Abstract
Systemic lupus erythematosus (SLE) is a complicated autoimmune disease with a definite genetic predisposition. Thrombocytopenia predicts severe disease and death in SLE, making the identification of the related genetic risk factors especially important. We selected the 38 pedigrees that had an SLE patient with thrombocytopenia (platelets, < 10 × 109/L [< 100 000/μL]) from a collection of 184 pedigrees multiplex for SLE. Linkages were established at 1q22-23 (maximum logarithm of odds [lodmax] = 3.71) in the 38 pedigrees and at 11p13 (lodmax = 5.72) in the 13 African American pedigrees. Nephritis, serositis, neuropsychiatric involvement, autoimmune hemolytic anemia, anti–double-stranded DNA, and antiphospholipid antibody were associated with thrombocytopenia. Other results show that SLE is more severe in the families with a thrombocytopenic SLE patient, whether or not thrombocytopenia in an individual patient is considered. These results are consistent with thrombocytopenia being a component of a severe familial form of SLE and with genes at 1q22-23 and 11p13 contributing to this severe phenotype and to the subsequent high mortality associated with thrombocytopenia in SLE.
Introduction
Systemic lupus erythematosus (SLE) is a clinically and immunologically complex disease, which can affect any organ system. The disease may be mild, or severe and life threatening. This variability is reflected in the classification system proposed by the American College of Rheumatology (ACR)1 in which patients with SLE may fulfill any 4 of 11 criteria. The presence of antinuclear antibodies (ANA) is the most sensitive of these criteria and is present in virtually all patients.
SLE also has complicated genetics. The disease tends to occur within families, but without a clear pattern of inheritance (reviewed in Sestak et al2). Familial aggregation has been reported as 2% to 5% among siblings of patients with SLE, whereas concordance among identical twins may be no more than 25%.3,4 Several genome-wide scans suggest that many genes are involved in the predisposition to SLE.5-10
Patients with SLE can have multiple hematologic abnormalities, including thrombocytopenia (< 10 × 109/L [< 100 000/μL]). Serious organ involvement in lupus is associated with thrombocytopenia. This involvement includes neuropsychiatric manifestations,11 hemolytic anemia,12,13 the antiphospholipid syndrome,14renal disease,13,15 and, most ominously, death.14-20 SLE patients with thrombocytopenia rarely die of bleeding complications. Nevertheless, thrombocytopenia forecasts a poor prognosis for SLE.14-20 Two large studies of SLE16,17 have shown that thrombocytopenia was the only, or virtually the only, independent risk factor for mortality in SLE. These studies involved European American, Hispanic, and African American patients with SLE.16,17 In addition, studies from Chile, Canada, and Southern China show poorer survival in SLE patients with thrombocytopenia. Although a few studies show no influence of thrombocytopenia on survival,13,21 most do show an effect with the relative risk of death during the period of follow-up being increased from 1.5- to 45-fold.14-18 20Clearly, thrombocytopenia is an important prognostic indicator for survival in lupus.
We used the presence of thrombocytopenia to stratify SLE families in an effort to increase homogeneity for genetic linkage studies. Families with thrombocytopenia in at least one SLE patient, taken from the Oklahoma collection of familial SLE,6,22 23 show sufficiently strong evidence to establish genetic linkages at chromosome 11p13 and 1q22-23. Other data demonstrate that thrombocytopenia identifies SLE families with a severe phenotype of the illness.
Patients, materials, and methods
Patients and pedigrees
SLE patients and their family members were enrolled in the lupus genetics studies previously described.6 Enrollment of a family required at least 2 members who fulfill the 1982 revised or modified criteria for SLE, as previously reported,1 24 and whose relationship was informative for linkage with an SLE phenotype. All patient material was gathered under protocols approved by the Oklahoma Medical Research Foundation and University of Oklahoma Health Sciences Center Institutional Review Boards at and after informed consent was obtained from participants. Clinical manifestations were defined as either present or absent based on the definitions in the SLE classification criteria. Categorical data were compared by using the chi-squared test.
