Abstract
P57KIP2 is a cyclin-dependent kinase inhibitor silenced in a variety of human malignancies. DNA methylation of a region surrounding the transcription start site of p57KIP2 was found in acute lymphocytic leukemia (ALL)–derived cell lines. Methylation of this region correlated with gene silencing, and treatment of methylated/silenced cell lines with 5-aza-2′-deoxycytidine resulted in gene re-expression. P57KIP2 was methylated in 31 (50%) of 63 patients with newly diagnosed ALL, and in 11 (52%) of 21 patients with relapsed ALL. In 5 of them (25%), methylation was acquired at relapse. No association was observed between methylation of p57KIP2 alone and clinical-biologic characteristics studied, including overall survival (OS) or disease-free survival. Methylation of multiple genes in a cell-cycle regulatory pathway composed of p73, p15, and p57KIP2 occurred in 22% of Philadelphia chromosome (Ph)–negative patients. Ph-negative patients with methylation of 2 or 3 genes of this pathway had a significantly worse median OS compared with those with methylation of 0 or 1 gene (50 vs 467 weeks, respectively;P = .02). Our results indicate that p57KIP2 is frequently methylated in adult patients with ALL, and that inactivation of a pathway composed of p73, p15, and p57KIP2 predicts for poor prognosis in Ph-negative patients.
Introduction
Aberrant methylation of promoter-associated cytosine guanine pair (CpG) islands (DNA methylation) is an epigenetic modification of DNA frequently observed in human malignancies,1 including leukemias.2 This phenomenon is particularly prevalent in acute lymphocytic leukemia (ALL) where it has been shown that 80% of patients have methylation of at least one promoter-associated CpG island both at initial presentation3 and at relapse.4
P57KIP2 is located on chromosome 11p15, a region frequently deleted in human cancers that has been associated with the Beckwith-Wiedemann syndrome.5-7 It encodes a cyclin-dependent kinase inhibitor (CDKI) that belongs to the CIP/KIP family and is considered a putative tumor suppressor gene. Evidence for this stands from experiments demonstrating that p57KIP2 inhibits cell transformation,8 and that lack of p57KIP2 results in altered cell differentiation and proliferation in mice.7Decreased expression of p57KIP2 has been observed in different epithelial malignancies,9-12 but p57KIP2 mutations are rare.13 This suggests that epigenetic alterations, including DNA methylation, may have a role in aberrant silencing of p57KIP2. Recently, several groups have shown that p57KIP2 is aberrantly methylated in hematopoietic malignancies.14,15 In particular, Kikuchi et al14 have shown that a region in close proximity to the transcriptional start site (− 300 to + 400 base pair [bp]) of p57KIP2 is aberrantly methylated in different neoplastic cell lines, including several of hematopoietic origin, but not in nonneoplastic tissues. Treatment of methylated/silenced cell lines with 5-aza-2′-deoxycytidine, an inhibitor of DNA methyltransferases with potent hypomethylating activity,16resulted in demethylation of that specific region and subsequent gene re-expression.
To study the prevalence and clinical and biologic implications of p57KIP2 methylation in ALL, we have analyzed its methylation status in a cohort of adult ALL patients whose methylation patterns have been previously characterized.3 Our results indicate that aberrant methylation of p57KIP2 is a frequent phenomenon in adult ALL both at initial presentation and at relapse in adult ALL, and that methylation of a cell-cycle regulatory pathway involving p73, p15, and p57KIP2 identified a subgroup of patients with Philadelphia chromosome (Ph)–negative disease with poor prognosis.
Patients, materials, and methods
Cell lines
The following 8 cell lines were studied: Jurkat, CCRF-CEM, BALL1, CCRF-HSB2, Malt4, SupT1, PEER, and TALL1. They were obtained from the American Type Culture Collection (Manassas, VA) or from the Japanese Collection of Research Bioresources (Tokyo, Japan). Cells were cultured under appropriate medium conditions until harvested for DNA or RNA extraction. SupT1 and PEER were treated with 5-aza-2′-deoxycytidine (Sigma, St Louis, MO) for 96 hours prior to extracting RNA.
