Abstract
Light chain–associated amyloidosis (AL) is a plasma cell dyscrasia in which the secreted monoclonal immunoglobulin (Ig) light chains form amyloid fibrils. There is considerable heterogeneity in clinical presentation, and prognosis of the disease relates to the severity of organ dysfunction induced by amyloid deposits. The mechanisms by which the amyloid fibrils are deposited as well as the predilection for specific organ sites have not been clearly elucidated. This study characterizes the repertoire of immunoglobulin light chain variable genes used by the clonal B cell in AL amyloid patients, and the association of light chain variable region (VL) genes with clinical presentation and outcome is assessed in 58 (32 λ and 26 κ) patients. A preferential use of VL germ-line genes was noted for both AL κ and λ patients. There was a significant correlation between the use of the Vλ VI germ-line donor, 6a, and renal involvement as well as the Vλ III gene, 3r, with soft-tissue AL. The use of a biased VL gene repertoire also correlated with clinical outcome, revealing important trends for predicting prognosis. The use of Vλ II germ-line genes was associated with cardiac amyloidosis and affected survival adversely. The presence of multiple myeloma also correlated with a poor prognosis. The presence of renal disease, on the other hand, was associated with improved survival. Therefore, identification of the clonal VL gene in AL has important implications in determining clinical outcome.
Introduction
Light chain–associated amyloidosis (AL) is an uncommon plasma cell disorder characterized by the transformation of immunoglobulin (Ig) light chains into amyloid, which can be deposited as fibrils, throughout the body.1 The clinical manifestations of the disease are caused by the presence of amyloid in the major organs of the body.2 The average age at diagnosis is 65 years3 with a marginal male predominance.3Once the disease becomes symptomatic, it can become rapidly progressive and is usually fatal, with a median survival of 12 to 18 months and less than 5% surviving 10 years or longer.4-6Only 22% of AL patients have more than 20% plasma cells in their bone marrow (BM), whereas most (∼ 80%) have less than 20% plasmacytosis.7 The degree of plasma cell clonality and the number of plasma cells have been inversely correlated with survival.8 There is also a preponderance of λ versus κ free light chains (FLCs) in AL (3:1) compared with healthy individuals (λ/κ = 1:2) or other plasma cell neoplasias, such as multiple myeloma (MM) (λ/κ = 1:2).9
It has been recently shown that VL gene usage may dictate which organs are preferentially affected by amyloid deposition,10suggesting an organ tropism for the light chains. Also, there appears to be a bias in germ-line donor use in AL patients10-12compared with the healthy population. Because there is a predominance of λ light chains in AL, most studies have either excluded or, at the most, included very few AL κ patients. In addition, there is a paucity of information on the association of VL gene usage with clinical presentation and survival in AL. This study attempts to address some of these issues by identifying the clonal VL gene in equal numbers of λ and κ patients and correlating light chain
Patients, materials, and methods
Patients
From a large AL database of 1962 patients seen at the Mayo Clinic 60 AL patients (32 λ and 28 κ) were chosen randomly. Frozen BM samples on these patients were stored in the Dysproteinemia Cell Bank. There were 40 male and 20 female patients in the group. The study was carried out under an institutional review board (IRB)–approved protocol and followed the Helsinki guidelines for research of human subjects.
Classification of clinical presentation
The extent of amyloid-related disease, organ impairment, and other clinical features were documented in the medical records. The presence of amyloid was visualized by Congo red staining with green birefringence under polarized light of fat aspirates, rectal or BM biopsies. For some patients where tissue biopsies were obtained, amyloid was detected by Congo red staining of the tissue. In addition to the Congo red staining, BM as well as tissue biopsies were stained with κ or λ light chain–specific antibodies. The presence of a monoclonal protein in serum or urine along with the detection of a monoclonal population in the BM formed the basis of classification of patients as having AL. These methods, however, do not allow a completely accurate determination of the nature of the protein forming the amyloid deposits, and it would be difficult to rule out the possibility of a sporadic serum A amyloid or transthyretin amyloid having similar clinical features being misclassified as AL amyloid. Mass spectrometric analysis of the amyloid deposits using material obtained from paraffin-embedded biopsy specimens13 permits accurate identification of the precursor protein, and we are in the process of implementing this method in the clinical setting. Dominant organ involvement was defined by using established criteria.1 14-16 Patients were classified according to their clinical presentation as having predominantly cardiac, renal, hepatic, neurologic, or pulmonary involvement. However, all organs affected by amyloid deposition were recorded for analysis. Gastrointestinal (GI) disease, myopathy, and vascular or asymptomatic AL were categorized in a separate group as “other.” Patients who had more than one major organ involved with amyloid deposition were listed under the clinically most prominent organ involved, which led to the formation of 3 categories—cardiac, renal, and “other.” Dominant hepatic, neurologic, and pulmonary AL were added to the latter category, to simplify analysis.