Serologic evaluation
ANA, anti–double-stranded (ds) DNA, anti-Sm, antiphospholipid immunoglobulin G (IgG) and IgM antibodies were extracted from the medical record and performed in the Oklahoma Medical Research Foundation (OMRF) Clinical Immunology Laboratory (anticardiolipin was measured in our laboratory) using the serum specimens obtained for this project by following standard procedures.25,26 Anti-Ro, anti-La, anti-P, and anti-nRNP (nuclear ribonucleoprotein) were also evaluated by using Ouchterlony immunodiffusion27 in each affected subject. Evidence for biologic false-positive test for syphilis and the lupus anticoagulant was extracted from the medical record.
Genotyping
Genomic DNA was isolated from peripheral blood mononuclear cells, buccal cell swabs, or lymphoblastoid cell lines by using conventional methods as previously described.22 A total of 307 microsatellite markers were typed from the Version 8 Weber screening set (http://research.marshfieldclinic.org/genetics/sets/Set8ScreenFrames.htm). Polymerase chain reactions were performed as previously described.22 Amplified fragments were detected by using 6% polyacrylamide gels electrophoresed on automated LiCor (Lincoln, NE) Model 4000 DNA sequencers. Gel images were collected by using Base ImagIR software, version 4.0, and alleles were determined by using Gene ImagIR, version 3.52. Twenty-nine pedigrees were initially genotyped by the Mammalian Genotyping Center in Marshfield, WI (http://research.marshfieldclinic.org/genetics/), using a fluorescent-based detection system.
Linkage analysis
Prior to any linkage analysis, sibling, half-sibling, and parent-offspring relationships were confirmed by using the statistics generated by RELTEST, a feature of the S.A.G.E. 4.0 package, version Beta 3.28 29 For all linkage analyses, the pedigrees were analyzed together and as subsets containing only African American or European American pedigrees.
Maximum-likelihood model-based linkage analysis
Two-point maximum logarithm of odds (lod) scores were calculated by using FASTLINK, version 4.1P, and the ANALYZE package.30-32 Lod scores were generated for each pedigree by using recombination fractions in increments of 0.05 from 0 (complete linkage) to 0.50 (no linkage). The final maximum likelihood estimate for each microsatellite marker was produced by summing each recombination fraction across all of the pedigrees. Six inheritance models were used for screening analysis as described elsewhere.6 SLE affects men and women at a different rate. We incorporated this difference into the models by using one penetrance function for men and another for women. For each marker, allele frequencies were estimated by allele counting using founders. Screening lod scores were calculated by using the MLINK subroutine of the ANALYZE package.
Model-free linkage analysis
Multipoint linkage analysis on concordant and discordant sibling pairs was performed at 2-cM increments for all 22 autosomes by using the new Haseman-Elston regression technique used by SIBPAL2, a subtest of the S.A.G.E 4.0 package, version Beta 3.29,33 The algorithm regresses the identity by descent sharing values against the mean corrected cross product of the sib-pair trait difference. In addition, we performed a multipoint affected-relative pairs (ARPs) analysis by using LODPAL, also a subroutine of the S.A.G.E. 4.0 Beta, version 7. This analysis models covariance of all affected relative pairs as a function of marker allele-sharing identical by descent using the conditional logistic model proposed by Olson34 and Goddard et al.35
Results
There were a total of 387 patients with SLE in the 184 families evaluated. Of these, 116 families and 233 patients with SLE were of European ancestry, whereas 63 families and 133 patients with SLE were of sub-Saharan African ancestry. A total of 47 (12.1%) of the 387 patients with SLE had definite thrombocytopenia as defined by the criteria (platelet count, < 10 × 109/L [< 100 000/mm3] without another cause), a frequency that did not vary among racial groups. Twenty-seven of those with thrombocytopenia were European American; among the African American patients, 15 patients from 14 families had thrombocytopenia, of which 13 pedigrees were useful for linkage. Five patients with thrombocytopenic SLE were from 5 different Hispanic families.
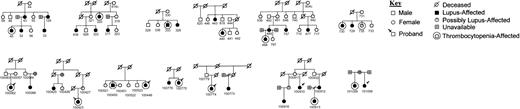
We examined the clinical characteristics of the SLE patients with thrombocytopenia. There were 43 women and 4 (8.5%) men, whereas 9.3% of the total SLE patient population was male. The average age of onset of SLE was 24.1 years (standard deviation = 8.3), which did not differ from the SLE patients without thrombocytopenia (not shown). The average lowest platelet count was 2.4 × 109/L (24 000) with a range of 1 to 90 × 109/L (1000-90 000/mm3). Among the families that had a patient with thrombocytopenic SLE, about one half constituted a sibling pair, whereas in the other families the relationships of the patients with SLE were more complicated (Figure1). The family structures of the African American families with thrombocytopenic SLE that were useful for linkage are given in Figure 1.
