Abstract
Activating length mutations in the juxtamembrane (JM) domain of the FLT3 gene (FLT3-LM) and mutations in the catalytic domain (FLT3D835/836) of this receptor tyrosine kinase represent the most frequent genetic alterations in acute myeloid leukemia (AML). Here, we describe a 6-bp insertion in the activation loop of FLT3 between codons 840 and 841 of FLT3 (FLT3-840GS) in 2 unrelated patients with AML. Screening for other activating mutations of FLT3, KIT, and NRASshowed no further genetic alterations in patients carrying the FLT3-840GS. In functional analyses we could show that this mutant is hyperphosphorylated on tyrosine and confers interleukin 3–independent growth to Ba/F3 cells, which can be inhibited by a specific FLT3 protein tyrosine kinase (PTK) inhibitor. Our results show for the first time that in addition to known mutations in the JM and the catalytic domain, further activating length mutations exist in theFLT3 gene.
Introduction
Recent advances in genetics have shown that not only chromosome abnormalities but also molecular alterations are useful to characterize and subclassify acute myeloid leukemia (AML). For example, a partial tandem duplication within the mixed lineage leukemia gene (MLL-PTD) has been shown to define a subgroup of AML patients with unfavorable clinical outcome.1
Activation of the FLT3 receptor tyrosine kinase (RTK) due to length mutations (LMs) in the juxtamembrane domain (JM) are found in 20% to 25% of AML cases.2-7 In addition, point mutations and deletions of codons 835/836 of FLT3, which are located in the activation loop (A-loop) of the protein tyrosine kinase (PTK) domain, have been described in about 7% of all AML cases.8 9
FLT3 has a high homology to other class III RTKs and plays an important role in the proliferation of hematopoietic progenitors and AML blast cells.10-12 Functionally, mutations in the JM region and also in the A-loop result in a constitutively active FLT3 kinase, which confers interleukin 3 (IL-3)–independent growth in Ba/F3 cells and activates the mitogen-activated protein kinase (MAPK) and signal transducer and activator of transcription 5 (STAT5) pathways.13-15 In the murine bone marrow transplantation model and in transgenic animals, active FLT3 mutants induce a myeloproliferative disease, but not acute leukemia, which shows that probably additional genetic alterations are necessary for the clinical phenotype in FLT3- LM+ AML.16 17
In the present study, we describe a recurrent activating LM in the A-loop of FLT3 in 2 patients with AML.
Study design
Patient samples
Bone marrow samples from 359 adult patients with newly diagnosed and untreated AML were analyzed for the new FLT3 mutation. The studies abide by the rules of the local internal review board and the tenets of the revised Helsinki protocol.
Patient 1, a 69-year-old woman, was diagnosed as having AML FAB M0/M1. The peripheral leukocyte count was 29 700/μL and the percentage of blasts in the bone marrow was 90%. The cytogenetic analyses showed the karyotype: 46,XX,der(7)t(1;7)(q25;q22)[13]/46,XX[2]. After treatment with a high-dose cytosine arabinoside (Ara-C)–based regimen, the patient died 4 weeks later with no response to therapy.
Patient 2, a 76-year-old man, was diagnosed with AML FAB M6 with the following cytogenetic findings: 44,X,−Y,der(7)t(7;8)(q22;?),t(8;11)(q11;q11),+11,der(12)t(12;18)(p12;?),−17,−18[2]/46,XY[11]. His peripheral leukocyte count was 1570/μL and the percentage of bone marrow blasts was 20%. Chemotherapy with low-dose Ara-C and idarubicin resulted in a complete remission, but an early relapse occurred and the patient died 3 months after diagnosis.
