Abstract
Current immunotherapeutic trials for patients with multiple myeloma (MM) focus on the idiotype (Id) as a tumor-specific antigen for active immunization. To bypass the need for the identification of shared MM-associated antigens and the characterization of possible immunogenic T-cell epitopes in a human leukocyte antigen (HLA) type–restricted manner, we focused on myeloma RNA transfection of dendritic cells (DCs). Total RNA encodes the whole antigen content of tumor cells, therefore allowing the transfected DCs to process and present the most relevant peptides and to induce a possible polyclonal cytotoxic T lymphocyte (CTL) response against different immunogenic antigens. We transfected monocyte-derived DCs with total RNA from the myeloma cell lines LP-1 and U266 by electroporation and investigated the potential of these DCs to induce myeloma-specific CTLs. We show that RNA-transfected DCs induce CTLs that lyse the LP-1 and U266 myeloma cells in an antigen-specific and major histocompatibility complex (MHC) class I–restricted manner, as demonstrated by cold-target inhibition and antibody-blocking studies. Interestingly, LP-1–specific CTLs showed no specificity for the idiotype. Consistent with studies demonstrating mucin 1 (MUC1) as a myeloma-associated antigen, we found MUC1 specificity of the CTLs induced with U266-derived RNA. As corresponding epitopes, we tested the described peptides M1.1 and M1.2 and found a striking fine specificity for M1.2, assuming a possible immunodominance of this peptide. This is the first report on the induction of myeloma-specific CTLs by RNA transfection of DCs.
Introduction
Immunotherapy has developed from a theoretical concept for the treatment of malignant disease to a feasible and promising means of tumor-specific therapy. Over the last decade, many strategies to induce tumor-specific immunity in the tumor-bearing host have been investigated, and several approaches have proved to be successful enough in vitro to lead to the initiation of clinical phase 1/2 trials.1-3 In multiple myeloma (MM), various investigators have focused on the myeloma-specific idiotype (Id) representing an individual, and therefore a truly specific target for immunotherapy. Unfortunately, the high immunological and clinical response rates to active Id vaccination seen in low-grade B-cell non-Hodgkin lymphoma4-6 have so far not been found in early clinical trials of Id vaccination in myeloma patients. Although investigators found mostly T-helper 1 (TH1) responses in Id/granulocyte-macrophage colony-stimulating factor (GM-CSF)–vaccinated patients, only single individuals were reported to develop a significant reduction of their monoclonal Id in the serum.7 Others described a high rate of Id-specific T-cell reactivity as measured by delayed-type hypersensitivity but were not able to find measurable tumor mass reduction.8 Even with the extended use of antigen-presenting cells, particularly dendritic cells (DCs) with the unique and very potent capacity to induce antigen-specific T-cell responses in naive T cells, modest success rates of Id-specific immune responses have been reported.9-11 These idiotype-based vaccine approaches in MM have so far not been able to significantly improve the clinical outcome as measured by prolongation of the time to disease progression.
For the number of known tumor-associated antigens (TAAs) in myeloma, no data have so far been published on active immunization strategies. Owing to the broader applicability of shared TAAs as compared with individual antigens such as Id, active investigation is under way to identify new immunological targets. Certainly, RNA array technology will substantially increase the number of potential targets in MM.12
Whole tumor–cell strategies that avoid the need for a defined TAA, including fusions of DCs with tumor cells, have been investigated recently in breast carcinoma models and very recently in a murine MM model system.13,14 The clinical applicability of this approach is being hampered by the need for a surgical procedure to obtain enough tumor cells for vaccine production. Another possibility allowing for the induction of unidentified tumor-specific or tumor-associated targets is the concept of tumor RNA transfection of DCs to induce tumor-specific T-cell responses. Studies in mouse models have shown that vaccination with RNA derived from plasmids or tumor cells elicits tumor protection and therapeutical benefit.15,16 Gilboa and colleagues have pioneered this approach in the human system and have successfully delivered RNA into DCs in different cancer systems. They have clearly shown the potential of transfected DCs to stimulate tumor antigen–specific cytotoxic T lymphocytes (CTLs) in colon, prostate, renal, and cervical cancers.17-23 We report here on the induction of myeloma-specific CTLs by transfection of major histocompatibility complex (MHC) class I–matched DCs with total myeloma RNA.