Cardiac AL was diagnosed if there was classic echocardiographic evidence of infiltrative cardiomyopathy.2 17 The presence of albuminuria higher than 1000 mg/24 h or a creatinine clearance of lower than 10 mL/min were considered indicative of renal disease. Neurologic disease was defined on the basis of orthostatic hypotension, GI motility abnormalities, or sensorimotor peripheral neuropathy. The sole presence of carpal tunnel syndrome was not considered indicative of a neurologic manifestation of AL. A diagnosis of hepatic involvement was made if a liver biopsy showed evidence of amyloid infiltration or the serum alkaline phosphatase level was 1 1/2 times greater than the normal and there was palpable hepatomegaly. The term “soft-tissue AL” has been used in this study to refer to patients who had evidence of macroglossia, skin involvement, or carpal tunnel syndrome without vital organ dysfunction due to amyloid deposits.
Identification of VL gene usage
BM aspirates were collected from patients with biopsy-proven AL who were seen in the Hematology Division at the Mayo Clinic. The marrow preparations were layered on Ficoll Paque (Amersham Biosciences, Uppsala, Sweden) to remove red blood cells, and the mononuclear cells were washed and frozen. Total RNA was extracted from approximately 107 BM cells using Trizol (Gibco-BRL, Gaithersburg, MD). The RNA obtained was used for the preparation of cDNA, using the Superscript reverse transcriptase (RT) kit (Life Technologies, Grand Island, NY).11,18 The cDNA was subjected to polymerase chain reaction (PCR) amplification using 5′ primers specific for the FR1 region of 7 Vλ (Vλ I, Vλ II/V, Vλ III, Vλ IVa, Vλ IVb, Vλ VI) and 4 Vκ (Vκ Ι/ΙΙΙ, Vκ ΙΙ Vκ ΙV) families, along with the 3′ constant region primer—Cκ or a pan-Cλ primer.19 Each patient sample was subjected to multiple cycles of amplification and PCR products were sequenced from 3 to 4 independent reactions. Each set of experiments was run with a positive λ and κ control (kindly provided by Drs R. L. Comenzo and Y. Zhang, Memorial Sloan Kettering Cancer Center, New York, NY). The appropriate band was cut and cleaned using centrifugal filter units (Millipore, Billerica, Spain). The PCR product was sequenced with forward and reverse primers at the Mayo Molecular Biology Sequencing Core Facility. The clonal VL gene used was determined if one gene was clearly overrepresented in each patient and the CDR3 region was identical for 3 to 7 separate products. Additional PCR amplification was done using the appropriate 5′ leader region primers (VL) along with the individual 3′ CL primers, because of the possibility of small errors in the sequence being introduced by the FR1 primers. VL genes with the correctly sequenced FR1 regions were evaluated for their homology to germ-line donor sequences, which also provided information on somatic hypermutation (R.S.A. et al, manuscript in preparation).
Classification of VL genes
The sequences obtained by the methods described in the previous paragraph were analyzed using the National Center for Biotechnology Information (NCBI) BLAST (Basic Local Alignment Search Tool) program and DNAPLOT (Hans-Helmar Althaus, Werner Muller, Koln, Germany), and assignation of germ-line donors was done using a database of rearranged immunoglobulin genes, V-BASE, based on comparison of sequences for maximum nucleotide homology. The homology analysis was undertaken for complete VL gene sequences with the exception of the codons that form the VJ junction and the last FR segment (FR4). All sequences obtained were submitted to GenBank (AF 490906-490969).
Statistical analysis
Patient selection.
Patients were selected based on a stratified sampling approach. The intention of this strategy was to select approximately equal numbers of both κ and λ patients for these analyses, recognizing the natural predominance of λ patients in this population. To ensure there was no selection bias, key clinical and biochemical parameters were compared between this sample and the remainder of AL patients in the overall database. In particular, Kaplan-Meier methods and log-rank tests were used to compare survival differences between patients in our research sample and in the overall database, and χ2 tests were used to compare sex, treatment status (treatment prior to being seen at Mayo Clinic), and light chain (κ vs λ) diagnosis. To compare the quantitative variables of hemoglobin, calcium, serum creatinine, β2-microglobulin, serum albumin, and plasma cell labeling index, 2-sample t tests were used.
Correlation of light chain V gene use with clinical presentation.
The correlation between VL gene usage and clinical presentation in AL patients was assessed by 2-sample t tests and Wilcoxon rank sum tests for continuous variables and χ2 and Fisher exact tests for relationships between categoric variables.
Survival analyses
Overall survival was defined as the time from the date of diagnosis of amyloidosis to the date of death due to any cause and was estimated using the Kaplan-Meier method. Survival differences for various factors were assessed univariately using the 2-tailed log-rank test. The criteria for evaluating survival in this cohort of AL patients included light chain diagnosis, VL gene usage, dominant clinical presentation, and sex. The survival analysis was not statistically significant in all the categories assessed, which was likely due to the small sample size. Survival analysis was also done for the larger group of patients in the overall database. These analyses had sufficient power to detect meaningful differences, but only on the limited number of variables of interest collected in the database.
All P values represented are 2-sided, and statistical significance was declared at a P value of .05 or less. Because these analyses were exploratory in nature, multiple comparison corrections were not used.
Results
Selection of AL patients
The 60 AL patients were selected from a larger group of 1962 patients representing the AL patients seen at the Mayo Clinic over a period of 19 years (1982-2001). To ensure the lack of a selection bias in choosing these patients the subset group was compared with the overall group for certain key clinical and biochemical criteria. The subset was not significantly different from the overall group except for light chain usage (P = .001), which was a reflection of the deliberate selection of patients with AL κ amyloid. In this study, equal numbers of κ and λ patients were chosen, as there is little information available on VL usage and clinical presentation in AL κ patients in the body of published literature. The subset and overall groups were compared for clinical presentation, frequency of MM, and overall survival (Tables1-3).
