Resistance to all-trans retinoic acid (ATRA) remains a clinical problem in the treatment of acute promyelocytic leukemia (APL) and provides a model for the development of novel therapies. Molecular alterations in the ligand-binding domain (LBD) of the PML/RARα fusion gene that characterizes APL constitute one mechanism of acquired resistance to ATRA. We identified missense mutations in PML/RARα from an additional ATRA-resistant patient at relapse and in a novel ATRA-resistant cell line, NB4-MRA1. These cause altered binding to ligand and transcriptional coregulators, leading to a dominant-negative block of transcription. These mutations are in regions of the LBD that appear to be mutational hot spots occurring repeatedly in ATRA-resistant APL patient cells. We evaluated whether histone deacetylase (HDAC) inhibition could overcome the effects of these mutations on ATRA-induced gene expression. Cotreatment with ATRA and TSA restoredRARβ gene expression in NB4-MRA1 cells, whose PML/RARα mutation is in helix 12 of the LBD, but not in an APL cell line harboring the patient-derived PML/RARα mutation, which was between helix 5 and 6. Furthermore, ATRA combined with TSA increases histone 4 acetylation on the RARβ promoter only in NB4-MRA1 cells. Consistent with these results, the combined treatment induces differentiation of NB4-MRA1 only. Thus, the ability of an HDAC inhibitor to restore ATRA sensitivity in resistant cells may depend on their specific molecular defects. The variety of PML/RARαmutations arising in ATRA-resistant patients begins to explain how APL patients in relapse may differ in response to transcription therapy with HDAC inhibitors.
Introduction
Acute promyelocytic leukemia (APL) is characterized by a translocation, t(15;17), between the promyelocytic leukemia (PML) gene on chromosome 15 and theretinoic acid receptor alpha (RARα) gene on chromosome 17. This translocation generates a PML/RARα fusion protein crucial to the pathogenesis of APL.1-8 A dominant-negative effect of the chimeric protein leads to the inhibition of PML and RARα pathways, a block of myeloid differentiation, and ultimately to the APL phenotype.9-11
The actions of retinoids are mediated by heterodimers of 2 classes of ligand-dependent transcription factors: the retinoic acid receptors (RARs) and the retinoid X receptors (RXRs).12 In the absence of all-trans retinoic acid (ATRA), RXR/RAR heterodimers interact with nuclear receptor corepressors (termed SMRT and NCoR), which recruit histone deacetylases (HDACs) to induce chromatin modifications and transcriptional repression.13-15 Binding of the ligand permits release of the corepressor complex and binding to coactivators, which in turn recruit histone acetylases that modify chromatin to increase promoter accessibility, leading to the activation of transcription.14,16 17
The PML/RARα protein retains most functional domains of RARα and behaves as an abnormal receptor with altered transactivation functions. In APL, the fusion protein can bind ATRA, but higher concentrations are required to activate transcription.18,19 This may be due to a more tightly associated PML/RARα-corepressor complex, in which the corepressor is not released at physiological ATRA concentrations.20-22The release of corepressor induced by higher ATRA concentrations allows recruitment of coactivators and may underlie the cytodifferentiation of APL cells.20-23
At pharmacologic concentrations, ATRA induces complete remission in a high percentage of APL patients.24,25 Nevertheless, APL cells develop resistance to ATRA in vitro and in vivo. Although combined cytotoxic chemotherapy with ATRA cures a high percentage of patients with APL, relapse with ATRA-resistant cells still occurs.26,27 Different mechanisms have been proposed to explain this clinical resistance, including altered pharmacokinetics and genetic changes.28-31
Much has been learned about response and resistance to ATRA by studying a cell line, NB4, derived from an APL patient.32 We and others have reported NB4 subclones that are highly resistant to retinoid-induced cytodifferentiation.33-35 We identified a point mutation in the LBD of the PML/RARα fusion gene of the subclone NB4-MR4, which abolishes the ATRA-binding capacity of the fusion protein and blocks the transcription of ATRA-responsive genes in a dominant-negative fashion.36 Additional mutations in the LBD of the PML/RARα have been reported in independently established ATRA-resistant NB4 subclones.37-39
A spontaneously ATRA-resistant APL cell line, called UF-1, has been established directly from an ATRA-resistant APL patient.40The PML/RARα gene of UF-1 cells harbors a point mutation in the LBD of its RARα portion replacing the arginine (Arg, R) at position 276 by a tryptophan (Trp, W).41
Similar mutations of the PML/RARα fusion gene have been reported in APL cells from a growing number of patients who developed ATRA resistance, demonstrating the clinical relevance of these genetic alterations as a mechanism of resistance.31,42-45 In a previous study, we analyzed in vitro expression of PML/RARα mutations from ATRA-resistant APL patients. We found that these amino acid changes cause a variety of abnormalities in the ligand-binding transactivation of RAREs and ligand-dependent interactions with the transcriptional coregulators SMRT and ACTR.46 This contrasts the nearly complete dominant-negative activity of mutations in PML/RARα previously characterized in cell lines developing ATRA resistance in vitro. This variability of binding to ligand and transcriptional coregulators suggests that strategies to overcome resistance might work in some cases but not in others. Recently, histone deacetylase inhibitors have been shown to restore ATRA-induced transcriptional activity and to induce the differentiation of patient leukemic blasts in vitro and in animal models of APL.47-49 Indeed, combined treatment with ATRA and an HDAC inhibitor led to a durable complete remission of one patient with ATRA-refractory APL.50
Here we report the presence of novel mutations in the LBD of thePML/RARα fusion gene in a newly established ATRA-resistant NB4 subclone, NB4-MRA1, and in an additional ATRA-resistant APL patient in relapse. Both PML/RARα mutant cells show impaired ATRA binding and transcriptional response to ATRA, as well as alterations in their ability to interact, in a ligand-dependent manner, with the corepressor SMRT and the coactivator ACTR. We characterized these mutations of PML/RARα for their response to the combination of ATRA with an HDAC inhibitor. We investigated whether the induction of histone acetylation by cotreatment with ATRA and trichostatin A (TSA) could restore transcriptional activity and expression of an ATRA-target gene, as well as granulocytic differentiation.
Materials and methods
Cell culture
NB4-MRA1 was generated by exposing NB4 cells to a stepwise increase from 1 × 10−8 to 1 × 10−5 M of ATRA (Sigma, St Louis, MO) for 3 months. Single clones were isolated with methylcellulose and expanded. The ATRA-resistant NB4-MRA1 cells were then propagated for 1 month in ATRA-free media and retested for resistance in 1 × 10−5 M ATRA. These cells are routinely cultured in ATRA-free RPMI 1640 medium (Life Technologies, [Gibco BRL], Burlington, ON, Canada) supplemented with 10% fetal calf serum (FCS; Wisent, St Bruno, QC, Canada) and remain ATRA resistant. In accordance with the proposed nomenclature of Roussel and Lanotte,51 the previously published NB4-R4 has been renamed NB4-MR4. The establishment of and conditions for cell culture of the ATRA-resistant APL cells NB4-MR4 and UF-1 have been published previously.35 40
Clinical history of the ATRA-resistant APL patient
The patient, a 35-year-old man with bleeding gums and bruises, received a diagnosis in May 1991, of APL with the presence of a typicalPML/RARα chimeric mRNA of the long type (L, Bcr1). He achieved complete clinical remission after chemotherapy with mitoxantrone 80 mg/m2, Ara-C 3 g/m2 × 5 and VP-16 150 mg/m2 × 3. He received consolidation therapy with ATRA (45 mg/m2 per day) for 10 weeks. In August 1993, in early clinical relapse, he received ATRA (45 mg/m2 per day) for 4 weeks and re-entered clinical remission. In February 1994, he underwent allogenic bone marrow transplantation without evidence of graft-versus-host disease. In July 1997, he had another relapse with progression of APL resistant to ATRA and chemotherapy.
Plasmid constructs
PML/RARα with either R276W or I410T mutations were cloned into the pSG5 mammalian expression vector harboring wild-type PML/RARα (L) cDNA using a QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). All the constructs were verified by sequencing analysis, using the dsDNA Cycle Sequencing System (Life Technologies). Construction of the PML/RARα (L) harboring the L398P mutation (PML/RARα–M4) was previously described.36
DNA sequencing analysis
One microgram total RNA was used for reverse transcription–polymerase chain reaction (RT-PCR) with random primers and Superscript II reverse transcriptase (Life Technologies). 2 μL of a 20 μL RT reaction were used for PCR amplification of the non-rearranged RARα with primers, oligo A: CAG CAC CAG CTT CCA GTT AG and oligo C: TGT CCG CTC AGA GTG TCC AG and of the RARα portion of the PML/RARα with oligo B: GTC TCC AAT ACA ACG ACA GC and oligo C. The PCR products were gel isolated and used as templates for direct sequencing using the dsDNA cycle sequencing kit (Life Technologies).
