Abstract
The formation of terminally differentiated plasma cells represents the critical final step in B-cell differentiation. In this study, utilizing oligonucleotide microarray analysis, we describe the highly specialized genetic profile exhibited by terminally differentiated plasma cells. A total of 1476 known genes were differentially expressed by plasma cells compared with B cells. Plasma cells displayed an up-regulation, induction, or a selective retention of a unique constellation of transcription factors, including members of the AP-1, nuclear factor–κB (NF-κB), nuclear factor of activated T cells (NFAT), and octamer binding factor families. Interestingly, plasma cells also displayed a down-regulation of several RNA polymerase I– related factors, consistent with terminal differentiation, and exhibited a down-regulation of the TATA box binding protein. Furthermore, plasma cells displayed alterations in multiple components of the Wnt and Notch signaling pathways and showed a unique pattern of apoptosis and proliferation-associated genes. Unexpectedly, plasma cells displayed an up-regulation of 2 factors normally associated with microenvironmental positioning of neuronal cells, reelin and neuropilin-1. These results supply insight into the developmental genetics of plasma cell differentiation and provide a foundation for further analysis of plasma cell biology.
Introduction
Plasma cells, which constitute the final stage of B-cell differentiation, are specialized terminally differentiated cells with one primary function, the secretion of antibody. Recent work has begun to reveal some of the factors that regulate plasma cell formation and function. The transcription factors XBP-1, Blimp-1, and IRF-4 have each been demonstrated to be critically involved in plasma cell differentiation.1-4 Furthermore, B-cell transcription factors Pax5 and BCL-6, which can repress transcription of XBP-1 and Blimp-1, respectively, are down-regulated during plasma cell differentiation.5-7 Recent analysis has provided important information regarding the critical role of Blimp-1 in regulating plasma cell gene expression, including, among other aspects, the repression of Pax5, BCL-6, and B-cell transcription factors Spi-B and Id3.8 9
Mice deficient in both E- and P-selectin display extremely elevated serum immunoglobulin G (IgG) levels and severe cervical lymphadenopathy consisting of expanded numbers of lymphocytes, including numerous plasma cells.10 Previously, we have demonstrated that the cervical lymph nodes of these mice provide a unique system for the investigation of B-cell differentiation. Furthermore, we described the purification of significant numbers of B220− terminally differentiated plasma cells as well as the distinct adhesion characteristics of these cells.11
Although recent advances have provided insight into plasma cell biology, many aspects of plasma cell function have yet to be determined, including many factors that modulate plasma cell differentiation and function in vivo. Plasma cell gene expression profiles potentially represent crucial information regarding unidentified transcription factors and signaling molecules important in plasma cells, the adhesion molecules, and secreted factors that regulate the interaction of plasma cells with their microenvironment as well as mechanisms of terminal differentiation. Here we report gene expression profiling of B220− terminally differentiated plasma cells. These cells displayed a distinct pattern of gene expression, including unique sets of transcription factors and molecules involved in signal transduction, survival, and cell-cell interactions. These findings provide important clues into the mechanisms underlying plasma cell function.
Materials and methods
Mice
The E-selectin/P-selectin double-deficient (E/P−/−) mice, backcrossed 5 generations to C57BL/6, were provided by Dr Dan Bullard (University of Alabama-Birmingham). Wild-type C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME) and bred in our mouse facility.
Cell isolation
B220− plasma cells were isolated from E/P−/− cervical lymph node cells from pooled mice, as previously described in detail,11 using a cocktail of anti-CD5, anti–Mac-1, and anti-B220 microbeads (Miltenyi Biotec, Auburn, CA). Approximately 20% of the mice used in these experiments displayed clear signs of infection such as skin lesions with hair loss. Previously we have demonstrated that these cells constitute a B220− noncycling population of plasma cells that spontaneously secrete antibody of each of the IgG subclasses but very little IgM.11 Furthermore, these cells were CD19−, CD38−, major histocompatibility complex (MHC) class II−, L-selectin−, CD43+, syndecan-1+, CD44hi, PSGL-1hi, LFA-1hi, and α4 integrinhi.11 Wild-type B cells were isolated from wild-type C57BL/6 cervical lymph nodes from multiple mice using anti-IgM microbeads and LS+ positive selection columns (Miltenyi Biotec). A purity of more than 96% was achieved for both cell types.
Complementary RNA probe preparation and chip hybridization
Total RNA was isolated from either plasma cells or B cells using Trizol (Life Technologies, Carlsbad, CA), followed by an additional purification with the RNeasy Mini Kit (Qiagen, Valencia, CA). Double-stranded cDNA was synthesized using a (dT)24 primer containing a T7 RNA polymerase initiation site (Genset, La Jolla, CA) and the Superscript Double Stranded cDNA Synthesis Kit (Life Technologies), with 5 to 10 μg total RNA as a template. Following phenol-chloroform extraction of the cDNA, biotinylated cRNA was generated using the Bioarray HighYield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY) and purified using the RNeasy Mini Kit (Qiagen). A total of 15 μg labeled cRNA was fragmented according to Affymetrix procedures, and the chip hybridization was performed as described.12 Briefly, the cRNA with hybridization controls was hybridized to the murine chip, MG-U74 AV2, according to the Affymetrix protocol. Staining was performed in the GeneChip Fluidics station, and chips were scanned in the Affymetrix GeneChip scanner. Three replicates were performed for both plasma cells and surface IgM+ B cells, each from independent cell preparations.
