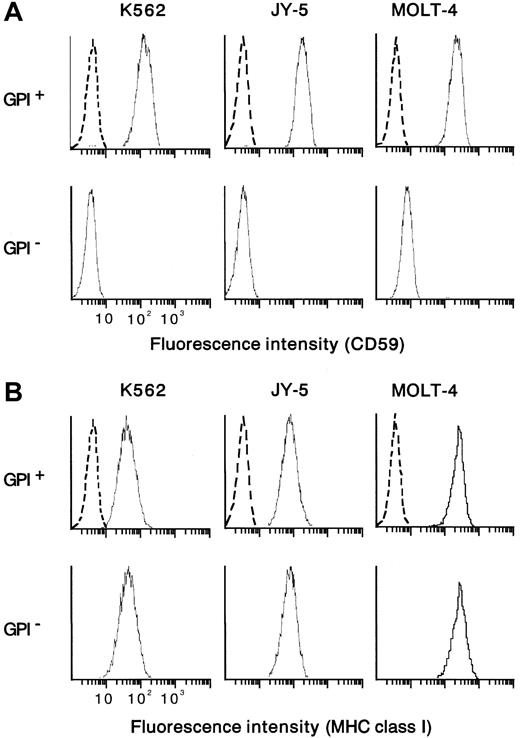
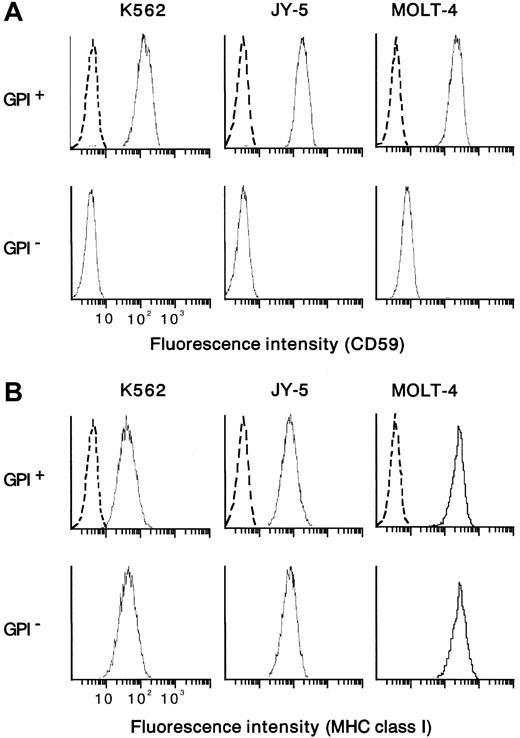
From the human myeloid leukemia cell line K562 bearing aPIG-A mutation (a gift from S. Hirose, Fukuoka University),48 a control counterpart with normal phenotype was prepared by PIG-A cDNA transfection.49 In parallel, mutant K562 cells were also transfected with a mock vector without PIG-A cDNA. Paired mutant K562 (mock-transfectant) and revertant (PIG-A-transfectant) cells were used as targets in cytotoxicity assay with NK cells. As another target, a human leukemic B-cell line (JY-5) with PIG-A mutation was transfected using a vector with and without PIG-AcDNA.49,50 Each control counterpart (revertant) restored the membrane expression of GPI-anchored proteins such as DAF (data not shown) and CD59 (Figure 1A). There was no difference in the expression of major histocompatibility complex (MHC) class I between each pair of PIG-A mutant and its control counterpart (Figure 1B). Contrary to wild-type K562, which is known to lack MHC class I expression, our mutant and control K562 cells expressed weakly but equally MHC class I (Figure 1B). As additional target cells, we isolated T cells of PNH phenotype from a human leukemic T-cell line (MOLT-4) by repeated negative selection using beads coated with antibodies to DAF and CD59.20 22 The mutant T cells were negative for both DAF and CD59, whereas their parent (MOLT-4) cells were positive for the proteins (Figure 1A). Both the mutant and parent T cells expressed MHC class I similarly (Figure1B). Cells were cultured in RPMI 1640 medium with 10% heat-inactivated fetal calf serum (FCS), 100 U/mL penicillin G, and 100 μg/mL kanamycin at 37°C in a humidified atmosphere of 5% CO2under mycoplasma-free condition. There was no difference in cell growth between mutant and control cells (data not shown).
Flow cytometric analysis of the expression of CD59 and MHC class I on the target cells.
Cells positive and negative for GPI were prepared from myeloid (K562) and lymphoid (B, JY-5; T, MOLT-4) cell lines. CD59 was missing from GPI− cells but appeared on the control counterparts (GPI+ cells) (A). The pair of the mutant (GPI−) and control (GPI+) cells of each cell line expressed the MHC class I equally (B). Dotted lines show nonspecific background staining with isotype-matched mouse Ig instead of MoAb.
Flow cytometric analysis of the expression of CD59 and MHC class I on the target cells.