The family structures of the African American families that were useful for linkage in which at least one patient with SLE had thrombocytopenia.
All patients with SLE are indicated (closed symbols) as are the SLE patients with thrombocytopenia (double symbol). / indicates separated; and //, divorced.
The family structures of the African American families that were useful for linkage in which at least one patient with SLE had thrombocytopenia.
All patients with SLE are indicated (closed symbols) as are the SLE patients with thrombocytopenia (double symbol). / indicates separated; and //, divorced.
Thrombocytopenia was associated with other manifestations of SLE within this cohort of patients (Tables 1 and 2), including autoimmune hemolytic anemia and lupus renal disease as defined by the ACR criteria.1 24 The relationship between thrombocytopenia and hemolytic anemia was present when those of European or African ancestry were evaluated separately (Table1). Neuropsychiatric disease is also associated with thrombocytopenia in SLE (Table2), most strikingly among the European Americans. Serositis, arthritis, and the cutaneous criteria for SLE classification (malar rash, discoid rash, photosensitivity, and oral ulcers) were not associated with thrombocytopenia (data not shown).
Anti-dsDNA and antiphospholipid antibodies were found more often in patients with thrombocytopenic SLE (Table 2), whereas antinuclear antibodies, anti-Sm, and anti-nRNP were not (data not shown). In addition, among black patients anti-Ro (or Sjögren syndrome A [SS-A]) was found much more commonly in those with thrombocytopenia of whom 9 (60%) of 15 had anti-Ro. Meanwhile, only 24 (20.5%) of the remaining 117 African Americans had anti-Ro (chi-squared test = 11.1, P < .001, odds ratio = 5.8; Table 2). Of course, each individual with SLE is related to at least one other subject with SLE in these families. So, the variables are not completely independent. However, analyses of all these clinical and laboratory manifestations of SLE using a single patient from each family showed similar results (data not shown).
We next tested whether or not thrombocytopenia in individuals or within families of the Oklahoma pedigree collection may identify a severe lupus phenotype, consisting of multiple thrombocytopenia-associated characteristics. Such a phenotype was arbitrarily defined as 2 or more of the autoantibodies (anti-ds DNA, anti-Ro, or antiphospholipid antibody) and 2 or more of the clinical manifestations that are associated with thrombocytopenia (serositis, nephritis, neuropsychiatric disease, and hemolytic anemia). Seventeen (36.2%) of the 47 patients with thrombocytopenic SLE had the phenotype as defined, whereas only 53 (15.5%) of 340 patients with nonthrombocytopenic SLE did so (odds ratio = 2.85, chi-squared test = 15.3,P < .0001).
Provocatively, this phenotype was of nearly identical frequency in SLE patients with thrombocytopenia and their nonthrombocytopenic SLE-affected family members (28 [31.8%] of 88 versus 17 [36.1%] of 47, chi-squared test = 0.46, P > .05). Moreover, the patients with nonthrombocytopenic SLE in the thrombocytopenia pedigrees have a much higher frequency of this severe phenotype than do patients with SLE in the pedigrees with no thrombocytopenia (45 [33.3%] of 135 versus 25 [9.9%], of 252, odds ratio = 4.5, chi-squared test = 23.8, P < .000 001). Thrombocytopenia in a patient with SLE, therefore, is associated with more severe disease in her or his relatives who also have lupus. A severe lupus phenotype is, therefore, not so much a characteristic of thrombocytopenic lupus, but instead a characteristic of patients with lupus from families identified by having one patient with thrombocytopenic lupus. Familial aggregation of the severe phenotype in these pedigrees predicts that the genetic linkages found in the pedigrees containing a SLE patient with thrombocytopenia will be associated with a more severe form of lupus. Patients with these SLE susceptibility genes would be expected to have increased morbidity and mortality from SLE.