None of the standard biochemical markers were significantly different between the groups indicating that the subset group, which was randomly chosen, was comparable with the total database. The subset was also evaluated for the presence of serum and urine M-protein by immunofixation with 51 and 54 of 60 patients, respectively, being positive. The patients were also stratified on the basis of the monoclonal protein they expressed either in serum or urine. Most patients (47%; 28/60) had less than 10% plasma cells in their BM, whereas 32% (19/60) had between 10% to 20% plasma cells and 22% (13/60) had more than 20% plasmacytosis.
Clinical features of the subset group
More than half the patients (63%; 38/60) had 2 or more organs affected by severe amyloid deposition, whereas 35% (21/60) had 1 organ involved. Only one patient had no evidence of systemic disease with only BM staining positive for light chain amyloid. The dominant clinical manifestation at diagnosis was renal disease with 43% of the patients affected. The remainder of the patients presented with cardiac (30%) or soft-tissue, hepatic, neurologic, pulmonary, or GI amyloid. The breakdown of the 60 patients into clinical categories revealed that approximately half the patients had renal (50%) and/or cardiac involvement (52%). The findings were consistent with MM in 18% (11/60) of patients. BM plasmacytosis higher than 10% and anemia, hypercalcemia, or lytic bone disease (9/11) was present. Pulmonary disease was present in only 2 AL patients.
Identification of the clonal VL gene in AL
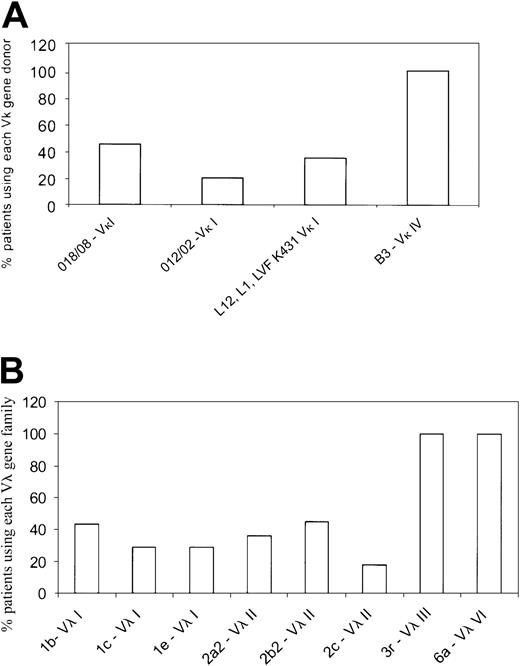
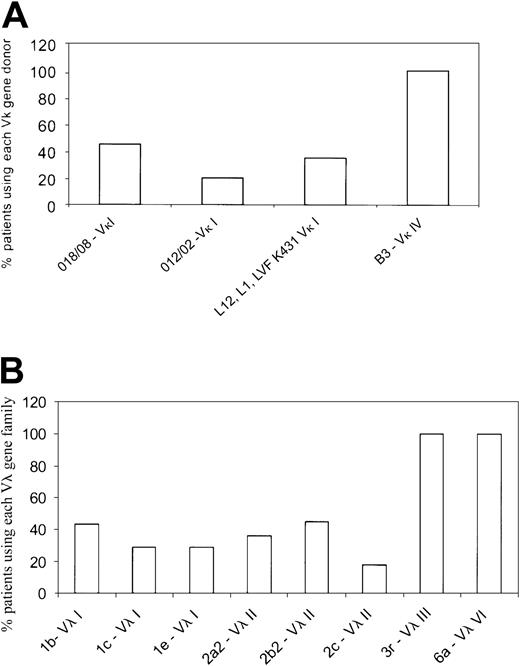
The dominant Vκ or Vλ gene family was identified as described earlier by RT-PCR and DNA sequencing using Vκ and Vλ family–specific primers. Of the 28 κ patients selected, we were unable to identify a dominant clonal Vκ family in 2 patients. This may have been because there were too few plasma cells in the sample, and consequently these patients were excluded from further analyses. Most κ patients used the Vκ I family (77%; 20/26), whereas the remainder (19%; 5/26) used the Vκ IV gene family. Only 1 patient of the 26 used the Vκ II family. In the κ group, the Vκ I patients primarily used 2 germ-line genes,O18/O8 (9 patients) and O12/O2 (4 patients) (Figure 1A; Table4). The Vκ I donor, LVFK 431, which has been reported as being commonly used in AL κ patients10 was used in only one patient in our cohort. The remaining 6 Vκ I patients used eitherL12 or L1 germ-line genes. The single Vκ II patient used the L5 gene, whereas the Vκ IV patients (5 patients) all used the B3 germ-line donor (Figure 1A; Table4). In the κ patients, the Jκ 2gene was used by most patients followed by Jκ4 (data not shown).
Germ-line donors used by the AL light chain variable gene families.