Materials and methods
Tumor cell lines
The following cell lines were purchased from ATCC (Manassas, VA): U266 (multiple myeloma), MCF-7 (breast cancer), A498 (renal cancer), and SK-OV-3 (ovarian cancer). LP-1 (multiple myeloma) and K562 (human chronic myeloid leukemia in blast crisis) were from DSMZ (Braunschweig, Germany), and Croft is an Epstein-Barr virus (EBV)–immortalized B-cell line and was kindly provided by O. J. Finn (Pittsburgh, PA). All tumor cell lines were maintained in RP-10 medium (RPMI 1640 with 2 mM glutamine [Invitrogen, Karlsruhe, Germany] supplemented with 10% heat-inactivated fetal calf serum [Invitrogen] and antibiotics).
Cell isolation and generation of DCs from adherent peripheral blood mononuclear cells (PBMNCs)
DCs were generated from peripheral blood monocytes as described previously.24-26 In brief, PBMNCs were isolated by Ficoll/Paque (Biochrom, Berlin, Germany) density gradient centrifugation of buffy coats from healthy volunteers of the blood bank of the University of Tübingen (Germany). Cells were plated in 6-well plates (Becton Dickinson Falcon, Heidelberg, Germany) at 1 × 107 cells per well in RP-10 medium in a final volume of 3 mL. After 2 hours of incubation at 37°C, nonadherent cells were removed, and the adherent blood monocytes were cultured in RP-10 medium supplemented with 100 ng/mL human recombinant GM-CSF (Leukomax; Novartis, Nuremberg, Germany) and 1000 IU/mL interleukin 4 (IL-4) (R & D Systems, Wiesbaden, Germany). For maturation of the DCs, 10 ng/mL tumor necrosis factor–α (TNF-α) (R & D Systems) was added to the culture on day 6.
DCs used as targets in the 51Cr-release assay were loaded with the purified LP-1 idiotype protein on days 1 and 5 of the culture to a final concentration of 50 μg/mL.
RNA isolation
Total RNA was isolated from tumor cells by means of the RNeasy Maxi Kit (QIAGEN, Hilden, Germany) according to the protocol for isolation of total RNA from animal cells provided by the manufacturer. Quantity and purity of RNA was determined by ultraviolet (UV) spectrophotometry. RNA samples were stored at −80°C in small aliquots.
RNA transfection of DCs with electroporation
At day 6 of culture, immature DCs were harvested, washed twice with serum-free X-VIVO 20 medium (BioWhittaker, Walkersville, MD), and resuspended to a final concentration of 2 × 107 cells per milliliter. Then, 200 μL cell suspension was mixed with 20 μg total RNA or 10 μg enhanced green fluorescence protein in vitro transcript (EGFP IVT) and electroporated in a 4-mm cuvette by means of an Easyject Plus electroporator (Peqlab, Erlangen, Germany). The physical parameters were as follows: voltage of 300 V, capacitance of 150 μF, resistance of 1540 Ω, and pulse time of 231 milliseconds. After electroporation, the cells were immediately transferred into RP-10 medium and returned to the incubator.
Induction of tumor-specific CTLs using DCs transfected with tumor RNA
DCs were transfected with LP-1 or U266 total RNA by electroporation on day 6 of DC culture. After transfection, DCs were incubated for 24 hours in RP-10 medium containing 10 ng/mL TNF-α for maturation. For CTL induction, 5 × 105 RNA-transfected DCs were incubated with 2.5 × 106 autologous PBMNCs in RP-10 medium. An aliquot of RNA-transfected DCs was stored at −80°C for later restimulation. After 7 days of culture, cells were restimulated with autologous RNA-transfected DCs and cultured with further addition of 4 ng/mL human recombinant IL-2 (R & D Systems) on days 1 and 3. The cytolytic activity of induced CTLs was analyzed on day 5 after restimulation by a standard 51Cr-release assay.
CTL assay
The standard 51Cr-release assay was performed as described.24 For peptide loading, tumor cells were incubated with 50 μg/mL peptide for 2 hours at 37°C. Idiotype-pulsed or unpulsed DCs used as targets were harvested on day 7 of the culture. Target cells were labeled with 51Cr–sodium chromate in RP-10 medium for 1 hour at 37°C. Then, 1 × 104 target cells per well were mixed with a varying number of effector cells in 96-well round-bottom plates in a final volume of 200 μL. After 4 hours of incubation at 37°C, 50 μL supernatants per well were transferred to a luma plate (Lumac*LSV, Groningen, The Netherlands) and counted in a β-plate counter (Wallac-ADL, Freiburg, Germany). The percentage of specific lysis was calculated as follows: 100 × (experimental release − spontaneous release/maximal release − spontaneous release). Negative specific lysis values (up to −8%) were depicted as 0% specific lysis. Maximal release was determined by adding 1% Triton X-100. Spontaneous release was lower than 31% of the maximum51Cr uptake.