Data analysis
Gene expression analysis was performed using Gene Spring (Silicon Genetics, Redwood City, CA). Per chip and per gene normalizations of the expression values (average differences) were executed with Gene Spring, and normalized expression values less than 0.01 were set to 0.01. Gene trees were generated by hierarchical clustering using the standard correlation (Pearson correlation around 0) with a separation ratio of 0.5 and a minimum distance of 0.001.
To eliminate genes from the analysis that were not expressed by either plasma cells or B cells, 3 categories of interest were formulated using the present/absent calls generated by the Affymetrix Microarray Suite 4.0. These categories were the following: present in plasma cells/absent in B cells, absent in plasma cells/present in B cells, and present in both cell types. A marginal call was considered absent for these purposes. The cutoff for inclusion in these groups was that 5 of 6 samples had to correspond with the category criteria. For example, to be included in the present in plasma cells/absent in B cells group, a present call in at least 2 plasma cell replicates with an absent call in all 3 B-cell replicates, or a present call in all 3 plasma cell replicates with an absent call in at least 2 B-cell replicates was required. A gene was classified as differentially expressed in plasma cells and B cells if it was (1) among one of these categories and (2) exhibited a statistically significant (P < .05, Mann-Whitney test) and (3) exhibited 2-fold or greater change in the mean expression level. When multiple oligo sets for one gene appeared on the chip, the set derived from the complete cDNA sequence was used. If multiple cDNA-derived sets appeared on the chip, a representative result is shown. A complete list of all the genes examined and all values for each replicate is available on the Blood website; see the Supplemental Data link at the top of the online article.
SDS-PAGE and Western Blot analysis
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis of plasma cell and B-cell lysates were performed as described.11 Cell lysates were made from equal numbers of cells with the following high-salt RIPA lysis buffer: 50 mM Tris (tris(hydroxymethyl)aminomethane) (pH 8), 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% SDS, and 1 mM EDTA (ethylenediaminetetraacetic acid), with protease inhibitors. Rabbit antimouse TBP antiserum was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Horseradish peroxidase–conjugated antirabbit and antimouse IgG antibodies were purchased from Biosource, and nitrocellulose membranes were developed using enhanced chemiluminescence reagents (Amersham Biosciences, Arlington Heights, IL). The murine plasmacytoma cell line, J558, was used as a positive control for TBP.
Real-time quantitative RT-PCR
Real-time quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) was performed using the 7700 sequence detector system as previously described.12 Twenty-five nanograms of total RNA isolated from plasma cells or B cells was used as a template in a 50-μL RT-PCR reaction with the following components: 1 × TaqMan buffer, 5 mM MgCl2, 0.3 mM deoxyadenosine monophosphate (dATP), 0.3 mM deoxycytidine triphosphate (dCTP), 0.3 mM deoxyguanosine triphosphate (dGTP), 0.6 mM deoxyuridine triphosphate (dUTP), 0.1 μM forward and reverse primers, 0.1 μM TaqMan probe, 1.25 units AmpliTaq Gold, 20 units RNase inhibitor, and 12.5 units murine leukemia virus (MuLV) reverse transcriptase. The primers and TaqMan probes were designed using Primer Express software (Perkin Elmer Life Sciences, Shelton, CT), and the sequences are listed in the methods document in the supplemental data. The primers were purchased from Integrated DNA Technologies, and the TaqMan probes were purchased from MegaBases. The cycling parameters were 48°C for 30 minutes, then 95°C for 15 minutes, followed by 40 cycles at 95°C for 15 seconds and 59°C for 1 minute. ΔRn represents the reporter signal with the baseline subtracted, and the threshold cycle (CT) is the cycle at which ΔRn crosses the threshold for the given sample. The default threshold was used, which is set at 10 standard deviations above baseline.
Results and discussion
Plasma cells and IgM+ B cells display highly distinct gene expression profiles
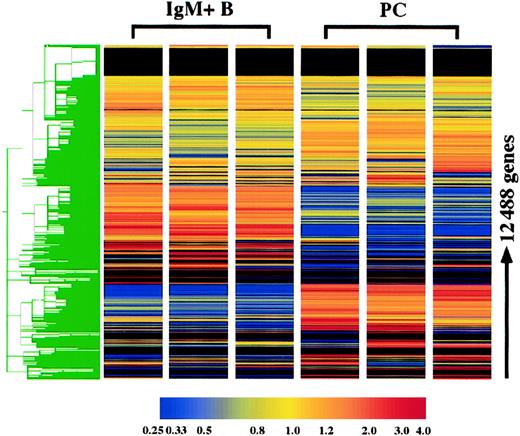
The gene expression profile of plasma cells was compared with surface IgM+ B cells using the Affymetrix murine oligonucleotide array, MG-U74 AV2. With this array, the expression of 12 422 genes was examined, and 3 replicates of each cell type were performed. Hierarchical clustering of these genes demonstrates the high degree of reproducibility of the replicates. As expected, the gene expression profiles of IgM+ B cells and plasma cells were extremely distinct (Figure 1). Furthermore, it was clear from initial observations that plasma cells in general lose expression of many more genes than they gain. Genes were initially categorized based on the present and absent determinations: 396 genes were consistently expressed in plasma cells although absent in B cells, whereas 1395 genes were absent in plasma cells and expressed in B cells, and 3676 genes were expressed in both cell types. Genes were considered altered in plasma cells if they were (1) among one of these categories and additionally displayed (2) a statistically different and (3) 2-fold or greater change in mean expression. With these criteria, 2469 genes were determined to be significantly altered in plasma cells, which included 1476 known genes and 993 unknown expressed sequence tags (ESTs). The altered expression in plasma cells of numerous genes, includingBlimp-1, IRF-4, XBP-1, Pax5, CIITA, BCL-6, syndecan-1, CD43, CD19, CD22, MHC class II, CD37, CD79A, L-selectin, CD38, andCD44, were all consistent with previous findings for plasma cells,9,11 13 confirming the fidelity of the analysis.