Cell differentiation
Cell differentiation was evaluated by direct immunofluorescence staining of CD11b (30455X; PharMingen, Mississauga, ON, Canada), Coulter Epics XL flow cytometer (Beckman Coulter, Miami, FL), and nitro blue tetrazolium (NBT) reduction assay as previously reported.52
Western blot analysis
Nuclear extracts (50 μg) were run on a 10% sodium dodecyl sulfate acrylamide gel and were transferred to a nitrocellulose membrane (BioRad Laboratories, Mississauga, ON, Canada). The membrane was blocked with 5% skim milk and 0.1% Tween 20 in phosphate-buffered saline (PBS) and was hybridized overnight with a RARα-specific antibody (SC-551; Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:500. After washing in 0.1% Tris-buffered saline, the membrane was hybridized with a secondary anti-rabbit antibody and developed with the enhanced chemiluminescence system (Amersham, Little Chalfont, Buckinghamshire, United Kingdom).
Assay for ligand-binding activity
Nuclear extracts were prepared and incubated for 18 hours at 4°C with 10 nM [3H]-ATRA (50.7 Ci/mmol; DuPont-NEN, Boston, MA) or with [3H]-ATRA in the presence of 200-fold excess of unlabeled ATRA. The extracts were subsequently fractionated by high-performance liquid chromatography (HPLC) as previously described.18
Limited proteolytic digestion of translated fusion proteins
Wild-type and mutant PML/RARα fusion proteins were in vitro synthesized in the presence of [35S]-methionine (NEN, Streetsville, ON, Canada), using a coupled transcription and translation reticulocyte lysate system as suggested by the manufacturer (Promega, Madison, WI). The radioactive fusion proteins were then analyzed in a limited proteolytic digestion assay as described previously.46
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays were performed using [35S]-labeled in vitro–translated wild-type and mutant PML/RARα fusion proteins and a direct repeat 5 (DR5) retinoic acid responsive element, as previously described.46 Where specified, bacterially expressed and purified GST fusions containing the receptor interactive domains of SMRT (GST-SMRT-ID II; amino acid 1073-116820) or ACTR (GST-ACTR–RID; amino acid 621-82117) were added.
In vitro interaction with GST-DRIP205
Glutathione-S-transferase (GST)-DRIP205 was kindly provided by Dr Leonard P. Freedman. In vitro interaction of the mutants PML/RARα with GST-DRIP205 was performed as described previously,53except that 150 000 cpm of [35S]-labeled in vitro–translated proteins were used.
Transient transfection experiments for transcriptional activity
APL cells (5 × 106 cells/transfection) were transfected by electroporation with 10 μg per transfection of the reporter plasmid DR5-tk-CAT54 and 10 μg per transfection of pCMV-βGalactosidase (β-Gal) as an internal control for transfection efficiency. Cells were electroporated, replenished in RPMI 1640 with 10% FCS, and grown for 48 hours in the absence or presence of different concentrations of drugs. Chloramphenicol acetyltransferase (CAT) counts were normalized with β-Gal activity to obtain the relative CAT activity.
Ribonuclease protection assay
Fifty micrograms total RNA was used for RNase protection analysis, as previously described.55 Hybridization of cRNA probes was performed at 50°C overnight, followed by the addition of 350 μL RNase digestion buffer (10 mM Tris-HCl, pH 7.5, 300 mM NaCl, 5 mM EDTA [ethylenediaminetetraacetic acid]) containing RNase T1 (Roche Diagnostics, Laval, QC, Canada). RNase digestion was performed at 30°C for 1 hour. RNase-resistant fragments were resolved by electrophoresis on 6% urea-polyacrylamide sequencing gels and visualized by autoradiography.
Chromatin immunoprecipitation
Two million cells were grown the day before treatment with 1 μM ATRA, 200 nM TSA (Sigma), or the combination for 1 hour. To measure histone acetylation levels, formaldehyde–cross-linked and sonicated chromatin was immunoprecipitated overnight with 5 μL antibody raised against the acetylated form of histone H4 N-terminal tail (Upstate Biotechnology, Lake Placid, NY) following the manufacturer's instructions. For PCR, 1 μL of 20 μL extracted DNA was used with the FastStart Taq DNA Polymerase kit (Roche Molecular Biochemicals, Laval, QC, Canada), and 28 to 35 cycles were allowed. Primers used for PCR detection of the RARβ promoter were: sense, 5′-TCC TGG GAG TTG GTG ATG TCA G-3′; anti-sense, 5′-AAA CCC TGC TCG GAT CGC TC-3′.
Results
RA-resistant NB4-MRA1 and relapsed patient APL cells harbor novelPML/RARα mutations in the LBD
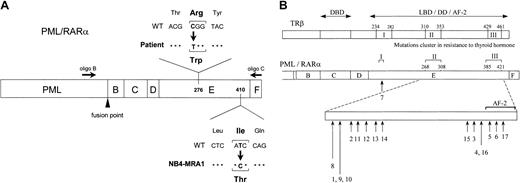
Our laboratory has developed several ATRA-resistant subclones of NB4 by selection in ATRA-containing media.35 The resistant subclone NB4-MRA1 and the leukemic cells from an ATRA-resistant patient with relapsed APL were subjected to DNA sequencing analysis of thePML/RARα gene. Two primers depicted in Figure1A were used in RT-PCR to specifically amplify the RARα moiety of the PML/RARα. We identified missense mutations in the LBD of PML/RARα in both the ATRA-resistant NB4-MRA1 and patient APL cells. A novel point mutation was detected in the LBD of the RARα portion of the PML/RARαgene in NB4-MRA1 cells. This mutation resulted in amino acid substitution of isoleucine (Ile, I) (ATC) for threonine (Thr, T) (ACC) in the long form (L, Bcr1) of PML/RARα protein, which corresponds to codon 410 (I410T) in wild-type RARα. APL cells from the ATRA-resistant patient in relapse expressed the long form (Bcr1) of PML/RARα, with a C-to-T substitution changing the codon specificity from arginine (Arg, R) to tryptophan (Trp, W) at position 276 (R276W) (Figure 1A). Locations of the mutations were described with reference to the normal amino acid sequence of RARα1.56 Sequencing both strands of NB4-MRA1 and patient cDNA confirmed the point mutations. No other mutations were found in the PML/RARα or the coexpressed wild-typeRARα of NB4-MRA1 or the resistant patient cells. Figure 1B and Table 1 present a summary of reported LBD PML/RARα mutations associated with ATRA resistance in cell lines (numbers 1-6) and in APL patients (numbers 7-17). Of note, the R276W mutation in our patient-resistant APL cells is identical to the previously reported PML/RARα mutation of the patient-derived APL cell line, UF-1.41 This allowed us to use the UF-1 cell line as an in vivo model to study the effects of the patient PML/RARα mutation on ATRA response.
Identification of novel point mutations in the LBD of the RARα moiety of PML/RARα in RA-resistant APL.
(A) Schematic representation of PML/RARα, describing point mutations identified in ATRA-resistant APL cells from a patient in relapse and from a newly established cell line, NB4-MRA1, showing the approximate positions of 2 primers used in RT-PCR. (B) Summary of the LBD PML/RARα mutations identified in ATRA-resistant APL cell lines (numbers 1-6) and patients in relapse (numbers 7-17). Numbers 5 and 9 indicate the novel mutations in the LBD PML/RARα of ATRA-resistant APL cells identified and characterized in the current study and correspond to NB4-MRA1 and the patient, respectively. Numbers 1 to 17 in Figure 1B correspond to numbers 1 to 17 in Table 1. The lengths of the arrows reflect the frequency of each mutation. The position of the mutations is described with reference to normal amino acid sequence of RARα1.56 The alignment of the PML/RARα E-domain and TRβ ligand-binding domain by sequence homology indicates that the mutations in ATRA-resistant patients with APL and cell lines cluster in accordance with the regions in RTH syndrome denoted as I, II, and III. DBD indicates DNA-binding domain; LBD, ligand-binding domain; DD, dimerization domain; AF-2, ligand-dependent activation function.