Cold-target inhibition assay and blocking studies
The antigen specificity of tumor cell lysis was further assessed in a cold-target inhibition assay by the addition of a 20-fold excess of unlabeled target cells to labeled target cells. MHC restriction was confirmed by blocking with an anti–MHC class I monoclonal antibody (clone W6/32). An isotype-matched irrelevant antibody (Jackson Immuno Research, West Grove, PA) was used as a control.
Synthetic peptides
The MUC1–derived peptides M1.1 (amino acids 950-958: STAPPVHNV) and M1.2 (amino acids 12-20: LLLLTVLTV) and the control peptide human immunodeficiency virus (HIV) (polymerase HIV-1 reverse-transcriptase peptide; amino acids 476-484: ILKEPVHGV) were synthesized with the use of standard N-(9-fluorenyl)methoxycarbonyl (Fmoc) chemistry on a peptide synthesizer (model 432A; Applied Biosystems, Weiterstadt, Germany) and were analyzed by reverse-phase high-performance liquid chromatography (HPLC) and mass spectrometry.
Isolation of idiotype protein
To isolate the secreted idiotype protein of the LP-1 cell line, cells were grown in large numbers, and the supernatant was collected. The immunoglobulin Gλ (IgGλ) idiotype protein was purified from the supernatant by protein A chromatography as previously described.9 The purity of the Id isolate was confirmed in an isotype enzyme-linked immunosorbent assay (ELISA) system, and no bovine immunoglobulin contamination could be detected in a species-specific ELISA.
Results
RNA isolation and transfection of DCs
Total RNA of the myeloma cell lines LP-1 and U266 was isolated by standard methodology. Typically, more than 250 μg total RNA could be harvested from 1 × 108 cells growing in log phase. Immature DCs were transfected by electroporation with purified total RNA by an optimized protocol27 yielding a transfection efficiency of up to 29%.
DCs transfected with LP-1 myeloma RNA induce LP-1–specific CTL responses
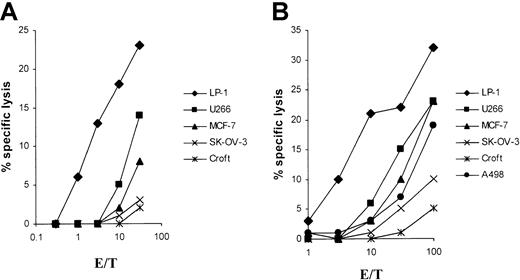
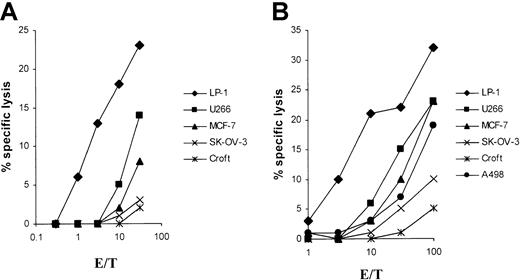
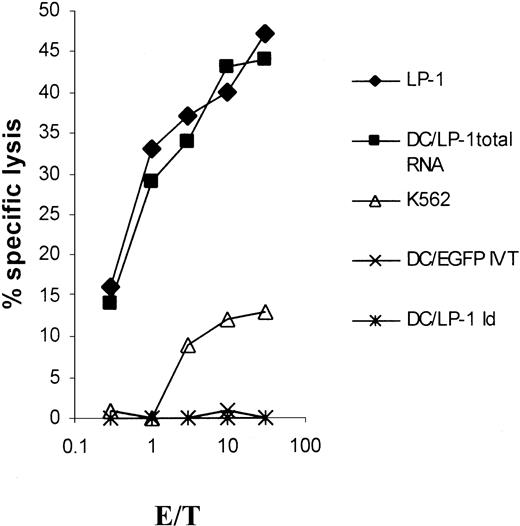
For the in vitro induction of LP-1 (human leukocyte antigen–A3/24 [HLA-A3/24], B7/18, Cw7,−)–specific CTLs, we generated an MHC class I–matched DC source from 2 different HLA-A3/24 buffy coats for RNA transfection. Already after a single in vitro restimulation with LP-1 RNA–transfected autologous DCs, specific lysis of native LP-1 myeloma target cells, could be detected in a51Cr-release assay with both CTL lines studied (Figure1). No significant cytolytic activity was seen with the use of the control cell lines SK-OV-3 and Croft. Despite HLA class I disparity, one CTL line induced by LP-1 RNA lysed the control renal and breast cancer cell lines (A498 and MCF-7), but mostly at a high effector-to-target (E/T) ratio as shown in Figure1B. Both CTL lines lysed the control myeloma cell line U266 at a lower level, indicating the potential existence of shared myeloma antigens in our system.