Genetic linkage of familial lupus among families with thrombocytopenia as a manifestation of lupus was assessed by using 6 screening models of inheritance (Table3). We used the microsatellite genotyping data to screen with 2-point lod scores by using a maximum likelihood model-based linkage analysis (Table4). The most interesting screening result demonstrates a lod score of 3.803 for marker D11s1392 on chromosome 11p13 among families of African American heritage (Table 3). This lod score exceeds the accepted threshold for established linkage.36 The best screening model for this effect is autosomal dominant with a penetrance of 97% in women and 42% in men.
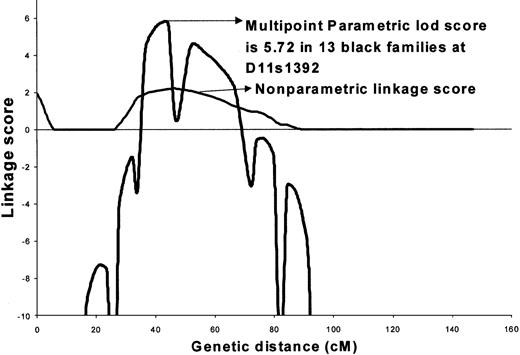
We next optimized this model based on penetrance for the 2 sexes and frequency of the disease allele (Figure2). The best model assumed that a rare, dominantly inherited factor segregated with the affected individuals (male or female). In this model, the disease allele frequency was pD = 0.001, where the 3 penetrances for men were Fdd, = 0.0, FDd = 0.39, and FDD = 0.39, and the 3 penetrances for women were Fdd = 0.0, Fdd = 0.99, and FDD = 0.99. The estimated prevalence of disease based on this model was 0.14%, which is very similar to the observed population prevalence of SLE. The maximum 2-point lod score of 4.89 was obtained with marker D11s1392 under complete homogeneity among African American families. The peak multipoint lod score of 5.72 under a dominant model was obtained at 41.7 cM between markers A34E8 and D11S1392 (Figure 2). The maximum nonparametric linkage score of 2.12 was obtained at the same position (P = .01) (Figure 2). Low informativity (0.34-0.45) is likely a major contributor to the low nonparametric linkage score.
Maximized multipoint and nonparametric analysis of linkage in 13 African American families on chromosome 11 showing a multipoint lod = 5.72.
Maximized multipoint and nonparametric analysis of linkage in 13 African American families on chromosome 11 showing a multipoint lod = 5.72.
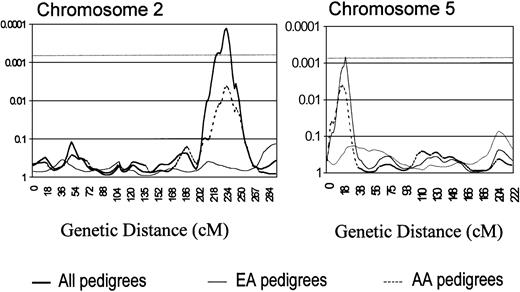
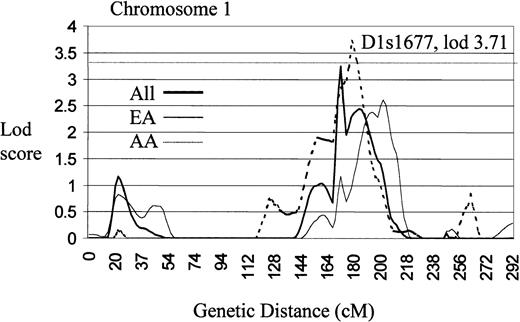
A model-free, multipoint linkage analysis of identity by descent sharing among related SLE-affected pairs (SIBPAL2) produced findings of interest on chromosomes 2 and 5 (Figure3), both meeting the threshold for a suggestive linkage (P ≤ .0007)36 (Table 4). The finding on chromosome 2 is also one of the more significant findings in the lod score analysis (Table 3). In addition, the chromosome 11 linkage presented above was also found by the sibpair analysis. In a second analysis (Table 5), which uses affected relative pairs (LODPAL), significant linkage on chromosome 1 at 1q22-24 was identified (Figure4).
Chromosome 2 genome scan analysis illustrating a region with SIBPAL2 significance of P = .0001 between Ds21 384 and D2s434 (top), along with a similar chromosome 5 analysis showing aP = .0007 nearest marker D5s807.
cM are plotted in the x-axis with P values shown on the y-axis. The dashed horizontal line indicates the P value for suggestive linkage (P ≤ .0007).