(A) Germ-line gene donors for the Vκ gene families. The Vκ category was represented by 4 Vκ families—Vκ I, Vκ II, Vκ III, and Vκ IV. The Vκ I family was the most widely used and represented by several germ-line genes, with O18/O8and O12/O2 being the most common. Only a single Vκ II patient was present of the 26 patients. A single donor B3represented the Vκ IV gene family. Each germ-line gene is represented as percent of patients using the specific gene within a given Vκ family. (B) The germ-line donors used by λ AL patients are shown. The Vλ I family was represented by germ-line genes1b, 1c, and 1e, whereas the Vλ II family primarily used 2a2, 2b2, and2c. Vλ III and Vλ VI were each represented as a single dominant germ-line donor, 3r and 6a, respectively. The Vλ II group was used most frequently in the λ category. Each germ-line gene is represented as percent of patients using the specific gene within a given Vλ family.
Germ-line donors used by the AL light chain variable gene families.
(A) Germ-line gene donors for the Vκ gene families. The Vκ category was represented by 4 Vκ families—Vκ I, Vκ II, Vκ III, and Vκ IV. The Vκ I family was the most widely used and represented by several germ-line genes, with O18/O8and O12/O2 being the most common. Only a single Vκ II patient was present of the 26 patients. A single donor B3represented the Vκ IV gene family. Each germ-line gene is represented as percent of patients using the specific gene within a given Vκ family. (B) The germ-line donors used by λ AL patients are shown. The Vλ I family was represented by germ-line genes1b, 1c, and 1e, whereas the Vλ II family primarily used 2a2, 2b2, and2c. Vλ III and Vλ VI were each represented as a single dominant germ-line donor, 3r and 6a, respectively. The Vλ II group was used most frequently in the λ category. Each germ-line gene is represented as percent of patients using the specific gene within a given Vλ family.
In the λ group, 34% (11/32) used the Vλ II family; 28% (9/32), the Vλ III; 22% (7/32), Vλ I; and 16% (5/32), the Vλ VI family. The Vλ I patients (7/32) used 3 different germ-line donors:1b (3 patients), 1c (2 patients), and1e (2 patients) (Figure 1B; Table 4); similarly, the Vλ II patients (11/32) used 2a2 (4 patients), 2b2 (5 patients), and 2c (2 patients). The Vλ III and Vλ VI families were represented by a single germ-line donor each—3r (9 patients) and 6a (5 patients) (Figure1B; Table 4). For the J regions in the λ patients, theJλ 3b andJλ 2/3a genes were used most frequently (data not shown).
Association between VL gene usage and symptomatic organ involvement
A natural corollary to the observation of a bias in VL gene usage leading to AL was the correlation with the clinical presentation, in terms of organs affected by amyloid deposition. There was a sex bias with 18 of the 26 κ and 21 of the 32 λ patients being male. On correlating organ involvement with VL gene usage, certain trends were observed. There were 3 major clinical categories identified—cardiac, renal, and “other,” comprising GI, pulmonary, hepatic, neurologic (autonomic), and soft-tissue (macroglossia, skin involvement, and carpal tunnel syndrome) AL.
Patients in each VL family subgroup were assessed for clinical presentation. Vλ II patients (73%; 8/11) showed dominant cardiac AL, whereas all Vλ VI (100%; 5/5) patients had renal disease (P = .045) (Table5). In addition, 2 of the 5 Vλ VI patients had early cardiac manifestations of amyloid-induced dysfunction. Among the Vλ III patients 22% (2/9) had cardiac AL, whereas 33% (3/9) had renal amyloid. The remaining 45% (4/9) of Vλ III patients were represented in the “other” category, and this difference was significant when compared with 14% of patients who did not use this Vλ family (P = .04) (Table 5). Additionally, MM was present in 18% (2/11) of Vλ II patients and 36% (4/11) of Vλ III patients.
Among the κ group, patients with Vκ I gene usage had primarily soft-tissue (macroglossia, skin involvement, and carpal tunnel syndrome) disease (70%), but 50% of the patients also had cardiac or renal AL (Table 5). The solitary Vκ II patient had cardiac and GI disease and MM. The Vκ IV gene family was associated with mainly cardiac (60%; 3/5) and renal (60%; 3/5) amyloid (Table 5).
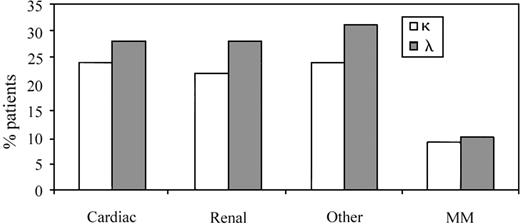
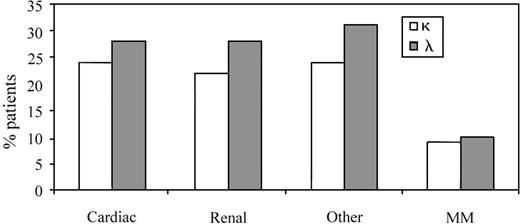
The data shown in Table 5 outline the proportion of patients in each specific light chain variable gene family (VL family or group) with defined organ involvement. However, it was also useful to assess clinical presentations in the λ and κ subgroups as a reflection of the total number of patients tested to determine if there was a bias in clinical presentation based on light chain usage per se (n = 58; clonal V gene identified in 32 λ and 26 κ patients) (Figure2). The κ group of patients, 24% (14/58) and 22% (13/58) presented with dominant cardiac and renal AL, respectively. The remaining κ patients (24%; 14/58) fell into the “other” category, defined previously. MM was present in 9% (5/58) of κ patients (Figure 2). In the λ group, a similar profile was observed with 28% of patients (16/58) having either cardiac or renal tissue as the primary organ site of amyloid deposition, whereas 31% (18/58) had either GI, pulmonary, neurologic, or other manifestations of disease. MM was seen in 10% (6/58) of AL λ patients (Figure 2).