Cytogenetics
PCR
Isolation of mononucleated leukemic cells, mRNA/DNA extraction, reverse transcription (RT), and polymerase chain reaction (PCR) was performed as described previously.5 RT-PCR was carried out using primers from exon 20 of the FLT3 gene: exon 20 forward: 5′-CCGCCAGGAACGTGCTTG-3′ (corresponding to nucleotides 2395-2412 of XM39994.1) and exon 20 reverse: 5′-ATGCCAGGGTAAGGATTCACACC-3′ (corresponding to nucleotides 2632-2610 of XM39994.1) resulting in a 238-bp amplification product. Genomic PCR was carried out with primers 20F: 5′-CCAGGAACGTGCTTGTCA-3′ (corresponding to nucleotides 76543-76526 of AL591024.14) and 20R: 5′-TCAAAAATGCACCACAGTGAG-3′ (corresponding to nuleotides 76349-76369 of AL591024.14) generating a 195-bp amplification product in normal controls.
Screening for further mutations
Antibodies
The following antibodies were used: anti-FLT3/flk2 (S18, sc-480, Santa Cruz, Heidelberg, Germany), anti-PY (PY99, Santa Cruz).
In vitro mutagenesis
The FLT3-840GS mutation, found in clinical samples, was introduced into the full-length human FLT3 wild-type cDNA using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions.
Cell culture and protein analyses
Cell proliferation of Ba/F3 cells, transient transfection of BOSC23 cells, transduction of Ba/F3 cells, flow cytometric analysis of FLT3 expression, immunoprecipitation, and Western blot analysis were performed as described previously.21
Results and discussion
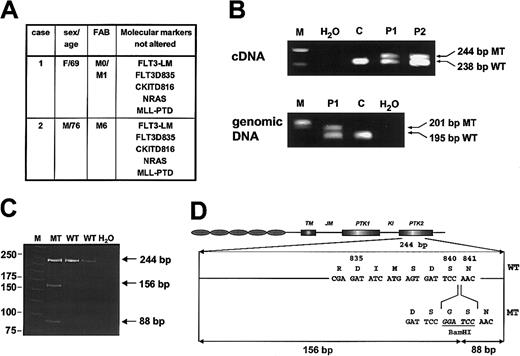
We used a PCR-based method to screen for mutations of codons D835/836 in patients with AML. After gel electrophoresis of the amplification products from exon 20 we observed an additional PCR fragment that was slightly larger than the PCR product of the wild-type allele in 2 patients (Figure 1B). RT-PCR was repeated more than 10 times with different aliquots of RNA as well as cDNA and always gave the same results. Sequencing revealed an insertion of 6 nucleotides between the codons S840 and N841 just 5 amino acids downstream of D835. These nucleotides generated aBamHI restriction site that was confirmed byBamHI digestion and gel electrophoresis (Figure 1C). The mutation resulted in the insertion of a glycine and a serine between amino acids 840 and 841 of FLT3 (Figure 1D). For patient 1 a DNA sample was available and the mutation could be shown also at the genomic level by PCR, confirming the presence of the mutation (Figure1B). The respective mutation was not detected in a further 357 unselected patients with AML. Additional screening for frequent genetic mutations in these 2 patients showed no further activating mutations ofFLT3, NRAS, KIT, or MLL (Figure1A).
Detection of the FLT3-LM in exon 20 in AML.
(A) Clinical data and alterations of other molecular markers in the 2 patients carrying the FLT3-840GS mutation. FLT3-LM indicates length mutations in the JM region of FLT3; FLT3D835, mutations in codons 835/836; KITD816, point mutations in codon 816; NRAS, activating point mutations in codons 12, 13, and 61; MLL-PTD, MLL-partial tandem duplication. (B) Detection of the LM in exon 20 after conventional agarose gel electrophoresis of PCR products from cDNA and gDNA. M indicates molecular weight standard; H2O, water control; C, patient without exon 20 mutation; P1 and P2, patients 1 and 2, respectively, with the length mutation in exon 20. (C) Confirmation of the mutation by BamHI digestion after polyacrylamide gel electrophoresis. M indicates molecular weight standard; MT, patient with mutation; WT, 2 patients without mutation; H2O, water control. (D) Schematic presentation of the FLT3-840GS mutation.