Specific lysis of the LP-1 cell line by CTLs induced by DCs transfected with LP-1–derived RNA.
DCs were generated from adherent PBMNCs of an HLA-A3/24, B62/50, Cw9/6 (panel A) and an HLA-A3/24, B35/60, Cw3/4 (panel B) buffy coat in RP-10 medium supplemented with GM-CSF and IL-4 and electroporated on day 6 with total RNA of the LP-1 cell line. After a 24-hour culture with TNF-α as maturation stimulus, the transfected DCs were used for the induction of CTLs. At 5 days after the first restimulation, the cytolytic activity of the cells was determined in a standard51Cr-release assay, with the use of the myeloma cell lines LP-1 (HLA-A3/24, B7/18, Cw7,−) and U266 (HLA-A2/3, B7/40, Cw3/7); the breast cancer cell line MCF-7 (HLA-A2,−, B18/44, Cw5,−); the ovarian cancer cell line SK-OV-3 (HLA-A3/68, B18/35); the renal cancer cell line A498 (HLA-A2,−, B8,−); and the EBV-immortalized B-cell line Croft (HLA-A2,−, B7/8, Cw7,−) (loaded with the HIV peptide) as targets.
Specific lysis of the LP-1 cell line by CTLs induced by DCs transfected with LP-1–derived RNA.
DCs were generated from adherent PBMNCs of an HLA-A3/24, B62/50, Cw9/6 (panel A) and an HLA-A3/24, B35/60, Cw3/4 (panel B) buffy coat in RP-10 medium supplemented with GM-CSF and IL-4 and electroporated on day 6 with total RNA of the LP-1 cell line. After a 24-hour culture with TNF-α as maturation stimulus, the transfected DCs were used for the induction of CTLs. At 5 days after the first restimulation, the cytolytic activity of the cells was determined in a standard51Cr-release assay, with the use of the myeloma cell lines LP-1 (HLA-A3/24, B7/18, Cw7,−) and U266 (HLA-A2/3, B7/40, Cw3/7); the breast cancer cell line MCF-7 (HLA-A2,−, B18/44, Cw5,−); the ovarian cancer cell line SK-OV-3 (HLA-A3/68, B18/35); the renal cancer cell line A498 (HLA-A2,−, B8,−); and the EBV-immortalized B-cell line Croft (HLA-A2,−, B7/8, Cw7,−) (loaded with the HIV peptide) as targets.
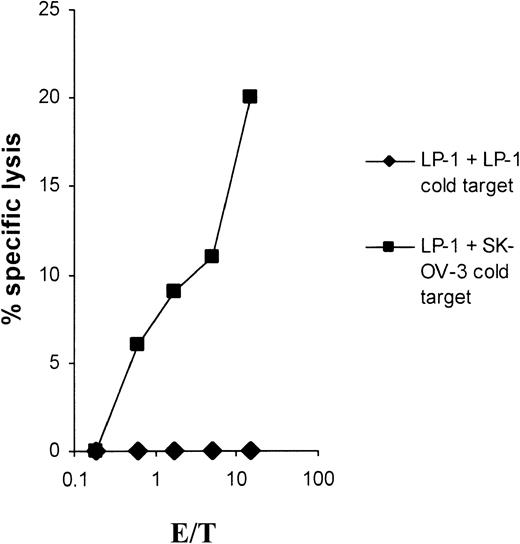
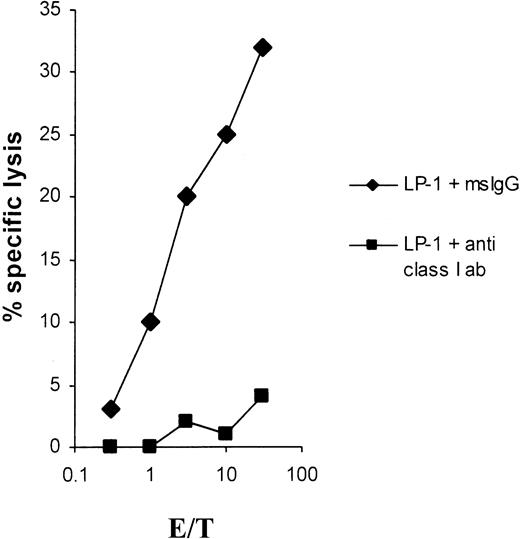
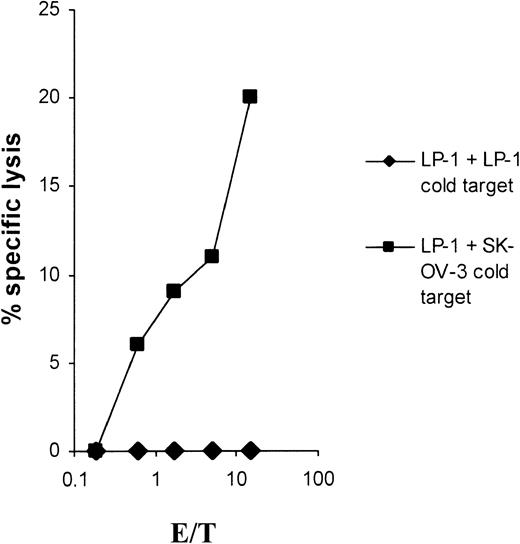
As shown in Figure 2, the lytic activity could be completely blocked in a cold-target inhibition assay emphasizing the specificity of the induced CTLs. Furthermore, the cytolysis was fully MHC class I restricted, as shown in Figure3.