The representation of organ involvement in AL κ and λ patients as a reflection of the total subset of 58 patients.
Cardiac, renal, and “other” clinical presentations were more dominant in the λ group, in keeping with λ bias in AL. The incidence of MM was comparable in the 2 groups.
The representation of organ involvement in AL κ and λ patients as a reflection of the total subset of 58 patients.
Cardiac, renal, and “other” clinical presentations were more dominant in the λ group, in keeping with λ bias in AL. The incidence of MM was comparable in the 2 groups.
Correlation of urine or serum M-protein with clinical presentation
Urine M-protein levels were significantly lower in patients with dominant cardiac presentation compared with those who did not present with cardiac disease (medians: 0.0915 and 0.4375 mg/dL, respectively;P = .0145). As expected however, urine M-protein levels were significantly higher in patients with dominant renal presentation than in those without primary kidney amyloid manifestation (medians: 0.5825 and 0.0605 mg/dL, respectively, P = .0004). In fact, patients who had any form of renal amyloid deposition, even if it was not the primary organ affected, had significantly higher urine M-protein levels than those who had no kidney disease (medians: 0.4700 and 0.0560 mg/dL, respectively; P = .0022). Interestingly, there was a significant difference between serum M protein levels in κ and λ patients (medians: 0.50 and 0.90 g/dL, respectively;P = .03) (data not shown). However, this difference may merely be a reflection of reduced renal clearance because λ light chains preferentially exist as dimers and, in some instances, can form even larger aggregates.20 The correlation between serum and urine M-protein with VL germ-line genes was analyzed and did not reveal any statistically significant findings (data not shown).
Correlation of organ involvement with overall survival
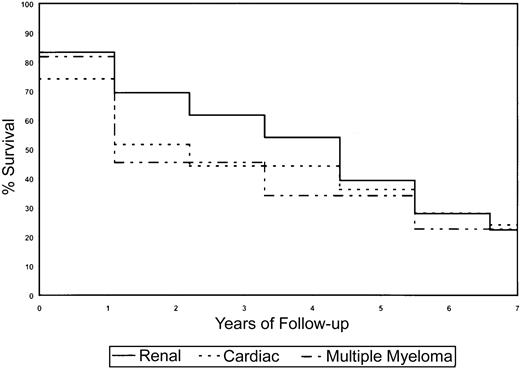
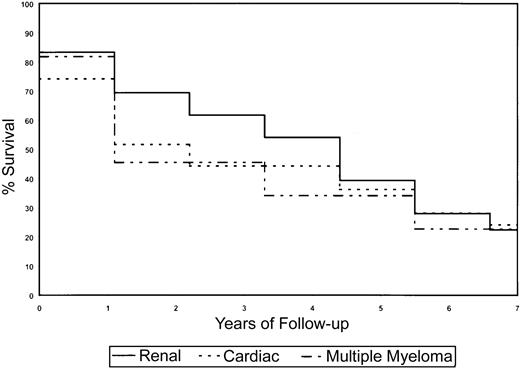
Survival analyses were done using log-rank statistics and Kaplan-Meier estimation methods to determine if sex, clinical presentation (organs affected by amyloid deposition), and Vκ or Vλ gene usage affected survival of AL patients. Because of the small numbers of patients in the various subcategories, the power to detect statistically significant differences was low, particularly with VL germ-line gene use and survival; however distinct trends were observed that may be of clinical relevance. In general, for the survival analyses on the subset of patients, we had at least 80% power to detect significant differences in survival if one group was at 3 times greater risk of death than another, assuming that the breakdown of risk groups was 20% to 80%. The power increased with more balanced numbers in the subgroups. Depending on the proportionality of patients with a marker of interest (eg, Vλ VI), as many as 100 to 150 patients would be required for at least 80% power to detect a 2-fold risk for one group with the marker of interest versus those without it. If the proportion of those with the marker is 35% to 50%, then it would be anticipated that 100 patients would be sufficient; otherwise, if the proportion is 20% to 35%, then at least 150 patients would be required. The use of κ or λ light chains did not significantly affect clinical outcome in the subset of AL patients (Figure3A). Despite the small numbers of patients using each germ-line VL gene, it was observed that patients using the Vκ IV gene family, which in our study was associated with cardiac AL, showed a reduction in overall survival (Figure 3B). The Vλ II family also associated with cardiac AL showed a similar decrease in survival (Figure 3C). There was evidence to suggest that use of the Vλ VI germ-line gene had a favorable prognosis as the majority of these patients had renal disease (Figure 3C). The Vλ I family of genes also correlated with stable disease and enhanced survival. Sex influenced survival, with females living longer than males (4.3 years vs 1.4 years,P = .02). Also, by univariate analysis, the presence of cardiac (1.9 years vs 3.0 years, P = .87) and renal (4.3 years vs 1.1 years, P = .44) involvement affected survival (Figure4). Patients who had coexisting AL amyloidosis with MM had a poorer prognosis, with a median survival of 1.1 years (versus 2.92 years, P = .81) (Figure 4).