Patients
Table 1 summarizes patient characteristics. Pretreatment initial presentation samples were obtained from 72 patients. All patients were subsequently treated with the hyper–cyclophosphamide, vincristine, adriamycin, and dexamethosone (CVAD) chemotherapy program at M.D. Anderson Cancer Center.17 Of the 72 patients, 35 (48%) relapsed. Paired matched samples obtained at initial presentation and at relapse from 21 of these 35 patients have also been analyzed. All samples were collected following institutional guidelines.
DNA extraction and bisulfite modification
Pretreatment samples consisted of cell suspensions obtained from bone marrow aspiration procedures. Median percentage of leukemic blasts was 89% (range, 22%-99%), indicating that the cell population analyzed is representative of the predominant malignant leukemic cell clone.
DNA was extracted from these samples and cell lines using standard phenol-chloroform methods. DNA from relapsed samples, and paired initial presentation samples, was extracted from paraffin-embedded bone marrow biopsies. Prior to DNA extraction, these samples were deparaffinazed using xylene and ethanol followed by digestion with proteinase K. After extraction, DNA was modified with bisulfite. Bisulfite induces deamination of unmethylated cytosines converting unmethylated CpG sites to uracil guanine pair (UpG) without modifying methylated sites. Bisulfite treatment of genomic DNA was performed as described.18 DNA (2 μg) was used. DNA was denatured in 0.2 N NaOH at 37°C for 10 minutes and incubated with 3 M Na-bisulfite at 50°C for 16 hours. DNA was then purified using the Wizard cleanup system (Promega, Madison, WI) and desulfonated with 0.3 N NaOH at 25°C for 5 minutes. DNA was then precipitated with ammonium acetate and ethanol, washed with 70% ethanol, dried, and resuspended in H2O. Further information can be obtained from http://www3.mdanderson.org /leukemia/methylation.19
Methylation analysis
To analyze the methylation status of p57KIP2, we have used 2 methods. First, we analyzed all cell lines and patient samples with bisulfite polymerase chain reaction (PCR) followed by restriction enzyme digestion (combined bisulfite restriction analysis; COBRA).20 This method allows the quantification of the methylation level, that is the ratio of methylated (restricted) to unmethylated (unrestricted) PCR products. PCR reactions were carried in 50 μL reactions. In each reaction, 2 μL bisulfite-treated DNA was used, as well as 1.25 mM deoxynucleoside triphosphate, 6.7 mM MgCl2, 5 μL PCR buffer, 1 nmol primers, and 1 unit Taq polymerase. Table2 summarizes PCR conditions, primer sequences, and GenBank sequence numbers. Figure1A shows a map of the p57KIP2 promoter–associated CpG island indicating the position of the primers used in relation to the transcription start site. PCR products were digested with EcoRI or HhaI and were separated in nondenaturing polyacrylamide gels. Gels were stained with ethidium bromide. The proportion of methylated versus unmethylated product (digested vs undigested, respectively) was quantitated by densitometric analysis, determining the level of methylation. Densitometric analysis was performed using a BioRad Geldoc 2000 digital analyzer (Hercules, CA) equipped with the Quantity One version 4.0.3 software (BioRad, Hercules, CA).
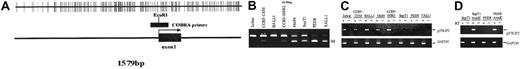
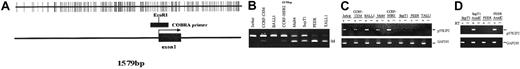
P57KIP2 methylation in ALL cell lines.
(A) Map of the promoter-associated CpG island of p57KIP2 in relation to exon 1 and its transcription start site (arrow). Each vertical line represents a CpG site. (B) Combined bisulfite restriction PCR analysis of p57KIP2 in ALL cell lines. The restricted band (M) represents the methylated allele. (C) RT-PCR analysis of p57KIP2 gene expression in methylated and unmethylated cell lines. + indicates with reverse transcriptase (RT); −, without. Bottom blots show GAPDH control. (D) RT-PCR analysis of p57KIP2 before and after 5-aza-2′-deoxycytidine (AzadC) exposure.