Identification of novel point mutations in the LBD of the RARα moiety of PML/RARα in RA-resistant APL.
(A) Schematic representation of PML/RARα, describing point mutations identified in ATRA-resistant APL cells from a patient in relapse and from a newly established cell line, NB4-MRA1, showing the approximate positions of 2 primers used in RT-PCR. (B) Summary of the LBD PML/RARα mutations identified in ATRA-resistant APL cell lines (numbers 1-6) and patients in relapse (numbers 7-17). Numbers 5 and 9 indicate the novel mutations in the LBD PML/RARα of ATRA-resistant APL cells identified and characterized in the current study and correspond to NB4-MRA1 and the patient, respectively. Numbers 1 to 17 in Figure 1B correspond to numbers 1 to 17 in Table 1. The lengths of the arrows reflect the frequency of each mutation. The position of the mutations is described with reference to normal amino acid sequence of RARα1.56 The alignment of the PML/RARα E-domain and TRβ ligand-binding domain by sequence homology indicates that the mutations in ATRA-resistant patients with APL and cell lines cluster in accordance with the regions in RTH syndrome denoted as I, II, and III. DBD indicates DNA-binding domain; LBD, ligand-binding domain; DD, dimerization domain; AF-2, ligand-dependent activation function.
Characterization of cell lines expressing novel PML/RARα mutants and their ATRA-binding activity
NB4-MRA1 cells were cultured in ATRA-free media for more than 6 months and remained resistant to ATRA-induced growth inhibition (data not shown) or cell differentiation (see “Results” and Figure8). The long-term maintenance of resistance in ATRA-free culture and the presence of a genetic change mediating resistance argue that this cell line is truly ATRA resistant, not transiently insensitive.57 Figure 2A confirms the expression of PML/RARα fusion protein in NB4-MRA1 cells and shows the ATRA-induced PML/RARα protein degradation in the ATRA-sensitive NB4 but not in the ATRA-resistant NB4-MRA1 APL cells. The lack of response of UF-1 cells to ATRA has previously been described.41
Characterization of cell lines expressing novel PML/RARα mutants and their ATRA-binding activity.
(A) ATRA induces PML/RARα protein degradation in the ATRA-sensitive NB4 cells, but not in the ATRA-resistant NB4-MRA1 APL cells. Cells were treated with 1 μM ATRA for 24 hours, and 50 μg of nuclear extracts were used in Western blot analysis for the expression of PML/RARα protein. NB4 and NB4-MRA1 expressed a long PML/RARα (L) isoform (110 kDa). The lower panel shows laminin B expression to confirm protein loading. (B) Specific HPLC ATRA-binding profiles of nuclear extracts from NB4 cells (●) compared with those from the ATRA-resistant NB4-MRA1 (○) and UF-1 (▾) APL cells. Nuclear extracts were incubated with 10 nM [3H]-ATRA (●, ○, ▾) or with [3H]-ATRA in the presence of 200-fold excess of unlabeled ATRA (▿). Extracts were subjected to HPLC analysis using a 6 HR 10/30 size exclusion column.
Characterization of cell lines expressing novel PML/RARα mutants and their ATRA-binding activity.
(A) ATRA induces PML/RARα protein degradation in the ATRA-sensitive NB4 cells, but not in the ATRA-resistant NB4-MRA1 APL cells. Cells were treated with 1 μM ATRA for 24 hours, and 50 μg of nuclear extracts were used in Western blot analysis for the expression of PML/RARα protein. NB4 and NB4-MRA1 expressed a long PML/RARα (L) isoform (110 kDa). The lower panel shows laminin B expression to confirm protein loading. (B) Specific HPLC ATRA-binding profiles of nuclear extracts from NB4 cells (●) compared with those from the ATRA-resistant NB4-MRA1 (○) and UF-1 (▾) APL cells. Nuclear extracts were incubated with 10 nM [3H]-ATRA (●, ○, ▾) or with [3H]-ATRA in the presence of 200-fold excess of unlabeled ATRA (▿). Extracts were subjected to HPLC analysis using a 6 HR 10/30 size exclusion column.
To evaluate how these point mutations of PML/RARα alter the function of the LBD, we examined their ATRA-binding activity (Figure 2B). The size exclusion HPLC profile of extracts from NB4 cells expressing the wild-type form of PML/RARα is characterized by the 50-kDa peak representing the binding of ATRA to the endogenous RARs and the approximately 670-kDa peak representing macromolecular complexes formed by the interaction of PML/RARα with itself or other nuclear proteins.18 The HPLC profiles of both NB4-MRA1 and UF-1 cells show an altered pattern of high-molecular–weight binding complexes. Nuclear extracts from NB4-MRA1 cells expressing the PML/RARα mutation I410T showed an HPLC profile consistent with ATRA binding to the mutated chimeric protein but at a much decreased level. Specific ATRA-binding activity was not detectable in nuclear extracts prepared from UF-1 cells harboring the PML/RARα mutation R276W, suggesting that this mutation completely prevents binding of the fusion protein to labeled ATRA.
Conformational change analysis of PML/RARα mutants
We predicted that modifications in the ligand binding of fusion proteins would correlate with altered conformations of the receptors. Because proteolytic analysis is a powerful method for analyzing conformational changes within proteins, we performed a limited trypsin digestion of the mutated fusion proteins in the absence and in the presence of ATRA. Distinct fragment patterns were observed for the wild-type and for mutant fusion proteins, especially after ATRA treatment (Figure 3). The distinction centered around a fragment at 32-kDa (lower asterisk in Figure 3) and a fragment at 37-kDa (upper asterisk in Figure 3). In the absence of ATRA, a 32-kDa fragment was more resistant to trypsin digestion. After 1 μM ATRA treatment, the 32-kDa fragment completely disappeared, whereas a fragment of 37-kDa was more resistant to protease treatment of the wild-type PML/RARα. The digestion pattern of the mutant R276W is different from that of the wild-type PML/RARα after ATRA treatment. The identical digestion patterns in the absence and in the presence of ATRA suggest that the mutant R276W fusion protein does not bind the ligand, which is consistent with our ligand-binding analysis (Figure 2B). Analysis of the mutation I410T showed that the trypsin digestion pattern is similar to that of the wild-type PML/RARα. This result indicates that ATRA can still bind to and alter the conformation of the mutated fusion protein LBD, even though binding is shown to be reduced by the HPLC studies (Figure 2B).
ATRA-induced conformational changes in wild-type and I410T mutant PML/RARα fusion proteins but not in R276W mutant.
Limited trypsin digestion analysis of wild-type and mutant PML/RARα proteins. In vitro [35S]-methionine synthesized PML/RARα proteins were incubated without (−) or with (+) 1 μM ATRA, and were subsequently treated with increasing trypsin concentrations (0-25 μg/mL). Digestion products were analyzed by denaturing electrophoresis. The arrows indicate the intact PML/RARα proteins. Lower and upper asterisks indicate resistant fragments at 32 kDa and 37 kDa, respectively.
ATRA-induced conformational changes in wild-type and I410T mutant PML/RARα fusion proteins but not in R276W mutant.
Limited trypsin digestion analysis of wild-type and mutant PML/RARα proteins. In vitro [35S]-methionine synthesized PML/RARα proteins were incubated without (−) or with (+) 1 μM ATRA, and were subsequently treated with increasing trypsin concentrations (0-25 μg/mL). Digestion products were analyzed by denaturing electrophoresis. The arrows indicate the intact PML/RARα proteins. Lower and upper asterisks indicate resistant fragments at 32 kDa and 37 kDa, respectively.
Loss of ligand-dependent association with transcriptional coregulators by mutant PML/RARα fusion proteins
Ligand-dependent activation of nuclear receptors is associated with displacement of corepressors and recruitment of coactivating proteins. Interactions of the PML/RARα mutants with the corepressor SMRT and the coactivator ACTR were tested in gel-shift assays. We first evaluated the binding of mutants to the receptor interacting domain (ID II) of the corepressor SMRT (Figure 4A). In vitro–translated wild-type and mutants PML/RARα bound a radiolabeled DR5 element in the absence of SMRT-IDII. Addition of purified GST-SMRT-IDII shifted the bound complex, as indicated by an arrow in Figure 4A. Binding of GST-SMRT-IDII to each of the mutants was similar to the wild-type PML/RARα in the absence of ATRA. As previously reported, the wild-type PML/RARα fusion protein completely dissociated SMRT-IDII at 1 μM ATRA. In contrast, the fusion proteins harboring the mutations R276W and I410T could not be dissociated from the corepressor SMRT-IDII, even at 10 μM ATRA.