Cells positive and negative for GPI were prepared from myeloid (K562) and lymphoid (B, JY-5; T, MOLT-4) cell lines. CD59 was missing from GPI− cells but appeared on the control counterparts (GPI+ cells) (A). The pair of the mutant (GPI−) and control (GPI+) cells of each cell line expressed the MHC class I equally (B). Dotted lines show nonspecific background staining with isotype-matched mouse Ig instead of MoAb.
Preparation of NK cells as cytotoxic effector lymphocytes
Peripheral blood mononuclear cells were isolated by centrifugation with Ficoll-Paque (Amersham Pharmacia, Uppsala, Sweden) from healthy volunteers.30 NK cells were enriched by negative selection with magnetic polystyrene beads coated with monoclonal antibodies (MoAbs) to CD4, CD8, CD14, and CD19 (Dynal, Oslo, Norway).22 NK cells were further concentrated using beads and MoAbs to CD3 (OKT3; American Type Culture Collection, Rockville, MD), CD41 (Beckman Coulter, Fullerton, CA), and glycophorin A (Beckman Coulter). The purity of isolated NK (CD2+, CD3−, CD16+, CD56+) cells exceeded 95% by flow cytometry; viability was more than 98% as assessed by trypan blue dye exclusion. In addition to peripheral blood NK cells, we obtained an interleukin-2 (IL-2)–dependent NK cell line with a p53 mutation (KHYG-1; Human Science Research Resources Bank, Osaka, Japan)51 and isolated a subclone (2E3) with high NK activity from KHYG-1 using beads coated with anti–CD56 antibody (B159, mouse IgG1; Pharmingen, San Diego, CA) and subsequently by limiting dilution. The cultured NK clone 2E3 was positive for CD2 and CD56 but negative for CD3 and CD4 (RPA-T4 MoAb, mouse IgG1; Pharmingen). Of note, KHYG-1 and 2E3 were negative for CD16 (FcγR type III) and Fas ligand.51 Cultured NK cells were maintained with 100 U/mL recombinant IL-2 (Sionogi, Osaka, Japan) and incubated in IL-2–depleted medium for 24 hours prior to cytotoxicity assay; under these conditions, NK cells remained viable.
Flow cytometry
Cell surface antigens were analyzed by flow cytometry as described previously.22 36 Briefly, cells were incubated for 30 minutes on ice with specific MoAbs, labeled with fluorescein isothiocyanate (FITC)–conjugated goat anti–mouse IgG or IgM (Zymed, San Francisco, CA), and then analyzed with a flow cytometer (Epics; Beckman Coulter). To identify the PNH phenotype, we used MoAbs to GPI-anchored proteins such as CD14 (M5E2, mouse IgG2a; Pharmingen), CD48 (Tu145, mouse IgM; Pharmingen), DAF (1A10, mouse IgG2a), and CD59 (5H8, mouse IgG1; provided by M. Tomita, Showa University). To determine cell purity, NK cells were analyzed with MoAbs to CD2 (RPA-2.10, mouse IgG1; Pharmingen), CD3 (OKT3, mouse IgG2a), CD16 (3G8, mouse IgG1; Pharmingen), and CD56. For cytotoxicity-associated molecules, we used MoAbs to CD58 (1C3, mouse IgG2a; Pharmingen), Fas (DX2, mouse IgG1; Pharmingen), Fas ligand (NOK1, mouse IgG1; Pharmingen), and MHC class I (W6/32, mouse IgG2a; donated by M. Takiguchi, Kumamoto University). Isotype-matched control mouse antibodies IgG1, IgG2a, and IgM were purchased from Beckman Coulter.
Cytotoxicity assay
Natural killer activity was determined by51Cr-release assay as described elsewhere.28,52 In brief, the number of target cells with and without PIG-A mutation was accurately adjusted to 5 × 105/mL with polystyrene fluorospheres (Flow-Count; Beckman Coulter) and a flow cytometer (Epics).53 Target cells (1 mL) were incubated with 3.7 MBq of Na251CrO4 (New England Nuclear, Boston, MA) for 3 hours at 37°C, 5% CO2, and washed twice with phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) (Sigma, St Louis, MO). After 10-fold dilution, target cells (5 × 103/100 μL per well) were mixed with NK cells at various ratios of effector and target cells (E/T ratio) in U-bottomed 96-well microtiter plates (Corning, New York, NY) in triplicate. Cells were then incubated for 3 hours at 37°C, 5% CO2 in RPMI 1640 supplemented with 10% FCS and 10mM HEPES (Sigma) at pH 7.2. Medium containing 51Cr liberated from target cells was collected using cellulose acetate absorption cartridges (Skatron, Lier, Norway). Radioactivity was determined with automated γ-counter (Minaxi 5530; Packard Instrument Company, Downers Grove, IL). There was no difference in the uptake and spontaneous release of 51Cr between target cells with and without PIG-A mutation (data not shown). Spontaneous 51Cr release was determined by adding culture medium instead of effector cells to target cells. Maximum release was determined by adding Triton X-100 (0.25% final concentration; Nacalai Tesque, Kyoto, Japan) to the target cells. Spontaneous release was less than 10% of maximum release. Percent cytotoxicity was calculated using the following formula: % specific cytotoxicity = (experimental release − spontaneous release) / (maximum release − spontaneous release) × 100.