Kaplan-Meier survival curves for the AL subset.
Panel A shows a comparison of the overall survival between the κ (thick line) and λ (thin line) groups within the selected subset. Panel B shows survival based on Vκ gene use (Vκ I [solid line] and Vκ IV [dashed line] gene families). The single Vκ II patient has not been represented, and because of the small numbers of patients in the subgroups, survival for individual germ-line genes has not been shown. The Vκ IV gene, which is associated mainly with cardiac and renal amyloidosis, showed reduced survival. Panel C shows survival analysis for the Vλ gene families. Vλ II patients present predominantly with cardiac infiltration and have reduced survival, whereas Vλ VI patients have comparatively better survival curves. The clinical categories documented are not mutually exclusive.
Kaplan-Meier survival curves for the AL subset.
Panel A shows a comparison of the overall survival between the κ (thick line) and λ (thin line) groups within the selected subset. Panel B shows survival based on Vκ gene use (Vκ I [solid line] and Vκ IV [dashed line] gene families). The single Vκ II patient has not been represented, and because of the small numbers of patients in the subgroups, survival for individual germ-line genes has not been shown. The Vκ IV gene, which is associated mainly with cardiac and renal amyloidosis, showed reduced survival. Panel C shows survival analysis for the Vλ gene families. Vλ II patients present predominantly with cardiac infiltration and have reduced survival, whereas Vλ VI patients have comparatively better survival curves. The clinical categories documented are not mutually exclusive.
Kaplan-Meier survival curves for the AL subset and clinical presentation.
The presence of MM and cardiac amyloidosis is negatively associated with survival, whereas renal AL improves survival.
Kaplan-Meier survival curves for the AL subset and clinical presentation.
The presence of MM and cardiac amyloidosis is negatively associated with survival, whereas renal AL improves survival.
As the Mayo AL database had a larger number of patients with the relevant clinical information, survival analyses were done on this group of patients using 3 major clinical presentations as a read-out. The presence of cardiac (0.9 years; P = < .0001), neurologic (1.4 years; P = .013), and renal (2.5 years;P = < .0001) disease significantly affected clinical outcome (data not shown). However, the use of either κ or λ light chain did not impact survival.
One of the most notable findings in our study was the strong correlation of the Vλ III and Vλ VI germ-line donors, 3r and 6a, respectively, with the presence of amyloidogenic light chains. There have been 2 previously published studies10 12 that have identified clonal VL genes in AL patients. However, our study additionally evaluated the correlation between VL gene usage, clinical presentation, and survival, which was not a focus for the previous studies. Although there were minor differences with regard to the proportion of patients represented in the various Vκ and Vλ families, between our study and the published studies (Table6), the data were concordant, in that most of the same germ-line genes were used in all 3 studies.
Discussion
The primary objective of this study was to analyze the immunoglobulin light chain variable gene (VL) repertoire used by the clonal B cell in AL κ and λ amyloidosis patients (n = 60) and correlate VL usage with clinical presentation (ie, organs affected by amyloid deposition leading to symptomatic organ dysfunction). In addition, we evaluated overall survival of patients with specific organ involvement to understand the clinical course of disease in this subset of AL patients. The clonal VL gene was identified by molecular analysis of BM samples in 58 of the original 60 AL patients (26 κ and 32 λ) using an RT-PCR–based assay with primers for the individual light chain variable gene families. DNA sequencing of several clones from each patient permitted identification of the dominant clonal population in the patient BM.
Clonal analysis of the most commonly used light chain V gene families in the 58 patients studied revealed a predominance of Vλ II (34% of the AL λ patients) and VκI (77% of the AL κ patients) family germ-line genes (Figure 1; Table 4). The 2 organs most frequently affected by amyloid deposition were the heart and kidney, with fewer patients having hepatic, neurologic, and GI infiltration. There were 18% of patients who had AL amyloid due to MM, which has been reported to increase the clonal burden10 and suggested as an independent risk factor for cardiac complications, although there is insufficient evidence to prove this theory. An early case report21 indicated that hepatomegaly was seen in an AL patient using the Vκ IV germ-line gene, and it was suggested that this clinical feature may be associated primarily with AL κ patients. However, a more exhaustive clinical analysis of hepatic AL amyloid in 2 studies of 77 AL patients22 and 80 AL patients23 revealed that the λ light chain bias in AL (λ/κ = 3:1) held true as well in the case of hepatic light chain amyloid deposition. These studies22 23 suggest therefore that the presence of an enlarged liver does not correlate uniquely with the presence of κ light chain amyloid deposits.