Alteration of the ligand-dependent association with transcriptional coregulators by mutant PML/RARα fusion proteins.
(A) Interaction of SMRT with wild-type and mutant PML/RARα proteins on a DR5 RARE in gel mobility shift assay. In vitro–translated PML/RARα fusion proteins were coincubated with the [32P]-labeled DR5 RARE, along with bacterially expressed GST-SMRT-IDII in the presence of increasing concentrations of ATRA. The position of the complex shifted by SMRT is indicated. (B) Interaction of ACTR with wild-type and mutant PML/RARα proteins on a DR5 RARE in gel mobility shift assay. In vitro–translated PML/RARα fusion proteins were coincubated with the [32P]-labeled DR5 RARE, along with bacterially expressed GST–ACTR-RID in the presence of increasing concentrations of ATRA. The position of the complex shifted by ACTR is indicated. (C) Interaction of mutant PML/RARα fusion proteins with the coactivator DRIP205 by GST pull-down analysis. In vitro–translated 35S-labeled wild-type and mutant PML/RARα fusion proteins were incubated with bacterially expressed and purified GST-DRIP205 in the absence and in the presence of increasing concentrations of ATRA as indicated. GST alone was included as the negative control.
Alteration of the ligand-dependent association with transcriptional coregulators by mutant PML/RARα fusion proteins.
(A) Interaction of SMRT with wild-type and mutant PML/RARα proteins on a DR5 RARE in gel mobility shift assay. In vitro–translated PML/RARα fusion proteins were coincubated with the [32P]-labeled DR5 RARE, along with bacterially expressed GST-SMRT-IDII in the presence of increasing concentrations of ATRA. The position of the complex shifted by SMRT is indicated. (B) Interaction of ACTR with wild-type and mutant PML/RARα proteins on a DR5 RARE in gel mobility shift assay. In vitro–translated PML/RARα fusion proteins were coincubated with the [32P]-labeled DR5 RARE, along with bacterially expressed GST–ACTR-RID in the presence of increasing concentrations of ATRA. The position of the complex shifted by ACTR is indicated. (C) Interaction of mutant PML/RARα fusion proteins with the coactivator DRIP205 by GST pull-down analysis. In vitro–translated 35S-labeled wild-type and mutant PML/RARα fusion proteins were incubated with bacterially expressed and purified GST-DRIP205 in the absence and in the presence of increasing concentrations of ATRA as indicated. GST alone was included as the negative control.
To test whether these mutants of PML/RARα could recruit the coactivator ACTR, the central receptor-interacting domain of ACTR (ACTR-RID) fused to the GST protein was used in a gel-shift study (Figure 4B). In the absence of ATRA, ACTR-RID did not form a complex with the wild-type or mutated fusion proteins. The wild-type PML/RARα–ACTR-RID complex was first noted at 0.01 μM and was maximal at 0.1 μM ATRA. The mutation R276W is fully associated with ACTR-RID only at 10 μM ATRA, which is 100-fold greater than for the wild-type association. PML/RARα harboring the mutation I410T showed only minimal recruitment of ACTR at 10 μM ATRA, indicating that the coactivator needs more than 100-fold ATRA concentrations for its association with this mutated fusion protein.
A distinct coactivator complex has been identified by 2 separate groups for its ligand-dependent interaction to the vitamin D3 receptor (DRIP) or to the thyroid hormone receptor (TRAP).58,59These studies suggest that the DRIP/TRAP complex acts as a ligand-dependent positive transcriptional regulator of various nuclear receptors. We previously showed that RARα and the wild-type PML/RARα chimeric protein interact directly with the DRIP complex.53 To assess whether PML/RARα mutants modify the interaction with the DRIP complex, in vitro–translated [35S]-labeled mutant PML/RARα proteins were allowed to interact with the purified GST-DRIP205 (Figure 4C). Wild-type PML/RARα showed a ligand-dependent interaction with the DRIP205 protein. As we previously reported, the M4 mutant PML/RARα of NB4-MR4 failed to bind DRIP205. In contrast, both mutants R276W and I410T showed an interaction with DRIP205 similar to that of the wild-type PML/RARα, though PML/RARα mutant I410T interacted less with DRIP205 in presence of 0.1 μM ATRA.
TSA cooperates with ATRA to induce transcriptional activity of a βRARE in ATRA-resistant APL cell lines
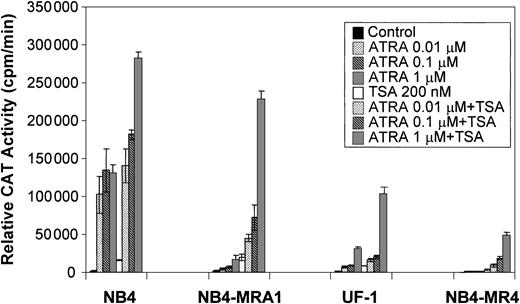
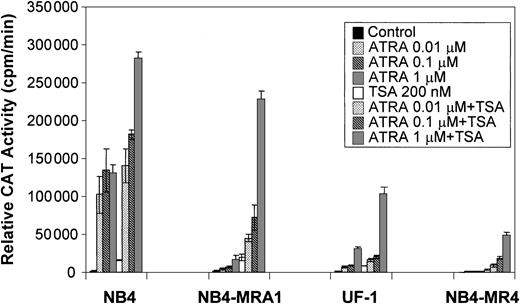
Because a histone deacetylase-dependent transcriptional repression of the ATRA-signaling pathway may underlie the differentiation block of APL, we evaluated the capacity of the HDAC inhibitor, TSA, to modulate the ATRA response of PML/RARα mutants in ATRA-resistant APL cell lines on a retinoid-responsive element. ATRA-sensitive NB4 and ATRA-resistant NB4-MRA1, NB4-MR4, and UF-1 APL cell lines were cotransfected with a tk-CAT reporter driven by a DR5 RARE and a β-Gal expression vector as a control for the efficiency of transfection. As shown in Figure 5, ligand-dependent transcriptional activity of the wild-type PML/RARα chimeric protein present in ATRA-sensitive NB4 cells was increased by concentrations of ATRA from 0.01 to 1 μM, and treatment with ATRA + TSA further increased transactivation. Figure 5 shows that ATRA-resistant NB4-MRA1, UF-1, and NB4-MR4 APL cells have significantly impaired ligand-dependent transcriptional activity. However, TSA cooperates with ATRA to increase transcriptional activity on a DR5-tk-CAT in ATRA-resistant APL cells. The combination of 200 nM TSA with 1 μM ATRA increased the DR5-tk-CAT activity in NB4-MRA1 and UF-1 cells to 160- and 67-fold, respectively, compared with a 160-fold induction in NB4 cells.
TSA cooperates with ATRA to induce transcriptional activity of a βRARE in ATRA-resistant cell lines harboring point mutations in the LBD of PML/RARα.
Relative CAT activity of ATRA-sensitive NB4 and ATRA-resistant NB4-MRA1, NB4-MR4, and UF-1 APL cells without (control) or with indicated concentrations of ATRA or 200 nM TSA is shown. A DR5-tk-CAT reporter was cotransfected with a β-galactosidase expression vector into the cells for normalization of the transfection. Each data point represents the mean of at least 3 independent transfections, and bars denote SD.
TSA cooperates with ATRA to induce transcriptional activity of a βRARE in ATRA-resistant cell lines harboring point mutations in the LBD of PML/RARα.
Relative CAT activity of ATRA-sensitive NB4 and ATRA-resistant NB4-MRA1, NB4-MR4, and UF-1 APL cells without (control) or with indicated concentrations of ATRA or 200 nM TSA is shown. A DR5-tk-CAT reporter was cotransfected with a β-galactosidase expression vector into the cells for normalization of the transfection. Each data point represents the mean of at least 3 independent transfections, and bars denote SD.