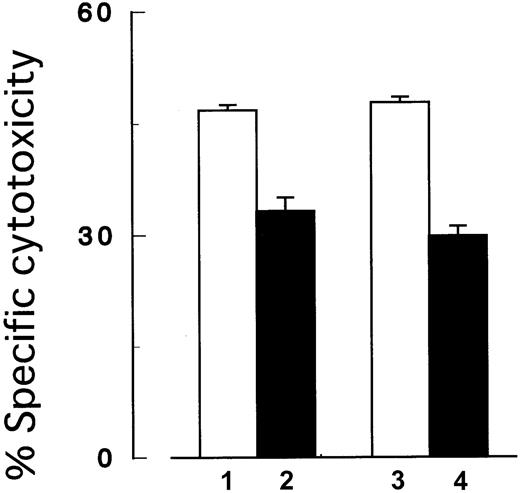
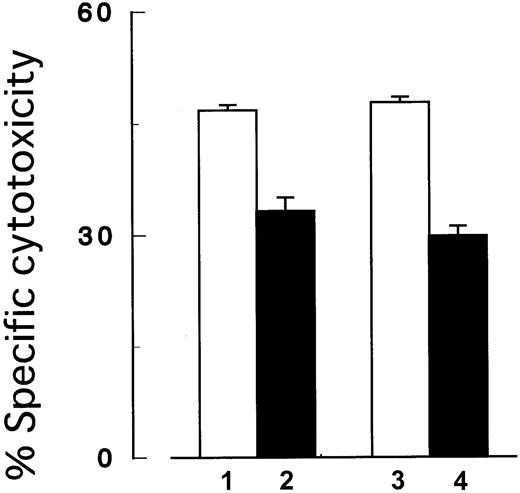
Next, we performed the cytotoxicity assay under the coexistence ofPIG-A mutant and control cells to mimic PNH patients' BM where blood cells with and without PIG-A mutation coexist. In brief, we prepared target cells by mixing the same number ofPIG-A mutant and control K562 cells in the same well. Either the mutant or control cells in the mixture were labeled with51Cr to assess the selection of target cells in the killing. Cells were then incubated with NK cells (2E3) at an E/T ratio (2:1) and cytotoxicity was assessed as described above.
We determined the sensitivity of target cells with and withoutPIG-A mutation to perforin. Perforin was purified from a mouse cytotoxic lymphocyte cell line (CTLL-R8.6) as reported previously.54,55 In brief, cells were disrupted using a nitrogen cavitation bomb in the presence of protease inhibitors. From the cell lysate, perforin-containing granules were collected by Percoll (Sigma) gradient centrifugation. Perforin was extracted from the granules by homogenization and purified by immobilized metal affinity column chromatography and hydrophobic interaction chromatography. Perforin activity was determined by cytolysis of sheep erythrocytes (1 unit corresponds to 50% hemolysis of 2 × 107 cells/200 μL). Cytotoxicity was determined by incubation of 6 U purified perforin with target cells for 1 hour in the absence of FCS. Other conditions were the same as those used for cytotoxic lymphocytes. (Mouse perforin exerts cytotoxicity to human cells as well as to mouse cells.56 57)
Inhibition of cytotoxicity
To exert cytotoxicity, lymphocytes usually use perforin/granzyme, Fas/Fas ligand, and tumor necrosis factor (TNF).58-60 Perforin-mediated cytotoxicity is inhibited with concanamycin A (CMA; Wako, Osaka, Japan) and a calcium chelate, ethylene glycol bis (β-aminoethylether)-N, N, N′, N′-tetraacetic acid (EGTA; Nacalai Tesque). CMA, a specific inhibitor of vacuolar type H+-adenosine triphosphatase (ATPase), accelerates degradation of perforin by an increase in the pH of lytic granules.61 To inhibit perforin-mediated cytotoxicity, killing was performed in the presence of EGTA or CMA at various concentrations. CMA required pretreatment of effector cells for 2 hours.
To determine the role of GPI-anchored proteins such as DAF and CD59 that are expressed only on the control cells, we tested cytotoxicity in the presence of 10 μg/mL MoAb to DAF or CD59. MoAbs to DAF were 1A10, BRIC 216 (mouse IgG1; Ylem, Roma, Italy), 1C6 (mouse IgG1; Wako), and MEM-118 (mouse IgM; Monosan, Uden, The Netherlands). MoAbs to CD59 were MEM-43 (mouse IgG2a; Monosan), MEM-43/5 (mouse IgG2b; Monosan), MEM-125 (mouse IgM; Monosan), 5H8, 2/24 (mouse IgG1; Chemicon, Temecula, CA), p282 (mouse IgG2a; Pharmingen), YTH53.1 (Rat IgG2b; Serotec, Oxford, United Kingdom), and BRA10G (mouse IgG1; Ancell, Bayport, MN). Isotype-matched control Ig was obtained from Beckman Coulter.