Hierarchical clustering of plasma cell and IgM+ B-cell replicates.
The expression of 12 422 genes and 66 hybridization controls were examined in 3 plasma cell (PC) and IgM+ B-cell replicates using Affymetrix oligonucleotide array, MG-U74 AV2. Hierarchical clustering was performed using Gene Spring following per gene and per chip normalizations of the average difference. As the color bar indicates, yellow represents the median expression of each gene, normalized to 1.0, and red and blue represent elevated and reduced expression relative to the median, respectively. For genes with a median raw average difference of less than 10.0, per gene normalizations were made relative to 10.0 instead of the median.
Hierarchical clustering of plasma cell and IgM+ B-cell replicates.
The expression of 12 422 genes and 66 hybridization controls were examined in 3 plasma cell (PC) and IgM+ B-cell replicates using Affymetrix oligonucleotide array, MG-U74 AV2. Hierarchical clustering was performed using Gene Spring following per gene and per chip normalizations of the average difference. As the color bar indicates, yellow represents the median expression of each gene, normalized to 1.0, and red and blue represent elevated and reduced expression relative to the median, respectively. For genes with a median raw average difference of less than 10.0, per gene normalizations were made relative to 10.0 instead of the median.
Plasma cells express a unique constellation of transcription factors
Among the many genes differentially expressed in plasma cells and IgM+ B cells were numerous transcription factors (Figure2). As expected and mentioned above, these included the transcription factors Blimp-1,IRF-4, and XBP-1, which were increased in plasma cells 29.4-, 4.0-, and 17.2-fold, respectively, and have been previously identified as important in plasma cell differentiation.1,2,4 In addition, the loss of multiple B-cell transcription factors in plasma cells, including CIITA, BCL-6, c-myc, EBF, Stat6, and Pax5 (Figure2), were all consistent with previous findings and with the role of Blimp-1 in repression of these genes.9,14 15 Alterations in expression of several genes were confirmed by quantitative RT-PCR, and these results are shown in Table 1 and Figure3A. These genes included, among others, XBP-1, Blimp-1, Stat6, and CIITA.
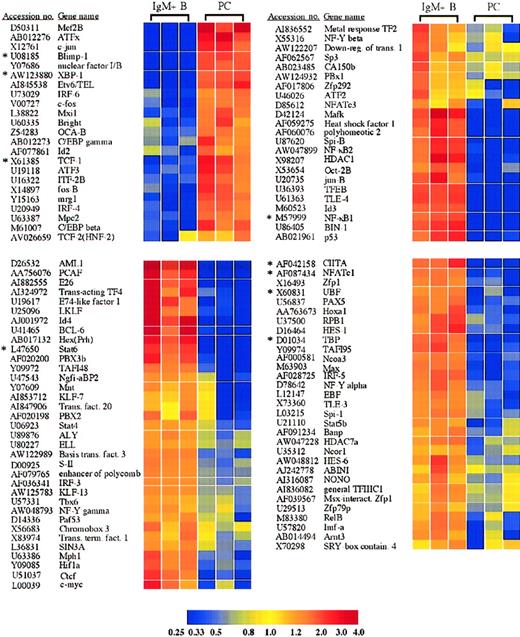
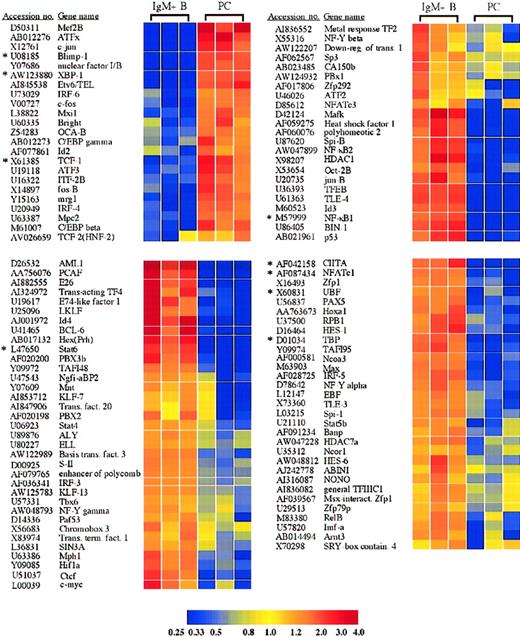
Transcription factors with significantly altered expression in plasma cells.
Twenty-three transcription factors were induced or up-regulated in plasma cells versus B cells, whereas 89 transcription factors were lost or down-regulated. The genes are listed according to the cluster analysis, which groups genes based on the relatedness of the expression pattern. Each of these genes displayed a statistically significant 2-fold or greater difference in expression between plasma cells and B cells. *Genes for which quantitative RT-PCR was performed (Table1).