Cold-target inhibition results.
Cold-target inhibition proves the specificity of the LP-1–specific CTLs. The myeloma cell line LP-1 was used as a target in a 51Cr-release assay. The antigen specificity of the CTLs was tested in the presence of unlabeled LP-1 or SK-OV-3 tumor cells as cold targets, with an inhibitor-target ratio of 20:1.
Cold-target inhibition results.
Cold-target inhibition proves the specificity of the LP-1–specific CTLs. The myeloma cell line LP-1 was used as a target in a 51Cr-release assay. The antigen specificity of the CTLs was tested in the presence of unlabeled LP-1 or SK-OV-3 tumor cells as cold targets, with an inhibitor-target ratio of 20:1.
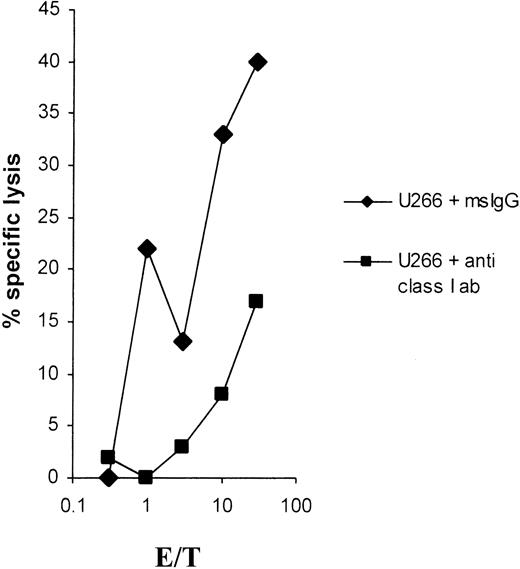
Effect of anti–MHC class I antibodies on LP-1 lysis.
Lysis of the myeloma cell line LP-1 by CTLs induced with LP-1 total RNA is blocked by anti–MHC class I antibodies. LP-1 cells were51Cr-labeled for 1 hour, washed, and then incubated for 30 minutes at 37°C with an anti–MHC class I antibody (clone W6/32) or with a mouse IgG isotype control antibody at a final concentration of 50 μg/mL and used as targets in a 51Cr-release assay.
Effect of anti–MHC class I antibodies on LP-1 lysis.
Lysis of the myeloma cell line LP-1 by CTLs induced with LP-1 total RNA is blocked by anti–MHC class I antibodies. LP-1 cells were51Cr-labeled for 1 hour, washed, and then incubated for 30 minutes at 37°C with an anti–MHC class I antibody (clone W6/32) or with a mouse IgG isotype control antibody at a final concentration of 50 μg/mL and used as targets in a 51Cr-release assay.
LP-1–specific CTLs induced by RNA-transfected DCs are not Id specific
We further analyzed whether LP-1 idiotype–specific cytotoxicity added to the reactivity of LP-1–specific CTLs. Autologous immature DCs were pulsed with purified LP-1 idiotype and matured by the addition of TNF-α for the last 24 hours before labeling with 51Cr. In the same experiment, native LP-1 targets and autologous DCs transfected with LP-1 total RNA or EGFP IVT were used as controls. While the native LP-1 myeloma cells and LP-1 RNA–transfected DCs were killed, autologous Id-pulsed DCs did not add to the lysis observed (Figure4). Under the presumption that Id protein processing and presentation were successful,28 the LP-1–specific cytolytic activity was not Id specific. No cytolysis toward the NK cell–sensitive K562 target cells was found.