Determination of cytokines by enzyme-linked immunosorbent assay
To characterize the activation of NK cells, we measured the concentrations of interferon γ (IFN-γ) and TNF-α released from NK cells in the coculture with target cells. We used peripheral blood NK cells isolated from 5 healthy volunteers. Targets were PIG-Amutant and control K562 cells. NK and target cells were incubated at an E/T ratio (32:1) for up to 12 hours. Cell culture supernatant was tested for cytokines using sandwich enzyme-linked immunoabsorbent assay (ELISA) kits (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions.62 Briefly, wells in a microtiter plate were coated with antibody specific for IFN-γ or TNF-α, incubated with standards or samples, labeled with an enzyme-linked antibody specific for each cytokine, and incubated with a substrate solution. The color developed and the intensity of the color was measured at 450 nm. Each assay was performed in duplicate. The minimum detectable doses of IFN-γ and TNF-α were less than 8 pg/mL and 4.4 pg/mL, respectively.
Analysis of perforin mRNA expression by reverse transcription–polymerase chain reaction
We performed reverse transcription–polymerase chain reaction (RT-PCR) to analyze the expression of perforin mRNA in peripheral blood NK cells obtained from 2 healthy donors, as described.63,64 After a 3-hour coculture with target K562 cells (E/T ratio, 32:1), NK cells were collected with magnetic beads coated with MoAb to CD2. From the NK cells, total RNA was isolated using a kit with a silica-gel membrane (RNeasy; Qiagen, Hilden, Germany) and converted to cDNA with a kit (ReverTra; Toyobo, Osaka, Japan) according to the manufacturer's instructions.64The coding region of the cDNA was then amplified by PCR with 0.5 U recombinant Taq DNA polymerase (ExTaq; Takara, Shiga, Japan), 200 μM dNTPs in a buffer (25 μL; 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin), and 10 pmol of a sense primer (5′-GAGGCCCAGGTCAACATAGGCA-3′) and an antisense primer (5′-TCACCACACGGCCCCACTCCG-3′).63 After initial denaturation at 94°C for 5 minutes, DNA amplification was performed with a PCR thermocycler (Astec, Fukuoka, Japan) for 30 cycles; each cycle consisted of denaturation at 94°C for 1 minute, annealing at 60°C for 1 minute, and extension at 72°C for 2 minutes with a final extention at 72°C for 10 minutes. PCR products were analyzed by 1% agarose gel electrophoresis and ethidium bromide staining.
Statistical analysis
Student paired t test was used to determine significance levels. P values less than .05 were considered significant.
Results
Decreased susceptibility of PIG-A mutant to NK cells
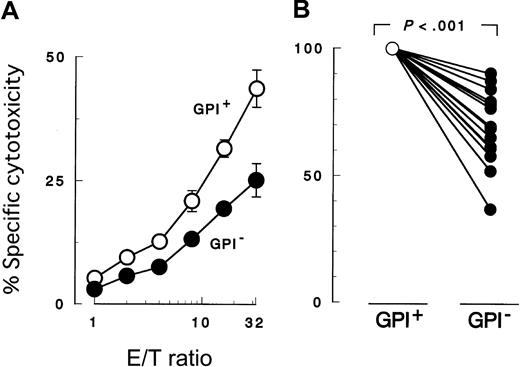
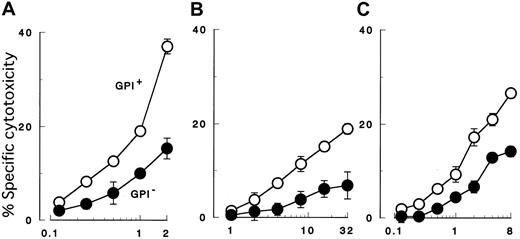
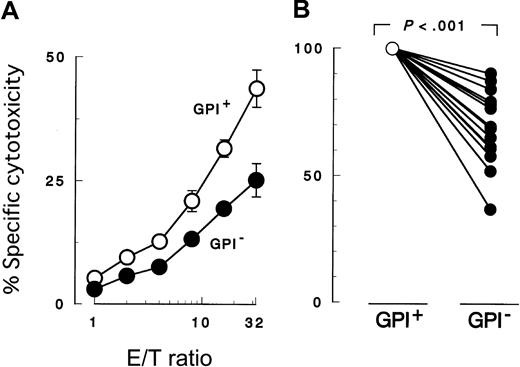
K562 cells with PIG-A mutation were less susceptible to NK cell killing than were control counterparts with a functionalPIG-A gene (Figure 2A). To confirm decreased sensitivity of the PIG-A mutant, we performed additional experiments using various effector (NK) and target cells. First we repeated the cytotoxicity assay using peripheral NK cells isolated from different individual healthy donors (Figure 2B). Each experiment showed decreased sensitivity of PIG-A mutant K562 cells. When cytolysis of control cells was normalized to 100%, cytolysis of PIG-A mutant cells ranged from 36% to 90% (mean 70%). The difference in lysis of K562 cells with and without aPIG-A mutation was significant (Student paired ttest, P < .000 01). Killing of our K562 cells was less than that of wild-type K562 cells (data not shown), probably due to the weak expression of MHC class I in our line inhibiting NK activity (Figure 1B).65 MHC expression on the target cells may also explain the variation in their killing by peripheral NK cells from the different healthy donors (Figure 2B).66 Further experiments, using a cultured NK cell line (2E3) (Figure3A,C) instead of peripheral NK cells or using lymphoid leukemic cell lines (B, JY-5; and T, MOLT-4) instead of K562 (Figure 3B-C), also showed decreased sensitivity ofPIG-A mutant to NK cells. Moreover, the decreased susceptibility of mutant cells to NK cells could also be observed in cytotoxicity assays combining mutant and control cells (Figure4).