P57KIP2 methylation in ALL cell lines.
(A) Map of the promoter-associated CpG island of p57KIP2 in relation to exon 1 and its transcription start site (arrow). Each vertical line represents a CpG site. (B) Combined bisulfite restriction PCR analysis of p57KIP2 in ALL cell lines. The restricted band (M) represents the methylated allele. (C) RT-PCR analysis of p57KIP2 gene expression in methylated and unmethylated cell lines. + indicates with reverse transcriptase (RT); −, without. Bottom blots show GAPDH control. (D) RT-PCR analysis of p57KIP2 before and after 5-aza-2′-deoxycytidine (AzadC) exposure.
To confirm the COBRA results we performed bisulfite sequencing in unselected cases. Bisulfite PCR fragments were cloned into pCR2 vector using a TOPO-cloning kit (Invitrogen, San Diego, CA). DNA sequencing was performed using an ABI377 automated sequencer (Applied Biosystems, Foster City, CA).
Analysis of gene expression
Gene expression was analyzed using reverse transcriptase (RT)–PCR. Total RNA (5 μg) was treated for 30 minutes with DnaseI, which was inactivated with DNA Free Kit (Ambion, Austin, TX). Reverse transcription was performed using Superscript II (Life Technologies, San Diego, CA), and it was followed by PCR using conditions summarized in Table 2. Glyceraldehyd-3-phospate dehydrogenase (GAPDH) was used as control as described.22 Of these reactions, 10-μL aliquots were run in 2% agarose gels and stained with ethidium bromide.
Statistical methods
To analyze association of methylation between p57KIP2 and clinical biologic characteristics, we used the Spearman rank correlation coefficient. Variables analyzed included age, sex, white blood cell count (WBC), hemoglobin levels, platelet count, albumin levels, β2-microglobulin levels, cytogenetics, immunophenotype, and systemic and central nervous system (CNS) risk at presentation.17 Complete remission (CR) rate, disease-free survival (DFS), and overall survival (OS) have been defined elsewhere.17 Estimates of 5-year DFS and OS were based on the Kaplan-Meier method, and differences were tested using the log-rank test. To perform this analysis a sample was considered methylated for p57KIP2 or p15 if the level of methylation was 15% or higher. For p73 we used methylation-specific PCR.3,23 For this gene, a sample was considered methylated if a band was observed in the methylated PCR reaction. We used at test for dependent samples to compare methylation of paired matched samples at initial presentation and at relapse. As in a previous study,4 methylation was considered stable if it was higher or lower than 5% both at initial presentation and at relapse. Increased methylation was considered if it was lower than 5% at initial presentation, and higher than that at relapse. Decreased methylation was considered if it was higher than 5% at initial presentation and lower than that at relapse.
Results
Methylation of p57KIP2 in ALL-derived cell lines
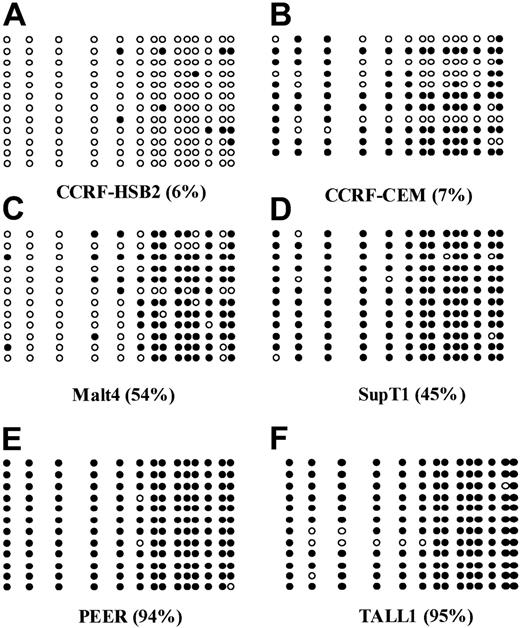
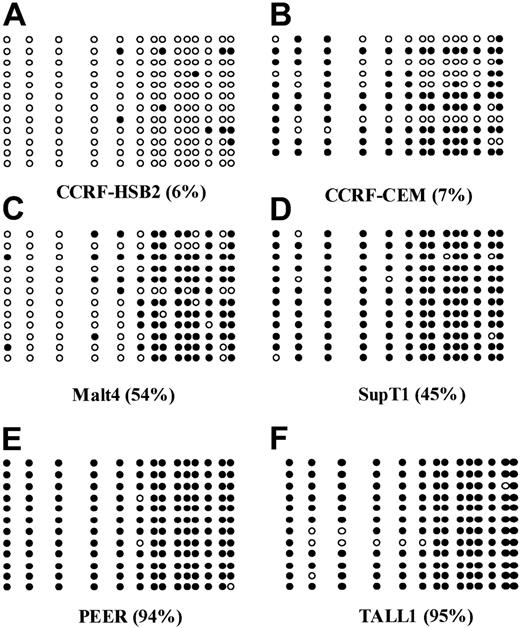