Detection of the FLT3-LM in exon 20 in AML.
(A) Clinical data and alterations of other molecular markers in the 2 patients carrying the FLT3-840GS mutation. FLT3-LM indicates length mutations in the JM region of FLT3; FLT3D835, mutations in codons 835/836; KITD816, point mutations in codon 816; NRAS, activating point mutations in codons 12, 13, and 61; MLL-PTD, MLL-partial tandem duplication. (B) Detection of the LM in exon 20 after conventional agarose gel electrophoresis of PCR products from cDNA and gDNA. M indicates molecular weight standard; H2O, water control; C, patient without exon 20 mutation; P1 and P2, patients 1 and 2, respectively, with the length mutation in exon 20. (C) Confirmation of the mutation by BamHI digestion after polyacrylamide gel electrophoresis. M indicates molecular weight standard; MT, patient with mutation; WT, 2 patients without mutation; H2O, water control. (D) Schematic presentation of the FLT3-840GS mutation.
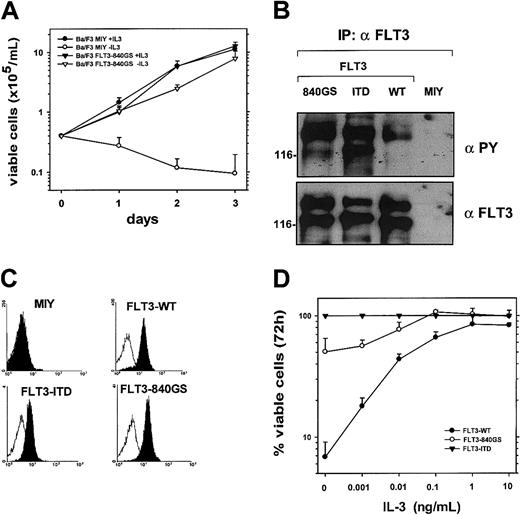
We generated FLT3-840GS– and FLT3–internal tandem duplication (ITD)–expressing Ba/F3 cells. In Western blot analyses, the FLT3-840GS and the FLT3ITD receptors were hyperphosphorylated compared to the FLT3WT receptor (Figure 2B). As shown in Figure 2A, the FLT3-840GS– as well as FLT3ITD-expressing Ba/F3 cells grew factor independently, whereas mock and FLT3WT-expressing cells were unable to proliferate in the absence of IL-3.
The FLT3-840GS mutant is hyperphosphorylated and induces IL-3–independent growth in Ba/F3 cells.
(A) Ba/F3 cells transduced with either pMSCV-EYFP-IRES-FLT3-840GS or empty vector (pMSCV-EYFP-IRES, mock) were grown in the absence or presence of IL-3 as indicated. Results represent means ± SD of 3 independent experiments. (B) Lysates from Ba/F3 cells were immunoprecipitated with αFLT3 antibody and analyzed by Western blot using an antiphosphotyrosine (αPY) antibody, stripped, and reblotted with αFLT3 antibody. (C) Ba/F3 cells expressing either FLT3WT, FLT3ITD, FLT3-840GS, or empty vector (MIY) were incubated with a mouse isotype-matched control antibody (open histograms) or CD135-phycoerythrin (filled histograms) antibody. Viable cells were analyzed using a FACSCalibur flow cytometer. (D) Ba/F3 cells expressing FLT3WT, FLT3ITD, or FLT3-840GS were grown for 72 hours in the absence or presence of different concentrations of IL-3 as indicated. The growth of FLT3ITD-expressing Ba/F3 cells at 72 hours was defined as 100%. Results represent means ± SEM of 3 independent experiments.
The FLT3-840GS mutant is hyperphosphorylated and induces IL-3–independent growth in Ba/F3 cells.