Cytotoxicity assay by 51Cr release.
Peripheral blood NK cells were cocultured for 3 hours with K562 target cells. PIG-A mutant cells were less susceptible to NK cells than normal control cells. GPI+ (○), control cells (revertant) expressing GPI; and GPI− (●),PIG-A mutant cells (mock transfectant) deficient in GPI. (A) Cytolysis by NK cells obtained from a representative healthy donor at various E/T ratios. (B) Relative decrease in the cytolysis of GPI− cells in 13 separate experiments with NK cells of 13 healthy individuals. In each experiment, the cytolysis of GPI+ cells was set at 100% and the E/T ratio was 32:1. Each value represents the mean (± SD) of triplicate assays.
Cytotoxicity assay by 51Cr release.
Peripheral blood NK cells were cocultured for 3 hours with K562 target cells. PIG-A mutant cells were less susceptible to NK cells than normal control cells. GPI+ (○), control cells (revertant) expressing GPI; and GPI− (●),PIG-A mutant cells (mock transfectant) deficient in GPI. (A) Cytolysis by NK cells obtained from a representative healthy donor at various E/T ratios. (B) Relative decrease in the cytolysis of GPI− cells in 13 separate experiments with NK cells of 13 healthy individuals. In each experiment, the cytolysis of GPI+ cells was set at 100% and the E/T ratio was 32:1. Each value represents the mean (± SD) of triplicate assays.
Cytotoxicity assay with target cells of various cell lineages.
GPI− cells (●) of 3 different cell lineages were less susceptible than control cells (GPI+, ○) to both peripheral and cultured NK cells. (A) Cytolysis of myeloid cells (K562) by cultured NK cells (2E3). (B) Cytolysis of B-lymphoid cells (JY-5) by peripheral NK cells of a healthy donor. (C) Cytolysis of T-lymphoid cells (MOLT-4) by cultured NK cells (2E3). Each value represents the mean (± SD) of triplicate assays. Each abscissa indicates E/T ratios.
Cytotoxicity assay with target cells of various cell lineages.
GPI− cells (●) of 3 different cell lineages were less susceptible than control cells (GPI+, ○) to both peripheral and cultured NK cells. (A) Cytolysis of myeloid cells (K562) by cultured NK cells (2E3). (B) Cytolysis of B-lymphoid cells (JY-5) by peripheral NK cells of a healthy donor. (C) Cytolysis of T-lymphoid cells (MOLT-4) by cultured NK cells (2E3). Each value represents the mean (± SD) of triplicate assays. Each abscissa indicates E/T ratios.
Cytotoxicity assay using a mixture of target cells with and without
PIG-A mutation. In the coexistence ofPIG-A mutant and control cells, NK cells preferably killed control cells than the mutant cells. Columns 1 and 2, control (GPI+) and PIG-A mutant (GPI−) K562 cells, respectively; columns 3 and 4, mixtures of the same number of control and the mutant cells. In the mixtures, only GPI+cells (column 3) and GPI− cells (column 4) were distinctively labeled with 51Cr. The target cells were then cocultured with NK cells (2E3) at E/T ratio (2:1). Each value represents the mean (± SD) of triplicate assays.
Cytotoxicity assay using a mixture of target cells with and without
PIG-A mutation. In the coexistence ofPIG-A mutant and control cells, NK cells preferably killed control cells than the mutant cells. Columns 1 and 2, control (GPI+) and PIG-A mutant (GPI−) K562 cells, respectively; columns 3 and 4, mixtures of the same number of control and the mutant cells. In the mixtures, only GPI+cells (column 3) and GPI− cells (column 4) were distinctively labeled with 51Cr. The target cells were then cocultured with NK cells (2E3) at E/T ratio (2:1). Each value represents the mean (± SD) of triplicate assays.