The following 8 cell lines have been studied here: Jurkat, CCRF-CEM, BALL1, CCRF-HSB2, Malt4, SupT1, PEER, and TALL1 (Figure 1B). There were 2 cell lines, Jurkat and BALL1, that had no methylation of p57KIP2. The other 6 cell lines were methylated. Median methylation level was 45.8% (range, 6%-95%). Of these 6 cell lines, 4 were densely methylated (Malt4, SupT1, PEER, and TALL1), median methylation level 65.5% (range, 28%-95%). Methylation results were confirmed by bisulfite sequencing (Figure 2). There were 4 densely methylated cell lines (TALL1, PEER, SupT1, and Malt4) that had methylation of the majority of CpG sites in the 12 alleles evaluated, whereas the 2 partially methylated cell lines, CCRF-HSB2 and CCRF-CEM, showed variable methylation.
Bisulfite sequencing of 6 ALL-derived cell lines.
(A) CCRF-HSB2; (B) CCRF-CEM; (C) Malt4; (D) SupT1; (E) PEER; and (F) TALL1. Each circle represents a CpG site. Open circles (○) indicate unmethylated CpG sites; black circles (●), methylated ones. Each row represents an allele. The numbers in parentheses indicate the level of methylation as determined by COBRA.
Bisulfite sequencing of 6 ALL-derived cell lines.
(A) CCRF-HSB2; (B) CCRF-CEM; (C) Malt4; (D) SupT1; (E) PEER; and (F) TALL1. Each circle represents a CpG site. Open circles (○) indicate unmethylated CpG sites; black circles (●), methylated ones. Each row represents an allele. The numbers in parentheses indicate the level of methylation as determined by COBRA.
As demonstrated by RT-PCR analysis, methylation was strongly associated with gene silencing (Figure 1C). Only 1 of the 4 densely methylated cells lines expressed p57KIP2, whereas the 2 unmethylated and the 2 partially methylated cell lines did. To further confirm that methylation of the region studied here was associated with gene silencing, we treated 2 densely methylated cell lines (SupT1 and PEER) with 5-aza-2′ deoxycytidine. Exposure of these 2 cell lines to this agent resulted in restoration of p57KIP2 gene expression (Figure1D).
DNA methylation of p57KIP2 in patients with ALL
We next studied p57KIP2 methylation in patients with ALL. There were 36 patients (50%) who had a p57KIP2 level of methylation higher than 5%. Mean p57KIP2 methylation was 14% (range, 0%-100%). Table 3 summarizes the range of methylation of p57KIP2. Figure 3A shows representative examples of p57KIP2 methylation at initial presentation.
p57KIP2 methylation analysis in patients with ALL.
(A) p57KIP2 methylation analysis at initial presentation. Restricted band (arrow) indicates methylated allele. The number underneath each lane represents the level of methylation. (B) Analysis of p57KIP2 methylation in paired matched samples obtained at initial presentation (c) and at relapse (d). The number underneath each lane represents the level of methylation. (C) Methylation of p21CIP1 in ALL patients. (D) Methylation analysis of p14ARF in ALL patients. None of the patients analyzed had methylation of p21CIP1 or p14ARF. (Con = control cell lines).
p57KIP2 methylation analysis in patients with ALL.