Idiotype nonspecificity of LP-1–specific CTLs.
DCs were generated from adherent PBMNCs of an HLA-A3/24, B44/60, Cw3/5 buffy coat in RP-10 medium supplemented with GM-CSF and IL-4. On days 1 and 5, the isolated idiotype of the LP-1 cells was given to the culture at a final concentration of 50 μg/mL, before the addition of TNF-α on day 6. After a culture period of 7 days, the Id-pulsed DCs as well as LP-1 myeloma cells and natural killer (NK)–sensitive K562 cells were used as targets in a 51Cr-release assay. Autologous DCs were transfected with LP-1 total RNA or control RNA (EGFP IVT) on day 6 by electroporation and used on day 7 as additional control.
Idiotype nonspecificity of LP-1–specific CTLs.
DCs were generated from adherent PBMNCs of an HLA-A3/24, B44/60, Cw3/5 buffy coat in RP-10 medium supplemented with GM-CSF and IL-4. On days 1 and 5, the isolated idiotype of the LP-1 cells was given to the culture at a final concentration of 50 μg/mL, before the addition of TNF-α on day 6. After a culture period of 7 days, the Id-pulsed DCs as well as LP-1 myeloma cells and natural killer (NK)–sensitive K562 cells were used as targets in a 51Cr-release assay. Autologous DCs were transfected with LP-1 total RNA or control RNA (EGFP IVT) on day 6 by electroporation and used on day 7 as additional control.
DCs transfected with U266 RNA induce U266-specific CTL responses
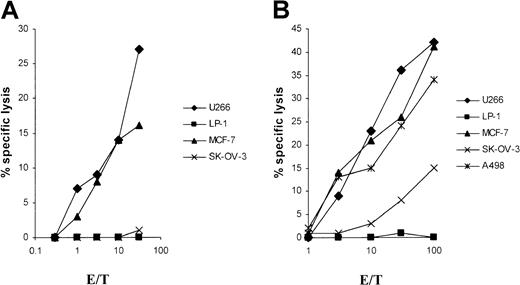
In the next set of experiments, we analyzed the feasibility of RNA transfection using the U266 myeloma cell line. DCs from 2 HLA-A2+ donors were generated for U266 (HLA-A2/3, B7/40, Cw3/7) RNA transfection. RNA-transfected DCs allowed the induction of U266-specific cytolytic T cells (Figure5). The control tumor cell lines, which matched in 1 or 2 HLA class I alleles, were tested for cross-reactivity. No cytolysis against the control myeloma cell line LP-1 was detected despite the sharing of 2 class I alleles: A3 and Cw7. Cytolysis at a significantly lower level was seen when SK-OV-3 cells were used as target cells in 1 of 2 experiments (Figure 5B). The induced CTLs, however, conferred lysis of the MCF-7 and A498 cell lines at levels comparable to that of U266. It could be speculated that, in these experiments, CTLs were generated that are preferentially HLA-A2 restricted, a characteristic that is shared by U266, A498, and MCF-7.
Induction of specific lysis of the U266 cell line by CTLs by the use of U266 RNA–transfected DCs.
Monocyte-derived DCs of an HLA-A2,−, B18/51, Cw7,− (panel A) and an HLA-A2/3, B35/8, Cw4,− (panel B) buffy coat, transfected with total RNA of the U266 cell line, were used as antigen-presenting cells (APCs) for the induction of CTLs. Cytolytic activity of the CTLs was determined in a standard 51Cr-release assay on day 5 after the first restimulation. Myeloma cell lines LP-1 (HLA-A3/24, B7/18, Cw7,−) and U266 (HLA-A2/3, B7/40, Cw3/7) and the tumor cell lines MCF-7 (HLA-A2,−, B18/44, Cw5,−), SK-OV-3 (HLA-A3/68, B18/35), and A498 (HLA-A2,−, B8,−) were used as targets.
Induction of specific lysis of the U266 cell line by CTLs by the use of U266 RNA–transfected DCs.