(A) Ba/F3 cells transduced with either pMSCV-EYFP-IRES-FLT3-840GS or empty vector (pMSCV-EYFP-IRES, mock) were grown in the absence or presence of IL-3 as indicated. Results represent means ± SD of 3 independent experiments. (B) Lysates from Ba/F3 cells were immunoprecipitated with αFLT3 antibody and analyzed by Western blot using an antiphosphotyrosine (αPY) antibody, stripped, and reblotted with αFLT3 antibody. (C) Ba/F3 cells expressing either FLT3WT, FLT3ITD, FLT3-840GS, or empty vector (MIY) were incubated with a mouse isotype-matched control antibody (open histograms) or CD135-phycoerythrin (filled histograms) antibody. Viable cells were analyzed using a FACSCalibur flow cytometer. (D) Ba/F3 cells expressing FLT3WT, FLT3ITD, or FLT3-840GS were grown for 72 hours in the absence or presence of different concentrations of IL-3 as indicated. The growth of FLT3ITD-expressing Ba/F3 cells at 72 hours was defined as 100%. Results represent means ± SEM of 3 independent experiments.
The detailed analysis of the growth characteristics of FLT3-840GS Ba/F3 cells showed a significantly slower growth rate compared to the FLT3ITD-expressing cells. For maximal proliferation, the FLT3-840GS–expressing cells required additional IL-3 at concentrations as low as 0.1 ng/mL (Figure 2D).
We next asked whether the FLT3-840GS mutant is sensitive to the growth inhibitory activity of SU5614, a FLT3 PTK inhibitor (K. S., unpublished data, October 2001). Our results clearly show that SU5614 induced a growth inhibition of the FLT3-840GS–transduced as well as of the FLT3ITD-transduced Ba/F3 lines in the absence, but not in the presence of IL-3 (data not shown).
The data presented here clearly indicate that activating mutations in the FLT3 gene do not only occur within the juxtamembrane domain and in codons D835/836 but also in other positions of the A-loop of the catalytic domain. The A-loop represents a hot-spot region for activating mutations in class III RTKs, which have been described for KIT (D816) and FLT3 (D835/836).8 These mutations induce a conformational change of the A-loop, which results in the opening of the catalytic pocket and a constitutive active kinase activity. Although no structural data on the catalytic domain of FLT3 are available that would allow the detailed structure-function analysis, the close proximity to the amino acids D835/836 suggests a similar mechanism of kinase activation by the FLT3-840GS mutant.
Activating mutations of the FLT3 gene provide an essential antiapoptotic and pro-proliferative signal in primary AML cells and cell lines.13,14 22 Our results clearly indicate that the FLT3840-GS mutant is hyperphosphorylated on tyrosine residues and induces IL-3–independent growth in Ba/F3 cells. These in vitro data underline the pathophysiologic role of this mutant for the leukemic phenotype in patients with AML.
Although the FLT3-840GS is probably a rare mutation, it clearly shows that activating mutations other than the FLT3-LM in the JM domain and FLT3D835/836 in the kinase domain exist in AML. These findings are of significant clinical importance because activating FLT3mutations could represent selective and specific molecular target structures for therapeutic strategies using PTK inhibitors in AML.23 24
The authors thank Inga Böll for help with mutational screening, cloning, and sequencing of constructs and Karin Schmieja, Ruth Schwab, Gudrun Mellert, Claudia Tschulik, and Tanja Skorupinski for excellent technical assistance. We greatly acknowledge all of more than 300 physicians for sending the patient samples.
Prepublished online as Blood First Edition Paper, July 5, 2002; DOI 10.1182/blood-2002-03-0953.
Supported by a grant from the Deutsche Krebshilfe (10-1562).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Karsten Spiekermann or Susanne Schnittger, Department of Medicine III, University Hospital Grosshadern, Ludwig-Maximilians University, Marchioninistraße 15, 81377 Munich, Germany; e-mail: spiekermann@gsf.de orsusanne.schnittger@med3.med.uni-muenchen.de.