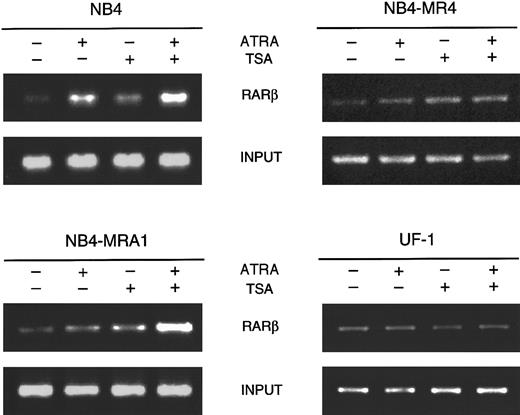
RA synergizes with the HDAC inhibitor TSA to induceRARβ gene expression only in NB4-MRA1 ATRA-resistant cells
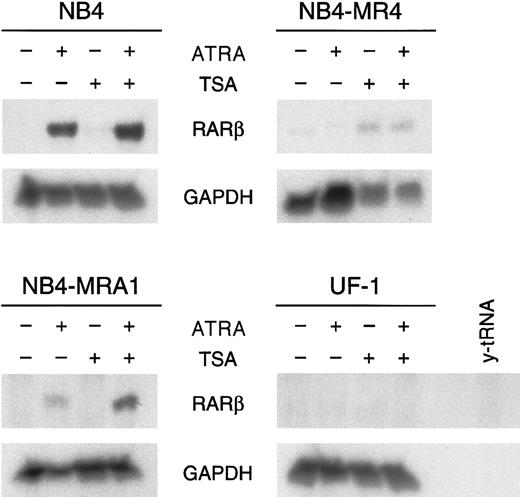
Given that these in vitro transcriptional activity data show that the PML/RARα mutants can respond to HDAC inhibitors, we next evaluated the capacity of ATRA and TSA to activate transcription of an endogenous ATRA target gene. RARβ is a direct ATRA target whose induction has been implicated in several tumor cell models in which retinoids inhibit growth and induce differentiation.12 We investigated the effects of ATRA alone and in combination with TSA on the expression ofRARβ in ATRA-sensitive NB4 and ATRA-resistant NB4-MRA1, NB4-MR4, and UF-1 cells by ribonuclease protection analysis (Figure6). No constitutive expression ofRARβ transcript in ATRA-sensitive or -resistant APL cell lines was observed. RARβ mRNA levels were markedly increased by the induction of differentiation of NB4 cells by ATRA. However, ATRA was unable to induce RARβ expression in ATRA-resistant NB4-MR4 and UF-1 cell lines, though a slight induction was detected in NB4-MRA1 cells. We found no modification of the levels of RARβ mRNA transcript by TSA alone in ATRA-sensitive NB4 and ATRA-resistant NB4-MRA1 and UF-1 APL cells and a very weak induction independent of ATRA treatment in NB4-MR4 cells. Treatment with ATRA and TSA produced a significant up-regulation of the levels ofRARβ mRNA in NB4 and, interestingly, in the ATRA-resistant NB4-MRA1 APL cells, but it failed to stimulate RARβ in UF-1 and NB4-MR4 cells.
HDAC inhibitor TSA potentiates ATRA induction ofRARβ mRNA expression in the ATRA-resistant NB4-MRA1 APL cells harboring the mutant I410T PML/RARα.
Ribonuclease protection analysis for RARβ expression of 50 μg total RNA isolated from ATRA-sensitive NB4 and ATRA-resistant NB4-MRA1, NB4-MR4, and UF-1 APL cells untreated or treated with 1 μM ATRA or 200 nM TSA and the combination of ATRA with TSA for 28 hours. The RNase-protected band corresponding to RARβ is identified, andGAPDH expression was used as a quantitative loading control. Yeast tRNA (y-tRNA) was included as a negative control to verify the specificity of the probes.
HDAC inhibitor TSA potentiates ATRA induction ofRARβ mRNA expression in the ATRA-resistant NB4-MRA1 APL cells harboring the mutant I410T PML/RARα.
Ribonuclease protection analysis for RARβ expression of 50 μg total RNA isolated from ATRA-sensitive NB4 and ATRA-resistant NB4-MRA1, NB4-MR4, and UF-1 APL cells untreated or treated with 1 μM ATRA or 200 nM TSA and the combination of ATRA with TSA for 28 hours. The RNase-protected band corresponding to RARβ is identified, andGAPDH expression was used as a quantitative loading control. Yeast tRNA (y-tRNA) was included as a negative control to verify the specificity of the probes.
HDAC inhibition with TSA potentiates ATRA-induced histone hyperacetylation on chromatin of RARβ promoter in ATRA-resistant NB4-MRA1 cells
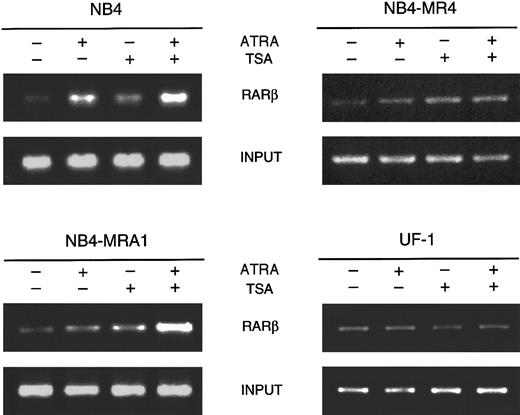
To directly assess whether the effects of TSA and ATRA induction on RARβ gene expression correlate with modifications of histone acetylation, we analyzed histone H4 acetylation at the receptor target gene RARβ by the chromatin immunoprecipitation (ChIP) assay. As shown in Figure 7, ChIP analysis with antibodies to acetylated H4 revealed that ATRA treatment of ATRA-sensitive NB4 cells induces the acetylation level on H4. The addition of TSA increases acetylation of histone H4 in the absence and in the presence of ATRA. Treatment of ATRA-resistant NB4-MR4 cells with TSA showed a very weak increase in the acetylation of H4 onRARβ, which was not changed by the presence of ATRA. Treatment of ATRA-resistant UF-1 APL cells with ATRA and TSA used as single or combined agents demonstrated no modification of the level of acetylation of histone H4 on RARβ. However, in NB4-MRA1 ATRA-resistant cells, consistent with our data on RARβ RNA expression, the combination of ATRA and TSA strongly induced the acetylation of H4 on the RARβ promoter.
HDAC inhibition by TSA intensifies ATRA-induced histone H4 hyperacetylation on chromatin of RARβ promoter in ATRA-resistant NB4-MRA1 APL cells.
Cross-linked chromatin preparations of ATRA-sensitive NB4 and ATRA-resistant NB4-MRA1, NB4-MR4, and UF-1 APL cells untreated or treated with 1 μM ATRA or 200 nM TSA, and the combination of ATRA with TSA were immunoprecipitated with an antibody against acetylated tails of histone H4. Immunoprecipitated and input material was analyzed by PCR using primers corresponding to the RARE region of theRARβ promoter.
HDAC inhibition by TSA intensifies ATRA-induced histone H4 hyperacetylation on chromatin of RARβ promoter in ATRA-resistant NB4-MRA1 APL cells.
Cross-linked chromatin preparations of ATRA-sensitive NB4 and ATRA-resistant NB4-MRA1, NB4-MR4, and UF-1 APL cells untreated or treated with 1 μM ATRA or 200 nM TSA, and the combination of ATRA with TSA were immunoprecipitated with an antibody against acetylated tails of histone H4. Immunoprecipitated and input material was analyzed by PCR using primers corresponding to the RARE region of theRARβ promoter.
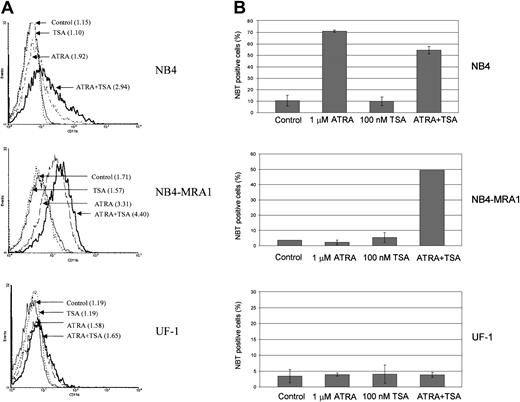
Combined effect of ATRA and TSA on differentiation of the ATRA-resistant NB4-MRA1 APL cells
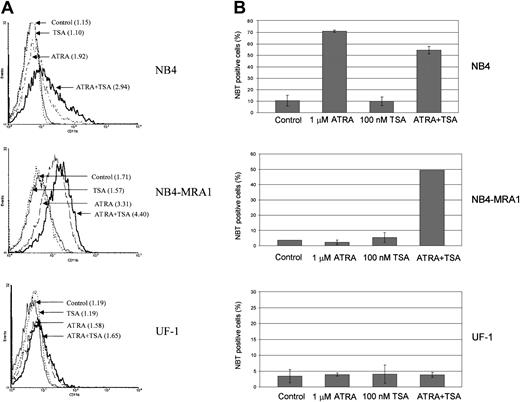
To determine whether the observed effects of ATRA and TSA on transcription and gene expression translate into induction of differentiation, we investigated the extent of differentiation response of the cell lines by CD11b staining and NBT reduction (Figure8). Although ATRA alone induces a moderate increase of the early differentiation marker, CD11b, in NB4-MRA1 (Figure 8A), there is no effect of ATRA alone on the terminal differentiation marker, NBT, in either NB4-MRA1 or UF-1 APL cells (Figure 8B). However, the combination of ATRA with TSA causes substantial differentiation in NB4-MRA1, but not in UF-1 APL cells, consistent with our data on transcription and gene expression.