Monocyte-derived DCs of an HLA-A2,−, B18/51, Cw7,− (panel A) and an HLA-A2/3, B35/8, Cw4,− (panel B) buffy coat, transfected with total RNA of the U266 cell line, were used as antigen-presenting cells (APCs) for the induction of CTLs. Cytolytic activity of the CTLs was determined in a standard 51Cr-release assay on day 5 after the first restimulation. Myeloma cell lines LP-1 (HLA-A3/24, B7/18, Cw7,−) and U266 (HLA-A2/3, B7/40, Cw3/7) and the tumor cell lines MCF-7 (HLA-A2,−, B18/44, Cw5,−), SK-OV-3 (HLA-A3/68, B18/35), and A498 (HLA-A2,−, B8,−) were used as targets.
The cytotoxic activity of the U266 RNA–induced CTLs could be blocked by means of a pan–HLA-class I monoclonal antibody, demonstrating that the U266-specific killing was typically class I restricted (Figure6).
Blockage of lysis of the myeloma cell line U266 by anti–MHC class I antibodies.
For blocking experiments, 51Cr-labeled U266 tumor cells were incubated for 30 minutes with an anti–MHC class I antibody (clone W6/32) or a mouse IgG isotype control antibody at a final concentration of 50 μg/mL.
Blockage of lysis of the myeloma cell line U266 by anti–MHC class I antibodies.
For blocking experiments, 51Cr-labeled U266 tumor cells were incubated for 30 minutes with an anti–MHC class I antibody (clone W6/32) or a mouse IgG isotype control antibody at a final concentration of 50 μg/mL.
To further evaluate the fine specificity of the cytolytic activity induced by U266 RNA transfection of DCs, we investigated the antigenic specificity of these CTLs.
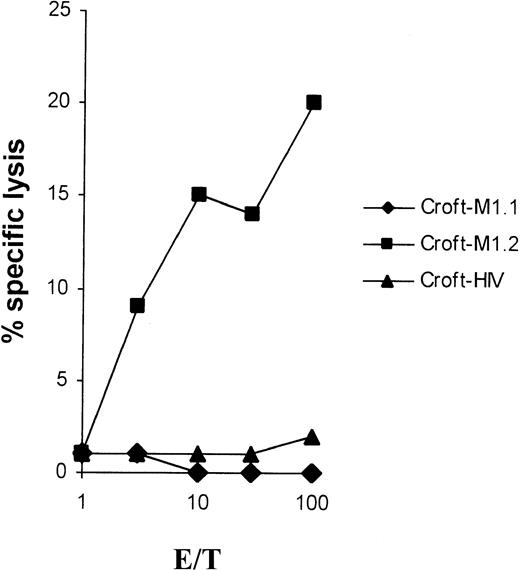
We and others have previously shown that breast and MM cancer cells express MUC1 and are recognized by MUC1 peptide–specific CTLs,29 suggesting that the cross-reactivity observed could be directed against MUC1 epitopes. Because U266 constitutively expresses MUC1, we investigated whether the RNA-induced CTLs were MUC1 specific. Using HLA-A2+ MUC1− target cells (Croft), we found a reproducible specificity of the U266 RNA–induced CTLs for the MUC1-derived peptide M1.2 (Figure7). However, no killing was found when the M1.1 peptide was used, indicating a preferential induction of CTL responses to a single peptide derived from a TAA when total myeloma RNA–transfected DCs are used as stimulators of CTLs.
U266-specific CTL reactivity against the MUC1-derived peptide M1.2.
To analyze the fine specificity of the U266-specific CTLs, Croft cells (EBV-immortalized B-cell line, MUC1−, HLA-A2+) were pulsed for 2 hours with 50 μg/mL MUC1-derived peptides M1.1 or M1.2, and used as target cells in a standard51Cr-release assay. As a negative control, Croft cells pulsed with the HIV peptide were used. Similar results were obtained in 2 separate experiments.
U266-specific CTL reactivity against the MUC1-derived peptide M1.2.
To analyze the fine specificity of the U266-specific CTLs, Croft cells (EBV-immortalized B-cell line, MUC1−, HLA-A2+) were pulsed for 2 hours with 50 μg/mL MUC1-derived peptides M1.1 or M1.2, and used as target cells in a standard51Cr-release assay. As a negative control, Croft cells pulsed with the HIV peptide were used. Similar results were obtained in 2 separate experiments.