Chromosome 2 genome scan analysis illustrating a region with SIBPAL2 significance of P = .0001 between Ds21 384 and D2s434 (top), along with a similar chromosome 5 analysis showing aP = .0007 nearest marker D5s807.
cM are plotted in the x-axis with P values shown on the y-axis. The dashed horizontal line indicates the P value for suggestive linkage (P ≤ .0007).
Genome scan of chromosome 1 illustrating a region of significant linkage (lod > 3.3) using an affected relative pairs analysis (LODPAL).
Centimorgans are plotted on the x-axis and lod scores on the y-axis. The dashed horizontal line represents the level of established linkage (lod ≥ 3.3).
Genome scan of chromosome 1 illustrating a region of significant linkage (lod > 3.3) using an affected relative pairs analysis (LODPAL).
Centimorgans are plotted on the x-axis and lod scores on the y-axis. The dashed horizontal line represents the level of established linkage (lod ≥ 3.3).
Discussion
We have examined the clinical associations and the genetics of thrombocytopenia in a relatively large collection of families multiplex for systemic lupus erythematosus. The data support thrombocytopenia-identified families tend to have a more severe lupus phenotype. In addition, new previously undetected linkages are found, also consistent with the possibility that thrombocytopenia in SLE identifies a group of pedigrees with increased genetic homogeneity. This result also supports the possibility that some of the heterogeneity of lupus clinical expression has a genetic origin.
Thrombocytopenia, a part of the widely accepted classification criteria, as proposed by the ACR,1,24 is one of the most interesting expressions of SLE because of its association with mortality and other serious clinical manifestations.14-21Low platelets occur in 10% to 25% of patients with SLE.15,17 Thrombocytopenia can be categorized in 2 subgroups. This complication may occur acutely in association with severe, multisystem SLE disease flares, or it may be a chronic condition that is present even when the disease is otherwise quiescent.13
Reveille et al16 first reported an association of mortality and thrombocytopenia among patients with lupus.16 In a multivariate analysis, thrombocytopenia was the only independent risk factor for death among the 389 patients with SLE followed for a mean of 8.8 years. Others have demonstrated similar findings.14-20 Other features of patients with SLE such as age of onset, presence of antiphospholipid antibody, renal disease, steroid dose, sex, and race have been found to be associated with risk of death in some studies. Nonetheless, even though patients with SLE rarely die of bleeding complications, thrombocytopenia is the clinical feature most consistently found to be associated with mortality in SLE.
Thrombocytopenia also has several other reported associations with other manifestations of SLE. For instance, 25% of SLE patients with neuropsychiatric manifestations had thrombocytopenia, whereas only 7% of nonneuropsychiatric patients did so.11 Autoimmune hemolytic anemia and thrombocytopenia have been found to be highly associated.12,15 Patients with one or both of these manifestations were also found to have more renal disease.12 Renal disease has been associated with thrombocytopenia in other studies.15 In the present study of 387 patients with SLE, we found that thrombocytopenia was associated with serositis, neuropsychiatric disease, renal disease, and autoimmune hemolytic anemia (Table 2). In addition to these clinical associations, we also found that thrombocytopenia was more common among patients with antiphospholipid antibody, anti-dsDNA or, in African Americans, anti-Ro (Table 3). A severe lupus phenotype consisting of at least 2 of the thrombocytopenia-associated clinical manifestations and 2 of the associated autoantibodies was much more common among patients with thrombocytopenia and their affected family members than among patients from families without thrombocytopenia. Thus, thrombocytopenia identifies not just patients, but also families, with a severe lupus phenotype, despite thrombocytopenia not often being a familial trait among patients with SLE.