Perforin-mediated cytotoxicity of NK cells
Because NK cells (2E3) exert cytotoxicity despite their lack of Fas ligand (Figure 3A,C), we first examined the effects of CMA and EGTA, which inhibit the perforin/granzyme pathway. Each reagent inhibited dose-dependently the killing of K562 cells with and withoutPIG-A mutation by either peripheral (Figure5) or cultured NK cells (data not shown). Killing was virtually abolished with either 5 nM of CMA or 1 mM of EGTA, and at these concentrations NK cells remained viable. These findings suggested that NK cells used only the perforin/granzyme pathway. Next, we replaced NK cells with purified perforin in the cytotoxicity assay (Figure 6). In the presence of soluble perforin, both mutant K562 and JY-5 cells were lysed similar to controls, indicating that the PIG-Amutation did not decrease sensitivity to perforin. There was a difference in susceptibility to perforin between K562 and JY-5.
Cytotoxicity assay with various inhibitors.
The killing of target cells by NK cells was perforin-mediated. Cytolysis by peripheral NK cells of K562 cells with (▪) and without (■) PIG-A mutation in the presence of a calcium chelate EGTA (A) and concanamycin A (B). E/T ratio was 64:1. Each value represents the mean (± SD) of triplicate assays.
Cytotoxicity assay with various inhibitors.
The killing of target cells by NK cells was perforin-mediated. Cytolysis by peripheral NK cells of K562 cells with (▪) and without (■) PIG-A mutation in the presence of a calcium chelate EGTA (A) and concanamycin A (B). E/T ratio was 64:1. Each value represents the mean (± SD) of triplicate assays.
Cytotoxicity assay with purified perforin.
PIG-A mutant and control cells showed similar sensitivity to purified perforin in vitro. Both K562 cells and JY-5 cells with (GPI−) and without (GPI+) PIG-Amutation were incubated for one hour with 6 U purified perforin. Each value represents the mean (± SD) of triplicate assays.
Cytotoxicity assay with purified perforin.
PIG-A mutant and control cells showed similar sensitivity to purified perforin in vitro. Both K562 cells and JY-5 cells with (GPI−) and without (GPI+) PIG-Amutation were incubated for one hour with 6 U purified perforin. Each value represents the mean (± SD) of triplicate assays.
Defective activation of NK cells by PIG-A mutant cells
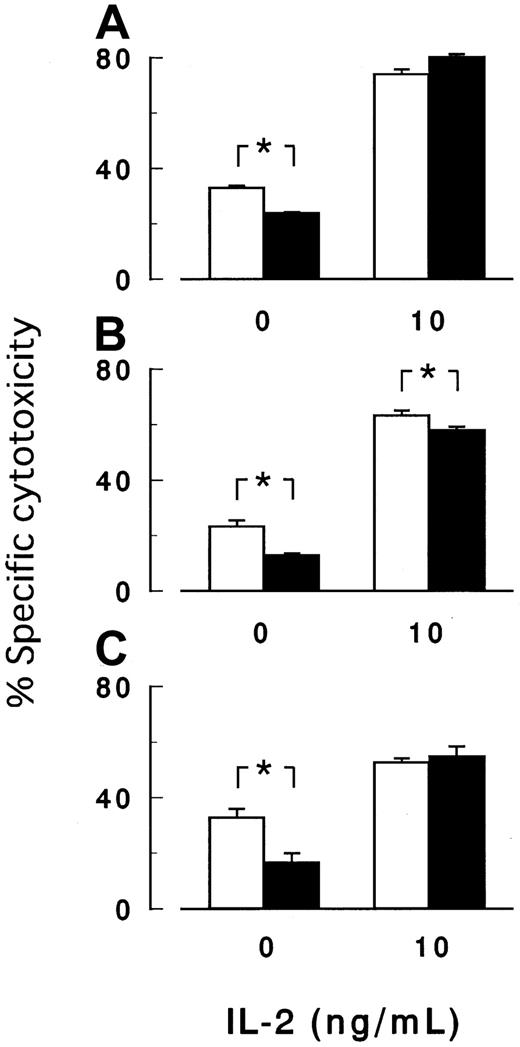
Figure 7 shows the 3-hour cytotoxicity assay in the presence of 10 ng/mL IL-2, a potent activator of NK cells. IL-2 enhanced the target cell (K562) killing by NK cells isolated from 3 healthy volunteers but diminished or abolished the difference in the sensitivity to killing between PIG-Amutant and control cells (Figure 7A-C). The mutant and control cells thus showed similar sensitivity to activated NK cells (Figure 7) as well as to purified soluble perforin (Figure 6). Additionally, killing was completely suppressed with perforin inhibitors (data not shown). We inferred that NK cells, once sufficiently activated with IL-2 to release perforin/granzyme, were equally effective againstPIG-A mutant and control cells.
Cytotoxicity assay in the presence of IL-2.