(A) p57KIP2 methylation analysis at initial presentation. Restricted band (arrow) indicates methylated allele. The number underneath each lane represents the level of methylation. (B) Analysis of p57KIP2 methylation in paired matched samples obtained at initial presentation (c) and at relapse (d). The number underneath each lane represents the level of methylation. (C) Methylation of p21CIP1 in ALL patients. (D) Methylation analysis of p14ARF in ALL patients. None of the patients analyzed had methylation of p21CIP1 or p14ARF. (Con = control cell lines).
To study the role of p57KIP2 methylation at relapse, we analyzed paired matched samples obtained at initial presentation and at relapse from 21 patients who initially had achieved CR. Thirteen patients (65%) had no change in the methylation status: 9 had no methylation (methylation level, < 5%) at both end points, and 4 were methylated (> 5%) at both end points. There were 5 patients (25%) who had increased methylation at relapse compared with initial presentation, and 2 patients (10%) had demethylation at relapse (P = .39). Figure 3B shows representative examples of p57KIP2 methylation at initial presentation and at relapse.
To confirm the results obtained with COBRA, we performed bisulfite sequencing of 9 cases (Figure 4). As in cell lines, we found an excellent concordance between COBRA and bisulfite sequencing results. There were 2 completely unmethylated cases who had methylation of none of the CpG sites involved, whereas a case that was completely methylated by COBRA was found to have all CpG sites analyzed methylated.
p57KIP2 bisulfite sequencing of ALL patients.
Each circle represents a CpG site. Empty circles (○) indicate unmethylated CpG sites; black circles (●), methylated ones. Each arrow represents a sequenced allele. The numbers on the left represent levels of methylation.
p57KIP2 bisulfite sequencing of ALL patients.
Each circle represents a CpG site. Empty circles (○) indicate unmethylated CpG sites; black circles (●), methylated ones. Each arrow represents a sequenced allele. The numbers on the left represent levels of methylation.
We also studied the methylation status of p21CIP1, another member of the CIP/KIP family, and p14ARF. Analyzed were 30 patients, none of whom had evidence of either p21CIP121 or p14ARF methylation (Figure 3C-D). Using a methylation level of 15% or methylation-specific PCR (MSP) positivity, p15 and p73 were methylated in 16 patients, respectively. No association was found between methylation of p73, p15, and p57KIP2. There were 33 patients (45%) who had methylation of 0 genes; 28 (39%), of 1 gene; 8 (11%), of 2 genes; and 3 (4%), of 3 genes.
Correlation between methylation of p57KIP2 and gene expression in ALL patients
To assess the impact of p57KIP2 methylation on gene expression, we performed RT-PCR using RNA extracted from cell suspensions obtained from bone marrow aspiration procedures obtained at the time of initial presentation. Analyzed were 11 patients. Of the 6 patients with methylated p57KIP2, only 1 expressed p57KIP2. In contrast, 4 of 5 unmethylated cases expressed p57KIP2 (P = .03) (Figure5).
RT-PCR analysis of ALL patients.
+ indicates with reverse transcriptase (RT); −, without RT. Bottom blots show GAPDH control.
RT-PCR analysis of ALL patients.
+ indicates with reverse transcriptase (RT); −, without RT. Bottom blots show GAPDH control.
Clinical and pathologic correlates
To assess the clinical impact of p57KIP2 methylation, we have analyzed the correlation of p57KIP2 methylation with the following variables: age, sex, WBC, hemoglobin levels, platelet count, albumin levels, β2-microglobulin levels, cytogenetics, immunophenotype, and systemic and CNS risk at presentation.17 No association was observed between any of these variables and p57KIP2 methylation. We have also analyzed the impact of p57KIP2 on DFS and OS. No differences were observed among patients with or without p57KIP2 methylation.