Transcription factors with significantly altered expression in plasma cells.
Twenty-three transcription factors were induced or up-regulated in plasma cells versus B cells, whereas 89 transcription factors were lost or down-regulated. The genes are listed according to the cluster analysis, which groups genes based on the relatedness of the expression pattern. Each of these genes displayed a statistically significant 2-fold or greater difference in expression between plasma cells and B cells. *Genes for which quantitative RT-PCR was performed (Table1).
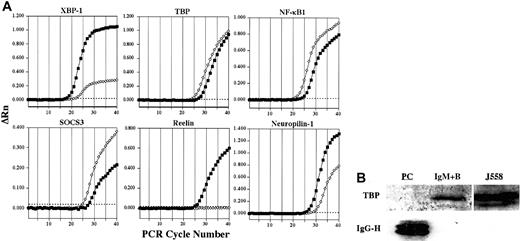
Representative confirmation analysis by real-time quantitative RT-PCR and Western blot.
(A) Representative plots confirming alterations in expression of select genes in plasma cells (▪) versus IgM+ B cells (⋄) by real-time quantitative RT-PCR are shown. ΔRn represents the reporter signal with the baseline subtracted. The threshold cycle (CT) values for these genes as well as additional genes are listed in Table 1. The threshold is indicated by the dotted line. (B) The expression of TBP protein in plasma cells and IgM+ B cells was determined by Western blot. The J558 murine plasmacytoma cell line was used as a positive control for TBP. To control for sufficient protein in plasma cell samples, lysates were examined for the presence of IgG using a polyclonal goat antimouse IgG antibody.
Representative confirmation analysis by real-time quantitative RT-PCR and Western blot.
(A) Representative plots confirming alterations in expression of select genes in plasma cells (▪) versus IgM+ B cells (⋄) by real-time quantitative RT-PCR are shown. ΔRn represents the reporter signal with the baseline subtracted. The threshold cycle (CT) values for these genes as well as additional genes are listed in Table 1. The threshold is indicated by the dotted line. (B) The expression of TBP protein in plasma cells and IgM+ B cells was determined by Western blot. The J558 murine plasmacytoma cell line was used as a positive control for TBP. To control for sufficient protein in plasma cell samples, lysates were examined for the presence of IgG using a polyclonal goat antimouse IgG antibody.
Additionally, these IgG plasma cells also displayed an induction, loss, or selective retention of specific members of multiple other transcription factor families. Among the AP-1 family, c-jun was induced 23.0-fold in plasma cells, and c-fos and fos-B were both up-regulated 5.1-fold and 4.5-fold, respectively (Figure 2). In contrast, jun-B was down-regulated 13.3-fold in plasma cells (Figure 2), whereas jun-D was retained equally in all samples (supplemental data). This altered expression of AP-1 proteins is particularly interesting considering recent data demonstrating that BCL-6 represses Blimp-1 by inhibiting AP-1 transcriptional activation of Blimp-1.16
Furthermore, the plasma cells demonstrated a selective down-regulation of several components of the nuclear factor–κB (NF-κB) pathway. These included NF-κB1(p50/p105), NF-κB2(p52/p100), and RelB (Figure 2). The down-regulation of NF-κB1 was confirmed by quantitative RT-PCR (Figure 3A and Table 1). In contrast, RelA(p65) was expressed equally in plasma cells and surface IgM+ B cells (supplemental data). Several regulatory components of the NF-κB pathway were also altered in plasma cells. These included 2 members of the IκB protein family, BCL-3 and IκBβ, which were lost in plasma cells (Figure 4). Furthermore, the IκB kinase (IKK) β subunit was lost in plasma cells, as was the NF-κB–inducing kinase (NIK) (Figure 4). However, like RelA, IKKα and IκBα were retained in plasma cells (supplemental data). Distinct roles for IKKα and IKKβ have been described.17 In addition, IgG+ B cells may exhibit alternative mechanisms of NF-κB activation, demonstrated by the nuclear localization of NF-κB components in the absence of IκB degradation in IgG+ but not IgM+ B cells.18 Taken together, although many NF-κB family members were lost in plasma cells, a regulatable and highly specific pathway consisting of IKKα, IκBα, and RelA remains intact.