Combined effect of ATRA and TSA on differentiation of the ATRA-resistant NB4-MRA1 APL cells.
(A) Expression of the differentiation marker CD11b by flow cytometry analysis. NB4, NB4-MRA1, and UF-1 APL cells were treated with 25 nM TSA, 1 μM ATRA, and the combination for 3 days. Numbers in parentheses indicate mean channel fluorescence. This experiment is representative of 3 that gave comparable results. (B) NBT reduction analysis of NB4, NB4-MRA1, and UF-1 APL cells treated with the indicated concentrations of agents for 5 days. Each data point represents the mean of 3 independent experiments, and bars denote SD.
Combined effect of ATRA and TSA on differentiation of the ATRA-resistant NB4-MRA1 APL cells.
(A) Expression of the differentiation marker CD11b by flow cytometry analysis. NB4, NB4-MRA1, and UF-1 APL cells were treated with 25 nM TSA, 1 μM ATRA, and the combination for 3 days. Numbers in parentheses indicate mean channel fluorescence. This experiment is representative of 3 that gave comparable results. (B) NBT reduction analysis of NB4, NB4-MRA1, and UF-1 APL cells treated with the indicated concentrations of agents for 5 days. Each data point represents the mean of 3 independent experiments, and bars denote SD.
Discussion
Different groups have identified specific genetic lesions associated with ATRA resistance, in cell lines and in patients. As shown in Figure 1B, several missense mutations in the LBD of the RARα moiety of the fusion gene PML/RARα have now been reported in APL cells, highlighting the role of the fusion protein in mediating the sensitivity and response to ATRA. Rather than being randomly distributed, the mutations cluster mainly in 3 areas, denoted by I, II, and III, corresponding to the amino-terminus (between H1 and H3), the central region (within or between H5 and H6), and the carboxy-terminus (within H11 and H12) of the LBD, respectively. Interestingly, these PML/RARα mutations can be compared to clusters of mutations in thyroid receptor beta (TRβ) associated with the syndrome of resistance to thyroid hormone (RTH).60-62
In this report, we characterize 2 novel missense point mutations in the E-domain of PML/RARα, associated with the development of resistance to ATRA, that cluster in regions II and III described above (Figure 1). The growing list of mutations identified in the LBD of PML/RARα is an indication of the importance of this mechanism for the development of ATRA resistance in APL cells in vitro and in vivo.
The R276W PML/RARα mutation was identified from leukemic cells of a patient in relapse. A spontaneously ATRA-resistant APL cell line, UF-1, established directly from an ATRA-resistant patient with APL, harbors exactly the same point mutation, R276W, in its PML/RARαgene41 as in the APL cells of our ATRA-resistant patient with APL. Importantly, this point mutation has been independently observed in a third ATRA-resistant patient with relapsed APL.43 Very recently, a different mutation at the same position, Arg276Gly, has been identified in 2 different ATRA-resistant patients.44 The identification of mutations of the arginine at position 276 by independent groups in cells of 5 different ATRA-resistant patients suggests that this amino acid is probably very important in the mechanism of ATRA response and, therefore, is a mutational “hot spot” involved in the mechanism of ATRA resistance.
The R276W mutation is centrally located in the LBD of PML/RARα at the end of H5. It has been shown in x-ray structure analysis that this arginine is conserved in the steroid nuclear receptor family and participates in a network of hydrogen bonds and salt bridges anchoring the carboxylate group of the ligand. This positively charged arginine may also serve as an electrostatic field guide for the ligand.63 64 This amino acid substitution, involving a radical shift from the most hydrophilic positively charged Arg to the nonpolar hydrophobic Trp, is likely to be responsible for the disruption of these bonds and the electrostatic field. The mutated PML/RARα chimeric protein of UF-1 cells completely lost its ATRA-binding capacity (Figure 2B). In vitro expression of this mutant PML/RARα exhibited no change in the pattern of resistant fragments in limited proteolytic digestion in the presence of ATRA (Figure 3). Consistent with these results, this R276W mutation severely impaired the ability of the PML/RARα protein to interact in a ligand-dependent manner with coregulators (Figure 4). These ligand-binding, conformational, and protein-interaction analyses are consistent with transient transfection studies in the ATRA-resistant UF-1 cells showing that this R276W mutant PML/RARα is minimally responsive to transcriptional activation by ATRA (Figure 5).
The importance of this amino acid as a target for mediating nuclear hormone resistance is further supported by the analysis of a natural TRβ mutant R320H from a patient with RTH.65 This amino acid corresponds in TRβ to the R276 in RARα. The R320H in TRβ has a reduced ligand-binding affinity and a decreased T3-dependent release of NCoR, and it mediates a weaker transactivation, but these effects are of a lesser degree than those we observed with R276W. Indeed, this difference may be explained by the more conservative amino acid substitution in the TRβ, where a polar positively charged arginine is replaced by an amino acid of the same group. This observation suggests that the type of amino acid substitution may influence interactions with ligands and nuclear coregulators.
NB4-MRA1 is a newly established ATRA-resistant NB4 subclone harboring a novel mutation, I410T, in the LBD of the RARα moiety of PML/RARα. This amino acid is one of 8 residues constituting the H12, which includes the AF-2 domain. H12 appears to be crucial for ATRA binding and transcriptional activity. In the presence of the ligand, the H12 of PML/RARα is repositioned, stabilizing the binding pocket and providing a surface for coactivators to bind and activate transcription.63,64 The isoleucine at position 410 produces an almost entirely apolar interaction by making Van der Waals contact with the beta-ionone ring of the ligand.66 The replacement of the nonpolar isoleucine at position 410 by a polar uncharged threonine considerably decreases the capacity of the fusion protein PML/RARα to bind ATRA (Figure 2B), probably by disrupting this apolar interaction. However, tryptic digestion analyses showed that ATRA can still bind to the mutated PML/RARα and induce conformational changes similar to those observed in the wild-type receptor (Figure 3). Although 2 assays show that the mutant I410T can bind ATRA, it also exhibited a decreased transcriptional response to ATRA (Figure 5), and ATRA regulation of binding to coregulators is completely lost (Figure 4). This suggests that a mutation in the AF-2 domain can have more prominent effects on binding to coregulators than to ligand. Further, observed differences between assays comparing PML/RARα mutations45 46 (Figures 3-5) suggest that no one in vitro assay will fully describe the altered biology of mutated PML/RARα.
Again, there is an interesting correlation with studies of TRβ mutations. Consistent with our data, the L454S TRβ mutant in RTH, which corresponds to the amino acid I410 in RARα, has been shown to bind ligand, whereas ligand-dependent release of the corepressor NCoR and the ability of the ligand to activate transcription are impaired.67 The natural occurrence of mutations in the amino acids of TRβ corresponding to each of the mutated residues of PML/RARα described here and the observed similarities in phenotype support their potential importance in mediating thyroid and ATRA resistance. Further comparison of mutations identified in ATRA-resistant APL with those found in TRβ with RTH syndrome may increase our understanding of the functional roles of these mutants.
Mutants R276W and I410T showed an interaction with DRIP205 similar to that of the wild-type PML/RARα protein, in contrast to the M4 mutant of NB4-MR4, which failed to interact with this coactivator (Figure 4C). Dilworth and Chambon68 propose a model for ATRA-induced initiation of transcription where DRIP/TRAP is directly involved in the activation pathway by RAR/RXR heterodimers, at a step that occurs after displacement of p160 proteins from AF-2. Consistent with this model, the interaction of R276W and I410T mutants with DRIP205 may explain the higher ATRA-induced transcriptional activity of UF-1 and NB4-MRA1 than of NB4-MR4 cells (Figure 5). Thus, our data suggest that the interaction of retinoid receptors with DRIP/TRAP may provide an additional regulatory step that can be involved in ATRA resistance. The data further support our hypothesis that ATRA resistance may be mediated by mutations that target different elements of the complex mechanism of ligand-dependent transcriptional activation by nuclear receptors.