Because thrombocytopenia identifies a severe subset of lupus patients and families, we hypothesized that this group might have more homogeneous genetics compared with lupus patients in general. This appears to be the case. By performing a lod score analysis and a related-affected pair analysis on the lupus families with thrombocytopenia as a manifestation, we have established genetic linkage not previously identified.6,22 On chromosome 11 at marker D11s1392 a lod score of 4.89 and a maximum multipoint lod score of 5.7 were obtained in only 13 African American SLE families (Figure2). Using SLE alone as the phenotype, we previously did not establish linkage at this genetic interval (lod = 1.87).6
There are a number of interesting candidate genes near this marker, including CD44 that is located 0.5 cM from D11s1392. We studied a microsatellite trinucleotide repeat within this gene and confirmed linkage with a screening lod score of 2.7. Thus, this new marker confirms linkage in this region. We also examined several single nucleotide polymorphisms within this gene and did not find association with lupus in the 13 African American families. Thus, these preliminary data imply that the gene responsible for the effect seen at this chromosomal location is notCD44.37 The genomic map for this region of chromosome 11 is incomplete, but there are several other genes of interest that are mapped to 11p13. Included among these areCD59, RAG-1, and the Wilms tumor suppressorWT1, all of which are involved in immune cell development. Also, present within the genetic interval are the fas-interacting serine/threonine kinase (FIST/HIPK3) as well as tumor necrosis factor (TNF) receptor-associated factor 6. We have also established linkage at 1q22-23. This region is very near other established linkages on chromosome 16 and may reflect the effect of the previously detected gene(s).
We have found evidence suggestive of linkage on chromosome 2, with the greatest effect at D2s1384. Similar to the chromosome 11 effect, no important effect was seen at this position when performing a genome-wide scan of all the pedigrees.6 Icelandic pedigrees collected in Uppsala, Sweden, show a linkage effect at 2q37.8 This is approximately 50 cM away from the 2q34 linkage described herein, which is probably too distant to be the same genetic effect. Affected pair analysis also supported an effect on chromosome 5 that exceeded the threshold for a suggestive linkage but not the threshold for established linkage (Tables 4 and 5). Another result also supports the hypothesis that this region of chromosome 5 contains a gene important for lupus. Gray-McGuire et al22used a revised multipoint Haseman-Elston regression technique along with a conditional logistic regression model–building technique. This technique found a novel linkage to chromosome 4p16-15.2 that is in epistasis with the same 5p15 marker reported herein.
Thus, multiple techniques of analysis support the findings of linkage of chromosomes 1, 2, 5, and 11 that are herein reported in SLE families in which at least one patient had thrombocytopenia. These are genetic linkages to lupus as a phenotype. Although their relationship to thrombocytopenia (or other associated findings) must be important, they are not necessarily direct nor are they revealed by this study. In any case, the success of this study provides another example of clinical features providing a stratification strategy for genetic analysis, one that increases statistical power through presumed increased genetic homogeneity. Clearly, stratification by clinical factors was critical for identification of the BCR1 gene38 as well as the gene for familial Mediterranean fever.39 We have also used this approach in studying SLE families with vitiligo40 or hemolytic anemia41 with resulting identification of genetic linkages distinct from the ones identified herein. For the 11p13 linkage, stratification by race and the presence of low platelets has produced one of the largest genetic effects reported to date in lupus (lodmax = 5.7). These data demonstrate the capacity of disease clinical manifestations to be informative for the genetics of SLE. Of course, stratification of our cohort may also lead to problems. By reducing the number of families, the potential for α error is increased. Therefore, we will need to confirm the present results in another cohort of patients with SLE.
The genetics of human SLE are obviously extremely complicated, which is likely the result of heterogeneity, low penetrance, and simultaneously operating protective genetic effects, among other impediments. Genes related to thrombocytopenia in lupus may also be related to clinically severe disease and early demise.11-20 If the pathology of the severe, deadly forms of this disease is ever to be understood and if genetic elucidation will generate new therapeutic options, then these are the genetic effects that require our most concentrated attention.
We thank the study participants for their help with this research effort. Some of the results presented herein were obtained with S.A.G.E. (RR03 655). Ninety pedigrees (cohorts A, B, and C) were obtained from the Lupus Multiplex Registry and Repository (AR-1-2253).
Prepublished online as Blood First Edition Paper, September 12, 2002; DOI 10.1182/blood-2002-04-1003.
Supported by the National Institutes of Health (AR-1-2253, AI24717, AR42460, AR45231, AI31584, and RR15577).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
R. Hal Scofield, 825 NE 13th St, Oklahoma City, OK 73104; e-mail: hal-scofield@omrf.ouhsc.edu.