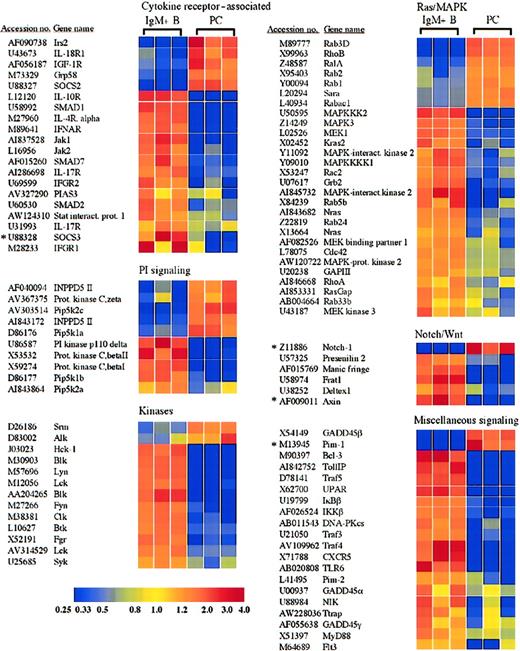
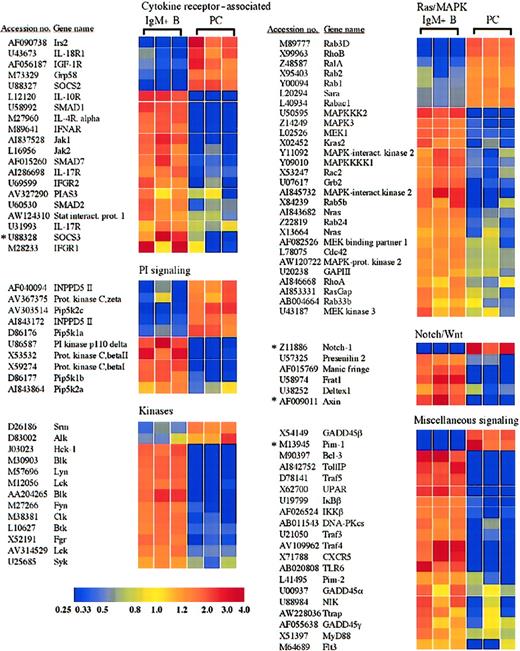
Receptors and signaling molecules differentially expressed in plasma cells and B cells.
Select receptors and signaling molecules representing several signaling families, which displayed significantly altered expression in plasma cells, are shown. A complete list of all of the genes examined is available in the supplemental data. *Genes for which quantitative RT-PCR was performed (Table 1).
Receptors and signaling molecules differentially expressed in plasma cells and B cells.
Select receptors and signaling molecules representing several signaling families, which displayed significantly altered expression in plasma cells, are shown. A complete list of all of the genes examined is available in the supplemental data. *Genes for which quantitative RT-PCR was performed (Table 1).
Examination of mice with lymphocytes deficient in both nuclear factor of activated T cells c1 (NFATc1) and NFATc2 suggested an intrinsic role for these transcription factors in B-cell function, with expanded numbers of plasma cells and elevated IgG1 and IgE serum levels, despite impaired T-cell effector function.19 The plasma cells examined here demonstrate a down-regulation of NFATc1 mRNA as well as NFATc3 (Figure 2). The down-regulation of NFATc1 was also confirmed by quantitative RT-PCR (Table 1). However, NFATc2 is maintained in plasma cells equal to surface IgM+ B cells (supplemental data). Although not required for plasma cell differentiation,19 20 the selective retention of NFATc2 mRNA described here suggests a possible role for NFATc2, nonredundant with NFATc1, in plasma cell function. Conversely, the selective loss of NFATc1 and NFATc3 may be important in plasma cell function through the loss of certain inhibitory functions.
The plasma cells demonstrated a loss in c-myc expression, consistent with Blimp-1 expression by these cells (Figure 2)11 and previous results that illustrate the role of Blimp-1 in c-myc repression.15 In addition to the loss of c-myc, the c-myc dimerization partner Max was down-regulated in plasma cells 5.9-fold, and Mxi1, a member of the Mad family of c-myc antagonists, was up-regulated in plasma cells 3.6-fold (Figure 2). The c-myc adaptor protein BIN1 was also lost in plasma cells (Figure 2). In contrast to the up-regulation of Mxi1, another c-myc antagonist, Mnt/ROX, was lost in plasma cells (Figure 2). Unlike Mxi1, which interacts only with Max, Mnt can interact with Max and the Max-like protein, Mlx.21Interestingly, Mlx and the Mad protein Mad4 were retained in plasma cells equal to B cells (supplemental data), suggesting that they could possibly play a role in plasma cell function. Although further analysis is certainly required to determine the mechanism, this expression profile suggests a cooperative effect of additional members of the Myc/Mad/Max system in the antagonism of c-myc function in plasma cells. Furthermore, as demonstrated previously, the repression of c-myc is necessary but not sufficient for plasma cell terminal differentiation.22 Accordingly, the plasma cells also displayed a down-regulation in many additional genes involved in cell cycle control and proliferation (Table2).
Octamer site binding transcription factors have been implicated in B-cell function, including the transcriptional activity of Ig promoters and the 3′ Cα enhancer,23 and B cells lacking octamer binding factor 2 (Oct-2) do not proliferate in response to mitogenic stimulation.24 Interestingly, the IgG plasma cells exhibited a loss of Oct-2 with a concurrent 2.4-fold up-regulation of the Oct-1/Oct-2 binding cofactor, OCA-B (Bob1, OBF-1) (Figure 2). The loss of Oct-2 in primary plasma cells is consistent with results from multiple myeloma cell lines and B-cell lines following introduction of Blimp-1.9 Oct-1 was expressed equally in both cell types (supplemental data). Mice deficient in both Oct-2 and OCA-B illustrate that these 2 transcription factors are not absolutely required for immunoglobulin transcription.25 However, in the production of an effective in vivo IgG response, a substantial role for OCA-B, with a lesser role for Oct-2, was demonstrated.25Therefore, in agreement with previous reports, Oct-1 is likely the octamer binding factor that plays a predominant role in immunoglobulin transcription.25 Up-regulation of OCA-B suggests at least a possible partial role for this cofactor.