The increased association that we found between mutants PML/RARα and the corepressor SMRT suggests an abnormal interaction with HDACs. An additional objective of this study was to determine whether increasing histone acetylation by the use of an HDAC inhibitor counteracts the effects of corepressors, thus allowing DNA to take a transcriptionally active conformation that would facilitate ATRA-induced gene expression. In the presence of TSA, ATRA induced transcription of a RARE almost to a normal level in NB4-MRA1 cells, and to a lesser extent in UF-1 and NB4-MR4 cells (Figure 5). Consistent with these data, we found that TSA synergizes with ATRA to induce the RARβ gene in the ATRA-resistant NB4-MRA1 cells but is less effective in the other ATRA-resistant APL cells, NB4-MR4 and UF-1 (Figure 6). ChIP analyses showed that HDAC inhibition with TSA potentiates ATRA-induced histone H4 hyperacetylation on chromatin of the RARβ promoter in NB4-MRA1 cells, but it has no effect in NB4-MR4 and UF-1 cells (Figure7), consistent with the data on RARβ gene expression. The biologic significance of these results is supported by our data that the HDAC inhibitor plus ATRA combination induces substantial differentiation of NB4-MRA1 but not of UF-1 (Figure 8). This suggests that ATRA plus HDAC inhibitor combination might not be effective in patients with this commonly occurring mutation.
We hypothesize that the increased ability of an HDAC inhibitor to synergize with ATRA in the I410T mutation is attributed to the location and the role of this residue in the AF-2 domain. The AF-2 residues (410-IQEML-414, underlined residues) resemble the sequence of the ϕXXϕϕ or theLXXLL motifs of corepressors or coactivators, respectively.69-72 In RARα, I410, M413, and L414 play the same role as the first, second, and third leucine residues, respectively, of the coactivator nuclear receptor box peptide and make similar interactions.73 74 Thus, we propose that the mutant I410T impairs the transcriptional properties of the PML/RARα by modifying the physical association of the fusion protein with corepressors and coactivators.
The data presented here support the hypothesis that resistance to ATRA in APL, both in vitro and in vivo, can be mediated by mutations in critical residues of the LBD of the PML/RARα fusion protein. An abnormal binding of nuclear coregulators of transcription to these mutated fusion proteins leads to transcriptional repression of PML/RARα target genes. Combined treatment with ATRA and HDAC inhibitors may represent a novel approach to reactivate target genes by blocking HDAC activity abnormally recruited to the promoter by the mutated fusion proteins, thus permitting ATRA-induced association of coactivators and differentiation. Interestingly, one patient with ATRA-refractory APL received combined treatment with ATRA and an HDAC inhibitor, leading to a durable complete remission.50Unfortunately, 4 other patients with ATRA-refractory APL failed to respond to this “transcription therapy.”75 We propose that differences in the molecular mechanism of resistance may determine whether a specific transcriptional therapy will be effective. For example, if a mutation of PML/RARα precludes effective interactions with coactivators, then inhibition of HDAC activity mediated by corepressors will not help the induction of ATRA-target gene and differentiation. Indeed, our study demonstrates that the ability of an HDAC inhibitor to restore ATRA sensitivity in resistant APL cells may depend on their specific molecular defects. Thus, the variety ofPML/RARα mutations arising in ATRA-resistant patients provides a model to study how patients with relapsed APL can differ in response to transcription therapy with HDAC inhibitors. Further investigation of the structural alterations caused by differentPML/RARα mutations and their interactions with modulators of histone acetylation, such as HDAC inhibitors, may provide new therapeutic strategies to overcome resistance to ATRA in APL.
This strategy may have more general application because oncoproteins that interfere with transcription, often by HDAC-mediated repression, have been discovered in an increasing number of malignancies.76-78 Indeed, recent studies have shown that HDAC inhibitors, in combination with ATRA or other differentiation-inducing agents, have activity against other leukemias. These include acute myeloid leukemia characterized by the chimeric protein AML1-ETO,48,78 APL harboring the PLZF/RARα fusion protein,49,79 and leukemia with the MLL–CBP chimeric protein.80 A recent report has shown a clinical response to an HDAC inhibitor in patients with T-cell lymphoma.81 Furthermore, a synergistic antitumor activity in prostate cancer cells and in neuroblastoma xenografts in vivo has been observed using a combination therapy of HDAC inhibitors with vitamin D382 and ATRA,83respectively. Thus, studies in APL may provide a model for the development of novel therapeutics in other malignancies.
We thank Dr Masahiro Kizaki for his generous gift of the ATRA-resistant APL cell line UF-1, Dr William W. Lamph for the RARβ and GAPDH probes, Dr Sylvie Mader for the DR5-tk-CAT response element, and Dr Ronald M. Evans for the GST fusions of the receptor interaction domain of SMRT and ACTR (GST-SMRT-IDII and GST-ACTR-ID).
Prepublished online as Blood First Edition Paper, June 7, 2002; DOI 10.1182/blood-2002-02-0614.
Supported by grants from the Canadian Institutes of Health Research and the Associazione Italiana per la Ricerca sul Cancro. S.C. is supported by Fonds de la Recherche en Santé du Québec and Israel Cancer Research Fund fellowships. A.B. is supported by a postdoctoral contract of the Università di Roma “La Sapienza.” W.H.M. is a Scientist of the Canadian Institutes of Health Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Wilson H. Miller Jr, Lady Davis Institute for Medical Research, Sir Mortimer B. Davis Jewish General Hospital, 3755 Chemin de la Côte Ste-Catherine, Montreal, Quebec, Canada H3T 1E2; e-mail: wmiller@ldi.jgh.mcgill.ca.


![Fig. 2. Characterization of cell lines expressing novel PML/RARα mutants and their ATRA-binding activity. / (A) ATRA induces PML/RARα protein degradation in the ATRA-sensitive NB4 cells, but not in the ATRA-resistant NB4-MRA1 APL cells. Cells were treated with 1 μM ATRA for 24 hours, and 50 μg of nuclear extracts were used in Western blot analysis for the expression of PML/RARα protein. NB4 and NB4-MRA1 expressed a long PML/RARα (L) isoform (110 kDa). The lower panel shows laminin B expression to confirm protein loading. (B) Specific HPLC ATRA-binding profiles of nuclear extracts from NB4 cells (●) compared with those from the ATRA-resistant NB4-MRA1 (○) and UF-1 (▾) APL cells. Nuclear extracts were incubated with 10 nM [3H]-ATRA (●, ○, ▾) or with [3H]-ATRA in the presence of 200-fold excess of unlabeled ATRA (▿). Extracts were subjected to HPLC analysis using a 6 HR 10/30 size exclusion column.