Discussion
We show in our study for the first time that myeloma RNA–transfected DCs can induce anti–myeloma-specific CTLs. RNA-transfected DCs process the encoded proteins and present the most relevant peptides in the context of class I and class II molecules. For defined TAAs, investigators have shown that mRNA-transfected DCs are as effective in their ability to induce antigen-specific CTLs in vitro as DCs loaded with antigen-derived peptides.15,18,20 A lower level of peptide surface concentration on DCs after RNA transfection as compared with concentrations achieved by exogenous peptide pulsing does not appear to limit the efficacy of CTL induction and may even favor the induction of higher-affinity CTLs, resulting in a higher specific tumor cell cytotoxicity. Given that the RNA-transfected DCs express the whole antigen repertoire of the tumor cell encoded by RNA, the induced CTLs may represent a polyclonal population with potentially different antigen specificities and therefore higher antitumor reactivity. Different studies investigated defined TAAs in human prostate and renal cancer and found an increased antitumor activity when CTLs were induced by DCs transfected with unfractionated tumor RNA as compared with DCs transfected with RNA of individual antigens such as prostate-specific antigen or telomerase reverse transcriptase (TERT).21-23
In our study, total RNA derived from the myeloma cell lines LP-1 and U266 was transfected into DCs by electroporation, and CTLs specific for these myeloma cell lines were generated. Interestingly, LP-1–specific CTLs failed to reveal idiotype specificity. Different parameters can influence the immunogenicity of idiotype peptide epitopes: for example, a low rate of protein processing by DCs or weak MHC-binding affinities of the resulting peptides. Successful induction of Id-specific CTLs with idiotype protein–pulsed DCs has been reported in vitro and has even been found in vivo.9,30,31 The corresponding CTL epitopes, however, have not been described so far. A systematic bioinformatically supported approach to search for HLA-A2– and HLA-A1–restricted T-cell epitopes in the Id-variable region of patients with B-cell malignancies revealed only a small number of epitopes with high MHC-binding affinity, and these were located mainly in the conserved framework regions of the Id.32 In our experiments, Id epitopes may be presented on the DC surface but are possibly masked by the presence of immunodominant epitopes of other tumor-associated antigens, resulting in the suppression of T-cell responses to the nondominant epitopes. Some dominant shared antigens might exist between LP-1 and U266 myeloma cells, as can be speculated by the results shown in Figure 1.
It has been shown that the tumor-associated antigen MUC1 is present on myeloma cell lines as well as on malignant plasma cells from myeloma patients.29 33-35 Within the presumably polyclonal U266 CTL reactivity responsible for the strong overall lysis, we could detect a significant reactivity of the U266 (MUC1+, HLA-A2+)–specific CTLs induced by tumor RNA–transduced DCs against the MUC1 antigen, specifically against the HLA-A2–restricted peptide M1.2. This finding is surprising for its fine specificity, but could explain the reactivity of these CTLs against other MUC1-expressing tumor cell lines of HLA-A2 haplotype. We observed a striking restriction to the HLA-A2 haplotype since HLA-A3+ control cell lines (LP-1 and SK-OV-3) were not recognized despite MUC1 expression (Figure 5; Table1).
The CTL reactivity specific for the MUC1-derived peptide M1.2 clearly contributed to the overall lysis, whereas no specificity to the MUC1-derived peptide M1.1 could be observed, thus suggesting an immunodominant role of the M1.2 epitope in myeloma-specific T-cell responses. We have recently shown that MUC1-specific CTLs induced with both MUC1 peptides (M1.1 and M1.2) recognized U266 cells, demonstrating that both peptides are presented by tumor cells.29Therefore, our data suggest that DCs and tumor cells might present different T-cell epitopes after processing of TAAs. This might result in the induction of CTL responses with a selective specificity for certain epitopes. Similar phenomena were recently described in a murine model of lymphocyte choriomeningitis virus (LCMV) infection.36
In conclusion, our data provide evidence that CTL induction by transfection of DCs with myeloma RNA is feasible and effective in eliciting antimyeloma cytotoxicity. Furthermore, we have revealed the fine specificity of a subpopulation of U266-specific CTLs recognizing the myeloma-associated MUC1 peptide M1.2; this subpopulation contributes significantly to the polyclonal CTL response targeting unidentified and immunogenic targets induced by myeloma RNA–transfected DCs.
This approach bypasses HLA restrictions of peptide-based vaccinations as well as the necessity of previous identification of immunogenic target antigens, therefore allowing a broad applicability for immunotherapy of myeloma patients.
Prepublished online as Blood First Edition Paper, September 19, 2002; DOI 10.1182/blood-2002-04-1273.
Supported in part by grants from Deutsche Krebshilfe (70-2287-Re1) and SFB 510 (Projekt B2).
C.M. and V.L.R. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Volker Reichardt, Department of Hematology, Oncology, Immunology, and Rheumatology, University of Tübingen, Otfried-Müller Str 10, D-72076 Tübingen, Germany; e-mail:volker.reichardt@med.uni-tuebingen.de.