In addition to the multiple specific transcription factors whose expression was altered in plasma cells, plasma cells also demonstrated expression changes in components of the basal transcription apparatus as well as in several transcriptional cofactors that play critical roles in the transcription of numerous genes. Among these were histone deacetylase-1 (HDAC1), the TATA box binding protein (TBP), the large subunit of RNA polymerase II (RPB1), and the coactivator PCAF, which all displayed reduced expression in plasma cells (Figure 2). Thus, consistent with the highly specialized function of the plasma cell, plasma cells may exhibit unique general transcription mechanisms. Although unexpected, a down-regulation of TBP mRNA in plasma cells was confirmed by quantitative RT-PCR (Figure 3A and Table 1), and TBP protein levels were sharply reduced in plasma cells as well (Figure3B). In contrast, mRNA encoding TBP-like factor (TLF) was expressed by both the plasma cells and B cells examined here (supplemental data), and TBP and TLF can differentially regulate the transcription of certain genes.26 Consequently, further analysis is required to elucidate the role of decreased TBP levels and the possible function of TLF in primary plasma cells. Additionally, 3 components of the RNA polymerase I complex important in rRNA transcription, the transcription factors UBF, TAFI48, and TAFI95, also showed a reduced expression in plasma cells (Figure 2). A down-regulation of UBF was also confirmed by quantitative RT-PCR (Table 1). Down-regulation of these components and concomitant decreases in rRNA transcription have been observed previously upon cellular differentiation with cell cycle exit. For example, UBF, TAFI48, and TAFI95 down-regulation was associated with the differentiation of F9 embryonal carcinoma cells into parietal endoderm.27 Strikingly, overall TBP protein was also reduced following F9 cell differentiation.27Furthermore, decreased rRNA transcription was correlated with the down-regulation of UBF during the terminal differentiation of skeletal muscle cells.28 Consequently, similar to other terminally differentiated cell types, the terminal differentiation of plasma cells is associated with a reduction in the expression of TBP and with a decrease in ribosomal RNA transcription.
Plasma cells show alterations in many receptors and signaling molecules, including cytokine receptor–associated factors and members of the Wnt and Notch pathways
Plasma cells and B cells displayed numerous differences in the expression of receptors and signaling molecules, and a selection of these genes is shown in Figure 4. For example, IRS2, which plays a role in the signaling via several receptors, including the interleukin-4 (IL-4) receptor and the insulin-like growth factor-1 (IGF-1) receptor,29,30 was induced in plasma cells (Figure 4). The IGF-1 receptor itself was also induced, consistent with results obtained with multiple myeloma cells.31 The presence of both the IGF-1 receptor and IRS2 suggests a possible role of this signaling pathway in primary plasma cell function. In contrast, the receptor for IL-4 as well as receptors for the cytokines IL-10, IL-17, and the α/β and γ interferons were lost or down-regulated in plasma cells (Figure 4). Among the Janus kinase (Jak) family, Jak1 and Jak2 were lost in plasma cells (Figure 4), whereas Jak3 was expressed equally in both cell types (supplemental data). In accordance with Stat6 as a target of Blimp-1–mediated down-regulation,9 Stat6 was lost in the plasma cells examined here (Figure 2 and Table 1). In addition, Stat4 and Stat5b were also lost (Figure 2), whereas Stat1 and Stat3 were retained at levels equal to B cells (supplemental data).
In addition to the selective retention of Stat3 itself, the plasma cells exhibited a loss in the expression of the protein inhibitor of activated Stat3 (PIAS3) (Figure 4). In contrast, the protein inhibitor of activated Stat1 (PIAS1) was expressed equally in plasma cells and B cells (supplemental data). Strikingly, mRNA encoding the suppressor of cytokine signaling-3 (SOCS3), which can inhibit IL-6 receptor signaling through Stat3,32 also displayed a reduced expression in plasma cells (Figure 4). This down-regulation of SOCS3 expression in plasma cells was confirmed by quantitative RT-PCR (Figure3A and Table 1). On the contrary, SOCS2, which can be induced by IL-6 signaling but does not effectively inhibit IL-6 signaling,32,33 was induced in plasma cells. Additionally, the chaperone protein Grp58, which has been shown to interact with Stat3,34 was up-regulated in plasma cells. The signaling component of the IL-6 receptor, gp130, was expressed by both B cells and plasma cells and displayed a 1.8-fold statistically significant increase in plasma cells (supplemental data). In contrast, the IL-6 receptor α chain was apparently absent from both B cells and plasma cells. However, IL-6 promotes the survival of these IgG plasma cells in vitro,35 suggesting that the IL-6 receptor α chain is expressed, although below the limit of detection of this analysis. Taken together, the retention of Stat3 and gp130, up-regulation of Grp58, and the down-regulation of PIAS3 and SOCS3 suggest that Stat3 controls important aspects of plasma cell biology. Furthermore, Stat1 and Stat3 can heterodimerize36 and can collaborate in mediating appropriate gp130 signaling in response to IL-6.37 Consequently, the loss of interferon receptors together with the retention of PIAS1 suggests that Stat1 is not retained as a constituent of interferon signaling but may display a novel cooperative role with Stat3 in plasma cell function.