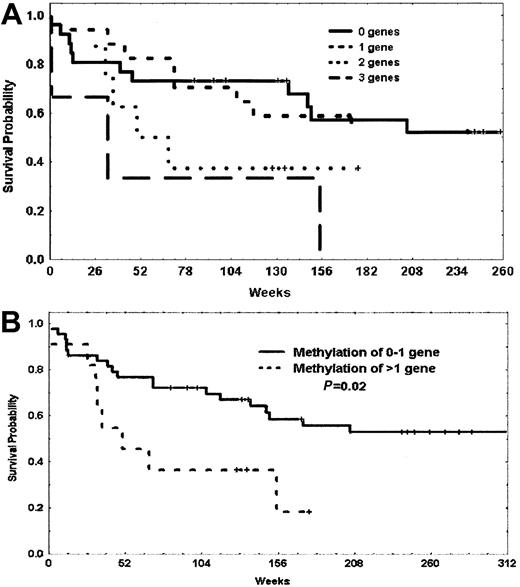
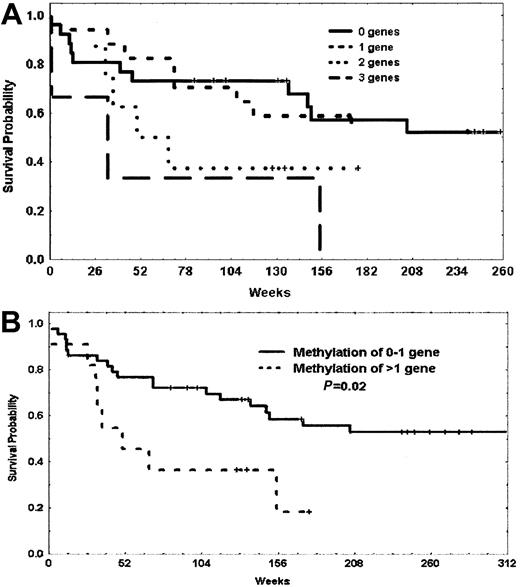
In a previous analysis,3 we observed a trend toward a worse prognosis in patients with methylation of p73 and p15. Because p57KIP2, p73, and p15 are all involved in cell-cycle regulation, we decided to investigate the prognostic impact of methylation of these 3 genes in our patients. Table 4 summarizes the impact of methylation of patients with methylation of 0 to 1 gene or 2 to 3 genes of this triad of p57KIP2, p73, and p15 including and excluding patients with Ph-positive disease. Although both OS and DFS were always worse for patients with methylation of at least 2 genes, this reached statistical significance for the subgroup of patients with Ph-negative disease. The median OS for patients with methylation of 0 to 1 gene was 467 weeks, whereas it was 50 weeks for those with methylation of 2 to 3 genes (P = .023) (Figure6). This last subgroup accounted for 22% of Ph-negative patients.
Probability of survival of patients with Philadelphia chromosome–negative ALL based on methylation of p73, p15, and p57KIP2.
(A) Probability of survival of patients with methylation of 0, 1, 2, or 3 genes. (B) Probability of survival of patients with methylation of 0 or 1 gene versus methylation of 2 or more genes. Vertical marks represent censored data.
Probability of survival of patients with Philadelphia chromosome–negative ALL based on methylation of p73, p15, and p57KIP2.
(A) Probability of survival of patients with methylation of 0, 1, 2, or 3 genes. (B) Probability of survival of patients with methylation of 0 or 1 gene versus methylation of 2 or more genes. Vertical marks represent censored data.
Discussion
We have identified that aberrant methylation of p57KIP2 is a common abnormality in ALL. First, we showed that this abnormality is frequent in ALL-derived cell lines. Methylation of a region in close proximity to the transcription start site of p57KIP2 was associated with gene silencing. Treatment of methylated/silenced ALL cell lines with a hypomethylating agent resulted in gene re-expression. As was originally found by Kikuchi et al,14 this finding strongly supports the notion that methylation of the promoter region studied here is important for the regulation of transcriptional activity of p57KIP2.