IL-2 enhanced the killing of target cells by NK cells, whereas IL-2 abolished or decreased the difference in the cytotoxicity betweenPIG-A mutant and control cells. Peripheral NK cells obtained from 3 healthy volunteers (A-C) were cocultured for 3 hours with K562 cells with (▪) and without (■) PIG-A mutation in the absence or the presence of 10 ng/mL IL-2. E/T ratio was 32:1. Each value represents the mean (± SD) of triplicate assays. *Statistical significance (P < .05).
Cytotoxicity assay in the presence of IL-2.
IL-2 enhanced the killing of target cells by NK cells, whereas IL-2 abolished or decreased the difference in the cytotoxicity betweenPIG-A mutant and control cells. Peripheral NK cells obtained from 3 healthy volunteers (A-C) were cocultured for 3 hours with K562 cells with (▪) and without (■) PIG-A mutation in the absence or the presence of 10 ng/mL IL-2. E/T ratio was 32:1. Each value represents the mean (± SD) of triplicate assays. *Statistical significance (P < .05).
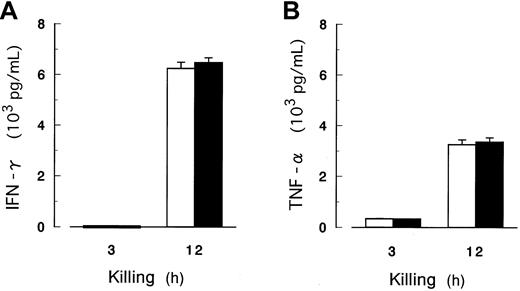
We also measured IFN-γ and TNF-α as other indicators of NK activation by coculture of peripheral blood NK cells with K562 cells in the absence of IL-2. Concentrations of these cytokines were very low within 3 hours and markedly increased within 12 hours; however, the amounts produced were equivalent for PIG-A mutant and control cells (Figure 8). During the cytotoxicity assay, the expression of perforin mRNA was persistent (data not shown; release of perforin was not determined because a reliable ELISA has not been established). Thus, PIG-A mutant cells were capable of inducing the release of IFN-γ and TNF-α, but possibly defective in their capacity to prompt sufficient release of perforin/granzyme from NK cells.
Release of IFN-γ and TNF-α in the cytotoxicity assay.
The concentrations of IFN-γ (A) and TNF-α (B) as indicators of NK activation were determined by ELISA in the culture supernate. NK cells were isolated from peripheral blood of 5 healthy volunteers and cocultured for up to 12 hours with K562 cells with (▪) and without (■) PIG-A mutation in the absence of IL-2. E/T ratio was 32:1. A and B show the data of representative of 5 donors. Each value represents the mean (± SD) of triplicate assays.
Release of IFN-γ and TNF-α in the cytotoxicity assay.
The concentrations of IFN-γ (A) and TNF-α (B) as indicators of NK activation were determined by ELISA in the culture supernate. NK cells were isolated from peripheral blood of 5 healthy volunteers and cocultured for up to 12 hours with K562 cells with (▪) and without (■) PIG-A mutation in the absence of IL-2. E/T ratio was 32:1. A and B show the data of representative of 5 donors. Each value represents the mean (± SD) of triplicate assays.
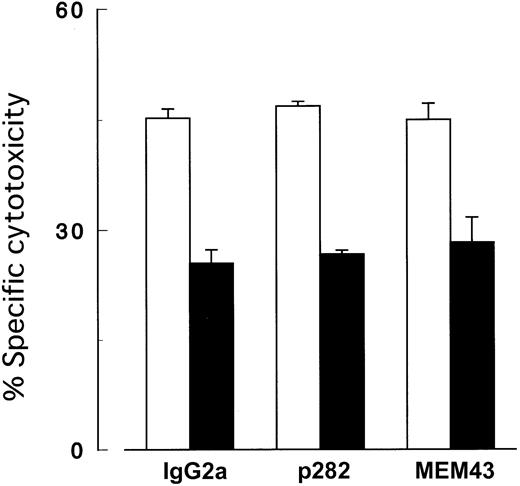
To further explore the possibility of impaired triggering of the perforin/granzyme pathway by PIG-A mutant cells, we screened GPI-linked membrane molecules missing from the mutant cells by flow cytometry. However, no MoAb clones to DAF (data not shown) and CD59 (Figure 9) exerted any effects in cytotoxicity assays with cultured NK cells. In these experiments, antibody-dependent cellular cytotoxicity was negligible because cultured NK cells lacked CD16 (FcγR).
Cytotoxicity assay with antibodies to CD59.
Killing of target cells by NK cells was not inhibited by MoAb to CD59. K562 cells with (▪) and without (■) PIG-A mutation were cocultured with NK cells (2E3) at E/T ratio (2:1) in the presence of 10 μg/mL MoAb to CD59. IgG2a, isotype-matched mouse Ig; p282 and MEM43, 2 clones (representatives of 8) of MoAb (IgG2a) to CD59. Each value represents the mean (± SD) of triplicate assays.