Several components of the Wnt signaling pathway also displayed an altered expression in plasma cells relative to B cells. In the absence of a Wnt signal, β-catenin is sequestered in a complex with axin, adenomatous polyposis coli (APC), and GSK-3β. Within this complex, GSK-3β phosphorylates β-catenin, thereby targeting β-catenin for proteasomal degradation.38 Wnt signaling results in the activation of the cytoplasmic protein dishevelled, which in association with Frat1/GBP inhibits the phosphorylation of β-catenin by GSK-3β, allowing β-catenin to accumulate and travel into the nucleus, where it can activate gene transcription in collaboration with the TCF/LEF family of transcription factors.39 Interestingly, axin and Frat1 showed reduced expression in plasma cells (Figure 4), whereas β-catenin and dishevelled were expressed equally in plasma cells and B cells (supplemental data). The down-regulation of axin in plasma cells was also confirmed by quantitative RT-PCR analysis (Table 1). Although Frat1 has been implicated with dishevelled in the inhibition of β-catenin phosphorylation as part of Wnt induced signaling,40,41 in the absence of sufficient axin, Frat1 may not be required. In fact, the directed loss of Frat1 could disconnect dishevelled and GSK-3β from the Wnt signaling pathway and allow them to participate in other cellular processes. Interestingly, the TCF/LEF family member TCF-1, which can interact with β-catenin and activate gene transcription following Wnt signaling, was previously demonstrated to be expressed in bone marrow plasma cells from healthy donors and was down-regulated in multiple myeloma samples.42 Conversely, APC was significantly up-regulated in myeloma versus normal plasma cell samples.42 Although a probe set derived from the TCF-1 cDNA sequence suggested an up-regulation of TCF-1 in plasma cells (Figure 2), another probe set derived from an EST representing TCF-1 showed opposite results (accession no. AI019193, Table 1, and supplemental data), and quantitative RT-PCR demonstrated a modest down-regulation of TCF-1 in plasma cells (Table 1). Taken together, the altered expression of Wnt signaling components suggests that this pathway may play a role in plasma cells, although further analysis is certainly required to determine the possible distinct function of the Wnt signaling pathway in primary plasma cells and myeloma cells as well as the role of TCF-1. Several components of another signaling pathway involved in cell fate decisions, the Notch pathway, were also altered in plasma cells (Figure4). For example, expression of manic fringe, which can modify Notch and subsequent signaling,43,44 was lost in plasma cells (Figure 4), but radical fringe was expressed in both plasma cells and B cells (supplemental data). Cleavage of the Notch receptor within the transmembrane domain that releases the Notch intracellular domain is mediated by presenilins and nicastrin.45 Presenilin 1 was expressed by plasma cells and B cells, but presenilin 2 was lost in plasma cells (Figure 4). Although both presenilin 1 and presenilin 2 have been implicated in the process of Notch cleavage,46differential functions have been described. For instance, presenilin 2 can enhance caspase 3 activation, thereby promoting apoptosis through a p53-dependent mechanism, as well as up-regulate p53 transcription and down-regulate presenilin 1 transcription.47 The microarray analysis suggested an induction of the Notch-1 receptor in plasma cells. However, quantitative RT-PCR failed to demonstrate a difference in mRNA expression between plasma cells and B cells (Table 1). Although the Notch-1 receptor may not be differentially expressed, the altered expression of other Notch signaling components suggests that this pathway may play a currently unidentified role in plasma cell function.
Interestingly, the plasma cells also demonstrated alterations in the mRNA levels of several secreted factors, including an induction of VEGF-A (Table 3). The expression of VEGF by plasma cells has previously been associated with multiple myeloma, where an autocrine loop of vascular endothelial growth factor (VEGF) signaling can stimulate myeloma cell proliferation.48 In contrast to myeloma, the VEGF receptor Flt-1 was not expressed by the primary plasma cells examined here (nor was the receptor KDR), suggesting that an autocrine loop is not present in primary plasma cells. VEGF expression by plasma cells has also been demonstrated in the lymph nodes of patients with Castleman disease but not in normal lymph nodes.49 Recently, it was demonstrated that VEGF gene expression can be induced in B-cell lines by the introduction of Blimp-1.9 Consequently, Blimp-1 may initially allow VEGF expression, which is then regulated by modifying factors in vivo. Perhaps VEGF expression by plasma cells in the E/P−/− mice is a result of the sustained immune response these mice clearly display.10 11 Further analysis of VEGF expression by plasma cells in the E/P−/− mice as well as other contexts will provide information into the possible role of this factor in plasma cell function.
Plasma cells up-regulate antiapoptotic factors, including Bcl-xL, Bcl-w, and XIAP, but lose Bcl-2
In accordance with other categories of genes, many more genes associated with apoptosis were lost rather than gained in plasma cells (Table 4). For example, Bcl-2 expression was lost in plasma cells. However, 2 other Bcl-2 family members, Bcl-xL and Bcl-w, were up-regulated in plasma cells 2.1-fold and 4.8-fold, respectively (Table 4). The higher expression of Bcl-xL versus Bcl-2 was reported previously in human plasma cells.50Additionally, several proapoptotic factors were lost or down-regulated in plasma cells, including, among others, the Bcl-2 family member BID, the Bcl-2 binding protein Bax, and caspases 6 and 2 (Table 4). Another factor that has been associated with B-cell survival is the Pim-1 kinase.51 Interestingly, although Pim-1 kinase activity in B cells has been attributed to BCR activation, CD40 signaling, or lipopolysaccharide (LPS) stimulation,51 the microarray analysis demonstrated a 21.2-fold up-regulation in Pim-1 expression in plasma cells (Figure 4), which have lost CD40 and multiple BCR signaling components (Table5 and Figure 4). However, Pim-1 can also be induced by Stat3-mediated transcription through gp130 signaling,52 a pathway that is retained in plasma cells (mentioned above). Furthermore, valosine-containing protein (VCP), which is a target of Pim-1 signaling and involved in Pim-1–mediated protection against apoptosis,52 is up-regulated in plasma cells 2.9-fold (Table 4). An up-regulation of Pim-1 mRNA expression in plasma cells was also confirmed by quantitiative RT-PCR (Table 1). Consequently, Pim-1 kinase expression resulting from IL-6 signaling and Stat3 activation may be a critical component in the protection of plasma cells against apoptosis.