To test our hypothesis and accurately evaluate correlations between light chain V gene use leading to AL and the organ specificity of amyloid deposition, it was important to determine first whether individual germ-line genes were overrepresented in the AL population. The frequency of use in the germ-line repertoire has been characterized in the circulating peripheral B-cell pool in healthy individuals24-27; however, this does not permit comparisons with the BM B-cell population in AL patients. More recently, however, the germ-line genes used by polyclonal B cells in normal BM has been described.12 From the normal BM analysis12 it appears that difference between the normal and AL B-cell repertoire lies not so much in the V gene family but in the specific germ-line donor used.12 There is considerable evidence suggesting a predominance in the use of the Vλ VI gene, 6a, in AL,28-30 although this gene is relatively rarely used in the healthy population. Data from a study done in AL patients showed that 37% of λ free light chains were derived from the 6a gene,30 confirming the propensity for amyloid deposition when this particular light chain is clonally expanded. There is more recent data, however, that suggest that light chains from other Vλ and Vκ gene families can also be amyloidogenic.12,31 Our data show that the Vλ I and Vλ II families used 3 to 4 different germ-line genes (1b, 1c, 1e,2a2, 2b2, 2c), whereas the Vλ III and the Vλ VI families used a single gene, 3r and 6a, respectively, (Figure 1; Table 4) a finding that has also been reported in other studies.10,12 Only 7% to 8% of normal B cells rearrange the 1c and 3r gene, whereas less than 5% rearrange the 6a gene.26 27
In the κ subgroup, the VκI patients showed dominant use of the common AL-associated O18/O8 gene,10 whereas the VκIV AL patients used the germ-line donor, B3. Interestingly, Perfetti et al32 have reported that there is an overrepresentation of the VκIV gene, B3, in patients with light chain deposition disease, in which there is excess production of free, monoclonal light chains with formation of amorphous aggregates, usually in the kidney.33,34
It is possible to speculate that there are a variety of factors that may account for the skewing of the VL genes used in AL, including events in the selection of the primary antibody repertoire. It has been shown that the early B-cell transcription factors, E2A andEBF, when expressed ectopically along with the recombination activating gene in nonlymphoid cells can induce rearrangement of both Ig heavy and light chain genes.35 The various gene families are interspersed throughout the Vκ and Vλ gene loci, but particular families and individual genes showed high levels of recombination after the ectopic expression of the early B-cell transcription factors, whereas other families in the same locus were not induced to rearrange.36 In light of this information, there could be at least 2 possible and not necessarily mutually exclusive explanations for a bias in VL gene use leading to AL. The first would suggest a dysregulation of the early B-cell–specific transcription factors that result in a change in accessibility of certain V gene families to recombination events, which is augmented by clonal expansion of cells expressing these rearranged genes. The second possibility relates to repertoire selection, with a skewing in V genes used, mediated either by the diversity-generating mechanisms alluded to earlier or by antigen.
The phenomenon of organ tropism has already been alluded to in a recent study on light chain variable gene use in AL amyloidosis, with a correlation suggested between V gene receptor specificity and the presence of amyloid in various organs.10 The data obtained in the present study support our initial hypothesis by delineating important trends with regard to light chain gene use and organ involvement; however as the numbers of patients in the individual gene groups were small, the data did not achieve statistical significance in all instances. There was a strong association between the use of the Vλ II family genes and dominant cardiac disease (Table 5). Likewise, all patients using the Vλ VI gene, 6a, had a renal amyloid-associated clinical presentation, similar to previously published data.29 Patients with Vκ IV gene,B3, had mainly cardiac and/or renal AL, in contrast to patients in the Vκ I group who had primarily nonvital organ amyloid infiltration, such as macroglossia, skin involvement, or carpal tunnel syndrome (Table 5).
One of the unusual features of AL and a key to understanding its pathophysiology is the diversity of amyloid deposition in various organs.9,37 The heterogeneity of amyloid in organs and, therefore, clinical presentation indicate that deposition may be primarily vascular or interstitial, or both. No apparent relationship has been established between the molecular mass of the protein deposit and the organ affected.38 It is important to bear in mind that despite a dominant organ involvement on presentation, usually other organs are affected by amyloid deposition over time.
It is still uncertain whether this selective affinity for tissues is a feature of the primary structure of the light chain inducing a local interaction with the tissue microenvironment or is due to an “antigen recognition” dictated by receptor specificity39 (of these light chains) for certain tissue components. Yet another possibility that may account for the tissue specificity of AL amyloid is the interaction of the amyloidogenic light chains with glycosaminoglycans, in particular heparan sulfate.40 It is interesting to speculate that this specificity may probably come from an association of the amyloid fibrils with tissue-specific isoforms of heparan sulfate. However, it is still unclear whether structural features of the protein, tissue affinity, or plasma cell homing and local production of light chains individually or collectively contributes to the clinical diversity of AL amyloidosis.
The effect of light chain use on survival was relevant only insofar as it correlated with the presence of cardiac and renal amyloidosis. The use of the Vλ II genes, which is associated with cardiac disease, adversely affected clinical outcome, although renal disease (and the use of the Vλ VI gene) correlated with a comparatively better survival. Survival analysis of the larger Mayo AL database with respect to the 3 major clinical categories of cardiac, renal, and neurologic presentations showed significantly adverse correlation with survival. The use of κ or λ light chain per se did not affect clinical outcome.