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/7/10.1182_blood-2002-02-0614/3/m_h81923183002.jpeg?Expires=1769201368&Signature=f5kMxuxesPee8pPO9v5bNk9sjSk4P01EOQwGoE1TVirEKz6Qs0ilA36KOMRxqEcDpuSOA0-cmgd-ZwqLRcxw0aJkbnd~XjylonAOCS2kgLzvU7B5s4gknwCdGrltalJ8P~ALn-tqDyK0TEfilgc6sl0qETdD6eS9oUNyBNs2yqBszR6fY7Tslf0rwx6GRTmVtpEOZF05pvOY3WSKIFnYKdk0iDIlekVVGQ339PTvSRr5RpROTYNM7lpUX7vjYbmshfDgNBS6Lmq1JozA9QxcFkqu27WBvsZ5FznuraHpZ5yEk5pQTOk90XSqI6heoN3FuFgUK7BzBZ70HFKCp5eLUQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. ATRA-induced conformational changes in wild-type and I410T mutant PML/RARα fusion proteins but not in R276W mutant. / Limited trypsin digestion analysis of wild-type and mutant PML/RARα proteins. In vitro [35S]-methionine synthesized PML/RARα proteins were incubated without (−) or with (+) 1 μM ATRA, and were subsequently treated with increasing trypsin concentrations (0-25 μg/mL). Digestion products were analyzed by denaturing electrophoresis. The arrows indicate the intact PML/RARα proteins. Lower and upper asterisks indicate resistant fragments at 32 kDa and 37 kDa, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/7/10.1182_blood-2002-02-0614/3/m_h81923183003.jpeg?Expires=1769201368&Signature=H1pZkDTziONezl4SDEjRqzw-9~XV4fh1R8kCSCwyzngCmHh7~U6Ckiqes-SzjSPYG3Houe2ZjgsHlOic05wjt1MRBSwtANY7oVDk4N4epQ0WpwMk-OV1J5ocVUIEhpd1pY90dm5RVYsg~jY~lei4jTiwbLJsxvo4HwYYenLz0UUntdYxqOKHKm3Y1Ed1fqxy-Lg0ewJEZmDJBoerCylYKsEL1s3a39OMmdtQJg-HhEQoAeV5eS6C2uMfB~Cz9ZZNb7GeaGC7qPUHfK8ayoNjqHcSNuwFjRjiW3E16Z0--UBfm8rWCsv8vgl8G-VKHGBi2s7449DT6Ut8yODta2ovvA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Alteration of the ligand-dependent association with transcriptional coregulators by mutant PML/RARα fusion proteins. / (A) Interaction of SMRT with wild-type and mutant PML/RARα proteins on a DR5 RARE in gel mobility shift assay. In vitro–translated PML/RARα fusion proteins were coincubated with the [32P]-labeled DR5 RARE, along with bacterially expressed GST-SMRT-IDII in the presence of increasing concentrations of ATRA. The position of the complex shifted by SMRT is indicated. (B) Interaction of ACTR with wild-type and mutant PML/RARα proteins on a DR5 RARE in gel mobility shift assay. In vitro–translated PML/RARα fusion proteins were coincubated with the [32P]-labeled DR5 RARE, along with bacterially expressed GST–ACTR-RID in the presence of increasing concentrations of ATRA. The position of the complex shifted by ACTR is indicated. (C) Interaction of mutant PML/RARα fusion proteins with the coactivator DRIP205 by GST pull-down analysis. In vitro–translated 35S-labeled wild-type and mutant PML/RARα fusion proteins were incubated with bacterially expressed and purified GST-DRIP205 in the absence and in the presence of increasing concentrations of ATRA as indicated. GST alone was included as the negative control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/7/10.1182_blood-2002-02-0614/3/m_h81923183004.jpeg?Expires=1769201368&Signature=STVeUrGK1Y8VbSw9Uqp8y4Pl77UrELDnfcetW3LzkQ9EnGmpTZgXPenn808Ttjc6Q~~qi2Rk0yV6hIZlq-rKZOEO0voGQVzsIYD8RXTR1TWYZl6QgO6UJ4e93s3wnDA5aPDE4Mevq~fcnTxD-ZR~mjoy1anJjD7QlgGB8dX3x4kqRKWOYcfLvk09aU3YKLqh9vJvrAWk11Ir7RsGBKYGtn~3aLya2Ueaeecp0ay5i8Nq38yTKToeZt4QIDfsJmokUATtL~C4x1uHeCkqUWPgSnLFe1n4zn8gDxzBpEO4H2leDnq1GfFo7DTN1lreAFlc3VSoQMyAnK2dvsjjEG6P-A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 2. Characterization of cell lines expressing novel PML/RARα mutants and their ATRA-binding activity. / (A) ATRA induces PML/RARα protein degradation in the ATRA-sensitive NB4 cells, but not in the ATRA-resistant NB4-MRA1 APL cells. Cells were treated with 1 μM ATRA for 24 hours, and 50 μg of nuclear extracts were used in Western blot analysis for the expression of PML/RARα protein. NB4 and NB4-MRA1 expressed a long PML/RARα (L) isoform (110 kDa). The lower panel shows laminin B expression to confirm protein loading. (B) Specific HPLC ATRA-binding profiles of nuclear extracts from NB4 cells (●) compared with those from the ATRA-resistant NB4-MRA1 (○) and UF-1 (▾) APL cells. Nuclear extracts were incubated with 10 nM [3H]-ATRA (●, ○, ▾) or with [3H]-ATRA in the presence of 200-fold excess of unlabeled ATRA (▿). Extracts were subjected to HPLC analysis using a 6 HR 10/30 size exclusion column.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/7/10.1182_blood-2002-02-0614/3/m_h81923183002.jpeg?Expires=1769645357&Signature=Cufv0ppTXnEuaSBTnRi~CYrX~55C3gmduQUGM7NB7q3vAoMcsVN89v2q7Re3XhfzdQ5071ns6czF2biGBjYSuLrqd~CSPDmJzm~YGeRvTZ7VEuD784w4ptkvYb-7iF6SOztJ5a3Q4AvGbL0KlwLbiQVoLBHVr~hZ4mTlu68xX4pCyC1BUWe7eWVZXo54IB1fWz~YoV36jhm6VbF2GlRWBpmtjb~vCBnMH~s3Pj15RGxs6YW2nGENJZBAF5VZOqmFeh~56sW~Bcc6PvBqR6lyZT6oCJC9qopv0mRal4kEkVdoA79fw7yPL2tIYOUQlDWgyOYfKIrD4p9xC37Z82WmZQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. ATRA-induced conformational changes in wild-type and I410T mutant PML/RARα fusion proteins but not in R276W mutant. / Limited trypsin digestion analysis of wild-type and mutant PML/RARα proteins. In vitro [35S]-methionine synthesized PML/RARα proteins were incubated without (−) or with (+) 1 μM ATRA, and were subsequently treated with increasing trypsin concentrations (0-25 μg/mL). Digestion products were analyzed by denaturing electrophoresis. The arrows indicate the intact PML/RARα proteins. Lower and upper asterisks indicate resistant fragments at 32 kDa and 37 kDa, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/7/10.1182_blood-2002-02-0614/3/m_h81923183003.jpeg?Expires=1769645357&Signature=AzBU1HjEfmBF-d8uWvk9fVdEuFXLNsSej4t~QKfEB9Oh~aXewnQpeY10pETlR0B6Hie5IxuQrA0g1zCxwXKAgqK-46tFdCgnLgAqv8pnLKOkH1tjtNuMcIPlMHVwkt5ker0NdTHX3PtUHd9sk5BudmT9Nt8Py2tNpk1NilwZ8H6FUJQtRiWBxs2ofZijjspuBbrGUivT74hVacZvEhCLdoMuJF9W4XDL16ppby1KZA0yjkVVLCURXAh50MkvwNuMjL7QefTpAmFFrbPu7lVgsZR1qEazQ9LITa5Lt0AVauJbdjgpx-WJjxIpYNq8qfOwKFufVhozrvCFbOFh3ZLgcw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Alteration of the ligand-dependent association with transcriptional coregulators by mutant PML/RARα fusion proteins. / (A) Interaction of SMRT with wild-type and mutant PML/RARα proteins on a DR5 RARE in gel mobility shift assay. In vitro–translated PML/RARα fusion proteins were coincubated with the [32P]-labeled DR5 RARE, along with bacterially expressed GST-SMRT-IDII in the presence of increasing concentrations of ATRA. The position of the complex shifted by SMRT is indicated. (B) Interaction of ACTR with wild-type and mutant PML/RARα proteins on a DR5 RARE in gel mobility shift assay. In vitro–translated PML/RARα fusion proteins were coincubated with the [32P]-labeled DR5 RARE, along with bacterially expressed GST–ACTR-RID in the presence of increasing concentrations of ATRA. The position of the complex shifted by ACTR is indicated. (C) Interaction of mutant PML/RARα fusion proteins with the coactivator DRIP205 by GST pull-down analysis. In vitro–translated 35S-labeled wild-type and mutant PML/RARα fusion proteins were incubated with bacterially expressed and purified GST-DRIP205 in the absence and in the presence of increasing concentrations of ATRA as indicated. GST alone was included as the negative control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/7/10.1182_blood-2002-02-0614/3/m_h81923183004.jpeg?Expires=1769645357&Signature=oreZ3fd68kd71Nt0Z7zwwNUzZmzWtnAUYOVch1GF24izRYdHA0sPMckjbXdbYx8eZgP2dVW5sbqSCDP0IOa338dqH-4715I3Qjv0zwcmzG-l32AILH5tuOxr1mPv0d8HbvyoRCr0tEFeI7aX4Z~6ipk9qK2m6ywisiPJHovS82AvnhBaq6uG-czkTDiqm7Lu9auvL7S8WqgoTWakc7arP4cZkXNeQ~NlDTh7FLNujSGcIKJLnD1yWTE4F1aAAJ2OQ85tdqaFJR7G2kRsNp~8OtMX77lHi2I2s1f~Wogl1Jqjxki2E1SmSkynWPsLCzOI5lRlA3A2fvLvo703YVB0Qw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)