Cytotoxicity assay with antibodies to CD59.
Killing of target cells by NK cells was not inhibited by MoAb to CD59. K562 cells with (▪) and without (■) PIG-A mutation were cocultured with NK cells (2E3) at E/T ratio (2:1) in the presence of 10 μg/mL MoAb to CD59. IgG2a, isotype-matched mouse Ig; p282 and MEM43, 2 clones (representatives of 8) of MoAb (IgG2a) to CD59. Each value represents the mean (± SD) of triplicate assays.
Discussion
Little is known about the mechanism responsible for expansion of PNH clones.15,21,22 The present work supports a selective survival advantage theory. In our experiments, leukemic cells withPIG-A mutations (GPI− cells) were less susceptible than were their control counterparts (GPI+cells) to killing by NK cells in vitro. The decreased susceptibility ofPIG-A mutant (GPI−) cells was confirmed by repeated experiments using cultured NK cells as well as peripheral NK cells, and using T- and B-lymphoid leukemic cells as well as myeloid leukemic cells as target cells. MHC class I expression, which inhibits NK activity,65 was not different between each of the pairs of PIG-A mutant (GPI−) and control (GPI+) cells. In addition, experiments with mixtures of GPI+ and GPI− targets, mimicking the coexistence of GPI− and GPI+ cells observed in the BM of PNH patients, gave similar results, suggesting that NK cells might discriminate PIG-A mutant cells in vivo as well as in vitro. The difference in sensitivity to 3-hour killing by NK cells in vitro between the PIG-A mutant (GPI−) and control (GPI+) cells was less than expected but might be sufficient over time to create a sustained advantage forPIG-A mutant cells. It is also possible that earlier negative data are at least partly attributable to the difficulty of detecting a small diminution in sensitivity of PIG-A mutant (GPI−) cells to cytotoxic lymphocytes.15,28,67 68 In the present study, this technical problem was overcome partly by the preparation of a pair of leukemic cells genetically identical except for PIG-A as targets for NK cells and further by accurate cell count by flow cytometry. Accordingly, we conclude that PIG-A mutation (intrinsic factor) decreases sensitivity to the killing by NK cells (extrinsic factor), thus allowing a PNH clone to escape immune attack, and confers a relative survival advantage to PNH clone in the context of BM injury by cytotoxic lymphocytes.
We investigated the mechanism for PIG-A mutant insensitivity to perforin-mediated killing by NK cells in their coculture. In general, perforin-mediated killing by NK cells consists of at least 3 discrete steps: first, stimulation of NK cells by direct contact between NK and target cells; second, signal transduction in NK cells to release perforin/granzyme; third, killing targets cells with the liberated cytotoxic molecules.69 In this regard, thePIG-A mutant and control cells showed similar sensitivity to soluble perforin used instead of NK cells (consistent with previous report67) or to perforin-mediated killing by NK cells driven by IL-2. Thus, our findings indicate that these later steps are not responsible for the difference in NK sensitivity between target cells. Rather, by exclusion, the first event appears important: resistance is manifest when NK cells are activated by contact withPIG-A mutant cells. It is thus conceivable thatPIG-A mutant cells are partly defective in their ability to signal the release of perforin (although we did not determine the relative concentrations of perforin liberated in these experiments). In further characterization of NK activation, coculture of NK cells with target cells induced marked release of IFN-γ and TNF-α, reliable markers of NK activation. However, there was no difference in their release between PIG-A mutant and control cells.
NK cell activation by contact with target cells is mediated or inhibited by cell surface molecules. PIG-A mutant cells distinctly lack GPI-linked membrane proteins. Although inhibition assays with MoAbs to DAF and CD59 gave negative results, unidentified GPI-linked proteins may explain the decreased sensitivity ofPIG-A mutant to NK cells. To date, 2 GPI-linked glycoproteins (mouse RAE-1 and human ULBP) have been shown to activate NK cells and to be inducible by transformation and viral infection.70-72 Of interest, such activation signals are dominant over inhibitory signals from MHC class I.72,73 NK cells killed leukemic cells despite their MHC class I expression in our assay. The identification of such putative membrane molecules suggested in the present study could provide a new strategy to control NK activity and also facilitate the unraveling of NK cell–associated pathophysiology of infection, cancer, autoimmune disease, and transplantation.60 74-76
We thank Masato Yagita of Osaka Kitano Hospital, Jaroslaw Maciejewski of National Institutes of Health, and Yasuharu Nishimura and Masafumi Takiguchi of Kumamoto University for their advice and help.
Supported by a grant from the Sagawa Foundation for Promotion of Cancer Research, and grants for scientific research and for cancer research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
S.N. and S. I. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hideki Nakakuma, Second Department of Internal Medicine, Kumamoto University, School of Medicine, Honjo 1-1-1, Kumamoto 860-8556, Japan; e-mail:nakakuma@kaiju.medic.kumamoto-u.ac.jp.