Recently, the induction of GADD45β by NF-κB, in particular RelA, was shown to protect cells from apoptosis by down-regulating JNK activation following tumor necrosis factor (TNF) receptor signaling.53,54 GADD45β was up-regulated in plasma cells 15.1-fold, whereas GADD45γ was down-regulated 2.2-fold and GADD45α was lost (Figure 4). Another NF-κB–dependent factor that was associated with the inhibition of JNK and protection from apoptosis was XIAP/MIHA.55 Interestingly, XIAP was also up-regulated (10.6-fold) in plasma cells (Table 4). The up-regulation of XIAP observed here is in contrast to the down-regulation of XIAP demonstrated for myeloma cell lines or following Blimp-1 introduction into B-cell lines.9 One possible explanation for this difference is the differential regulation of XIAP in cell lines versus primary cells. NF-κB plays a role in the survival of myeloma cells,56 and the inhibition of JNK/c-jun through an IL-6 signaling mechanism can also protect multiple myeloma cells from apoptosis.57 58 Consequently, the retention of the IL-6 signaling pathway as well as select NF-κB family members by primary plasma cells reported here (Figures 2 and 4) may be crucial in the protection of plasma cells from apoptosis, possibly through the inhibition of JNK. Taken together, these results demonstrate that plasma cells display a distinct spectrum of apoptosis-related factors. Further analysis will be focused on determining the profile of apoptotic mediators in short-lived versus long-lived plasma cells as well as the anatomic factors that regulate plasma cell survival.
Plasma cells express the nervous system genes, reelin and neuropilin
Select additional alterations in plasma cell gene expression are displayed in Table 5. Unexpectedly, the plasma cells expressed factors that have previously been associated with nervous system function. One such gene that was highly induced in plasma cells encodes the protein reelin (Table 5), which regulates the migration of neurons in the developing cortex and is mutated in the reeler mouse strain.59 The induction of reelin in plasma cells was confirmed by quantitative RT-PCR (Figure 3A and Table 1). Receptors for reelin include the very low density lipoprotein receptor (Vldlr), the apolipoprotein E receptor-2 (ApoER2), and the integrin α3β1. ApoE can block receptor binding of reelin and subsequent signaling60 and, interestingly, ApoE expression was lost in the plasma cells examined here (Table 5). In contrast, mRNA for ApoE has been reported previously to be expressed by bone marrow plasma cells and significantly down-regulated in myeloma.42Consequently, the expression shift from ApoE to reelin upon plasma cell differentiation may be important in regulating the migration of plasma cells or an associated cell type with which plasma cells interact within distinct anatomic sites. Further analysis into reelin expression by plasma cells could provide insight into additional physiological roles of reelin and the possibility that analogous to its function in the nervous system, reelin could function in cell positioning within lymphoid organs.
Another factor up-regulated in plasma cells that plays an important role in the organization of cells within the nervous system was neuropilin-1 (Table 5),61,62 and this up-regulation was also confirmed by quantitative RT-PCR (Figure 3A and Table 1). Neuropilin-1 has also been shown to bind VEGF and modulate VEGF signaling through the VEGF receptor, KDR.63 Recently, neuropilin-1 was shown to be expressed by dendritic cells and resting T cells and to be involved in a homotypic interaction between these 2 cell types, suggesting that neuropilin-1 plays a role in the initiation of the immune response.64 Furthermore, neuropilin-1 expression was described in bone marrow stromal cell lines as well as bone marrow adherent cells.65 Collectively, these findings strongly suggest that neuropilin-1 may be involved in positioning of plasma cells within appropriate bone marrow microenvironments.
In summary, by utilizing oligonucleotide microarray analysis of primary plasma cells and B cells, we have demonstrated that plasma cells exhibit multiple genetic alterations specific to plasma cell differentiation. The plasma cells displayed a unique pattern of expression of transcription factors, signaling molecules, as well as genes involved in the regulation of proliferation and apoptosis. Additionally, the comparison of plasma cells and B cells provides important information regarding not only gene expression alterations but also genes that are selectively retained during plasma cell differentiation. In conclusion, these results provide insight into the gene expression changes underlying the mechanisms of plasma cell terminal differentiation and the factors that govern plasma cell function in vivo. Furthermore, the plasma cell profile of gene expression described here provides a solid foundation for future research into plasma cell biology.
We thank Dr Susan Winandy for critical review of this manuscript.
Prepublished online as Blood First Edition Paper, January 23, 2003; DOI 10.1182/blood- 2002-08-2673.
Supported by National Institutes of Health grant HL58710 (G.S.K.). The microarray facility at the Children's Memorial Institute of Education and Research is supported by the Falk Foundation.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Geoffrey S. Kansas, Department of Microbiology-Immunology, Northwestern Medical School, 303 E Chicago Ave, Chicago, IL 60611; e-mail: gsk@northwestern.edu.