Further, we have found that methylation of p57KIP2 occurs in 50% of adult patients with ALL, with 30% of them having dense methylation (> 20%). The results obtained with the bisulfite PCR method were confirmed in a subset of patients by bisulfite sequencing. This last method allows the analysis of the methylation status of each CpG site in a given DNA sequence.18 As was shown in cell lines, methylation of the region studied here inversely correlated with the level of gene expression. This evidence strongly suggests that methylation of the region analyzed here is important for the control of p57KIP2 gene expression also in vivo. It is important to note that several lines of evidence indicate that p57KIP2 is an imprinted gene,5,6 but the role of promoter DNA methylation in the control of imprinting is unknown. Indeed, the evidence shown here and that of 2 other groups14 15 indicate that p57KIP2 is expressed in nonneoplastic tissues and argue against any role of DNA methylation in the regulation of p57KIP2 imprinting.
It has been proposed that aberrant methylation of target genes may have a role in resistance/relapse mechanisms in leukemia.2 We had previously studied the methylation patterns at initial presentation and at relapse in a cohort of patients with ALL.4 Our results demonstrated that methylation patterns were stable in a majority of patients, but that a significant minority, 28%, acquired methylation changes at relapse, affecting, in particular, cell-cycle regulatory genes such as p15 and p16. Methylation of p57KIP2 followed the same pattern as described for p16 and p15,4 remaining stable in 65% of patients, increasing in 25%, and decreasing in 10%.
We had previously observed a trend toward a worse prognosis for patients with methylation of both p73 and p15.3 p73 is a p53 family member that is frequently methylated and silenced in lymphoid malignancies.23,24 By acting as a p53 homologue, activation of p73 may lead toward cell-cycle arrest.25This phenomenon is mediated in part by CDKIs, such as p57KIP2.25,26 Indeed, p57KIP2 has been shown to be up-regulated by p73.27 Based on this knowledge, we studied the impact of methylation of p73, p15, and p57KIP2 in this cohort of homogeneously treated ALL patients.17 Results of this analysis revealed that methylation of 2 or 3 genes of this triad occurs in close to 25% of patients with Ph-negative disease, and that patients with this abnormality had a very poor prognosis. It could be argued that this finding is an artifact based on arbitrary gene selection. Several facts argue against this. First is the lack of an association between methylation of these 3 genes (data not shown), and the fact that other genes, including p14ARF, p21CIP1,20and p16,3 that are also involved in this cell-cycle regulatory pathway are not methylated in these patients. Second is the need for methylation of more than one gene to alter the prognosis of these patients. This cooperative effect could be explained by the redundancy in the function of these genes,25,26 and is also supported by the lack of a fully malignant phenotype in animal models in which these genes were individually deleted.7,28 29 This observation suggests that at least 2 of these genes are required to induce a more aggressive disease phenotype.
The implications of these findings are several. First, methylation of p57KIP2 characterizes a molecular pathway prevalently altered in adult ALL patients. Second, methylation of 2 or 3 genes of this pathway may allow the identification of a subgroup of patients with Ph-negative ALL and extremely poor prognosis analogous to Ph-positive disease. It should be noted that due to the relatively small sample size, these results should be considered preliminary and need to be confirmed in a larger cohort of patients. The early identification of these patients may allow the development of targeted therapies for this subgroup of patients. These could include the incorporation of hypomethylating agents, such as 5-aza-2′deoxycytidine, to chemotherapy programs in ALL, and the introduction of early allogeneic stem cell transplantation in first complete remission for these high-risk patients, as is recommended for patients with Ph-positive disease.
In summary, we have identified methylation of p57KIP2 as a common abnormality in adult patients with ALL, and a cell-cycle regulatory pathway involving p73, p15, and p57KIP2 that when methylated confers poor prognosis to patients with Ph-negative disease.
Prepublished online as Blood First Edition Paper, February 13, 2003; DOI 10.1182/blood-2002-08-2466.
G.G.-M. is the recipient of a Career Development Award from the American Society of Clinical Oncology and the Physician-Scientist Program Award. M.T. is supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sport, Science, and Technology of Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Guillermo Garcia-Manero, Department of Leukemia, M.D. Anderson Cancer Center, Box 428, 1515 Holcombe Blvd, Houston, TX, 77030; e-mail: ggarciam@mdanderson.org.