The data presented in this study showed strong concordance with the 2 previously published reports on light chain variable gene use in AL amyloidosis,10 12 suggesting that across different ethnic AL patient groups, there is a large uniformity in the representation of specific germ-line genes in the clonal light chain amyloid B-cell repertoire. The findings reported in this paper as well as the published studies clearly provide evidence of the importance of identifying the clonal light chain variable gene used in AL patients as a tool that could aid in the medical management of this disease. The strong association between VL germ-line genes and specific organ involvement could be used as an adjunct to the chemical analysis of amyloid deposits to determine the clinical course of disease and therefore the nature of therapeutic intervention. The organ specificity of the VL gene usage also suggests that some light chains may be more pathogenic than others and therefore may be appropriate targets for the development of new therapeutic strategies. We are in the process of characterizing the intrinsic ability of light chains to induce organ dysfunction in an organ-specific model system, as well as determining other cellular components of amyloid deposits that may be responsible for the pathogenic changes involved in light chain amyloidosis.
The authors wish to acknowledge Drs Raymond L. Comenzo and Yana Zhang, Memorial Sloan Kettering Cancer Center, New York, for their assistance in the project as well as providing reagents. We also wish to thank Dr Comenzo for critical reading of the manuscript.
Prepublished online as Blood First Edition Paper, December 19, 2002; DOI 10.1182/blood-2002-09-2707.
Supported by the CI-5 Damon-Runyon Clinical Investigator Award of the Damon-Runyon-Walter Winchell Foundation and a Leukemia and Lymphoma Society Translational Research Award (R.F.). This work was also supported in part by Public Health Service grant no. RO1 CA83724-01 (R.F.), the Hematologic Malignancies Fund, Mayo Clinic, and the National Cancer Institute, CA-62242.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Rafael Fonseca or Roshini S. Abraham, Division of Hematology, Stabile 6-28, Mayo Clinic, 200 1st St SW, Rochester, MN 55905; e-mail: fonseca.rafael@mayo.edu,abraham.roshini@mayo.edu.

![Fig. 3. Kaplan-Meier survival curves for the AL subset. / Panel A shows a comparison of the overall survival between the κ (thick line) and λ (thin line) groups within the selected subset. Panel B shows survival based on Vκ gene use (Vκ I [solid line] and Vκ IV [dashed line] gene families). The single Vκ II patient has not been represented, and because of the small numbers of patients in the subgroups, survival for individual germ-line genes has not been shown. The Vκ IV gene, which is associated mainly with cardiac and renal amyloidosis, showed reduced survival. Panel C shows survival analysis for the Vλ gene families. Vλ II patients present predominantly with cardiac infiltration and have reduced survival, whereas Vλ VI patients have comparatively better survival curves. The clinical categories documented are not mutually exclusive.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/10/10.1182_blood-2002-09-2707/4/m_h81034327003.jpeg?Expires=1767958198&Signature=hTxnLQT0XhiqSmNXPs9mb~sRcHzTsJ-2c8JHajp0PExZXvG4gOquEh6oQySGrfe6EwkVyC26Rt6P4SGT7x8w-IWxc6rJM65OqAwavPDRtPN9C1camyuBgo6PBaq5zXHU8PDZ3mz0brLcE-XMOZyGYAw7ZOnFfo3M9kPbsoc14NGdy3I9rpcS1qB3byhgDdhN6W-J~YSSKc~InsyIK3WmMzzfA1mMYTZV-BDcnv~HRX8NZbv3W6hSDFRunpwj2~YvJBWbIL~6QkA3SzvZ8whEiECCJmRrdmZ2HddmRRgInC8lfbIhlacZBTwZGrhkYKTLDEzI5xWnF4iU~E-b8bz6wQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 3. Kaplan-Meier survival curves for the AL subset. / Panel A shows a comparison of the overall survival between the κ (thick line) and λ (thin line) groups within the selected subset. Panel B shows survival based on Vκ gene use (Vκ I [solid line] and Vκ IV [dashed line] gene families). The single Vκ II patient has not been represented, and because of the small numbers of patients in the subgroups, survival for individual germ-line genes has not been shown. The Vκ IV gene, which is associated mainly with cardiac and renal amyloidosis, showed reduced survival. Panel C shows survival analysis for the Vλ gene families. Vλ II patients present predominantly with cardiac infiltration and have reduced survival, whereas Vλ VI patients have comparatively better survival curves. The clinical categories documented are not mutually exclusive.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/10/10.1182_blood-2002-09-2707/4/m_h81034327003.jpeg?Expires=1768167766&Signature=o4t7j60nDW1nMEyOhoFky8f5LW29LF1keQ1E69dz~OF1wu3lRsrsL3UDBhGMHuKaK~~YO88QJwlo9CX0vSqbeZj1oCTK4GKq2I5EVXZc-8ceEiPviFfElmtchFjzPDvHUgE-fl7HyFH0olHmsODM5lGho4rLGRfDVw9~qf-jGRwQ5~KvzIHhNTT68ZgrdLwQEbljL6RkFFP2cV7jNJC7q3MGWAzxxkMzrLhXkNDJd54XxetImtcxJBjUUy2QI0zX461UtcbOC9MVSgmCMgH5vnOFWfVL0WECEN9YAWzVJjzpOMjhm0XZeU-L16dJ2MzXznggAuIjPMJggamlnywgyA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)