Severe combined immunodeficiency disease (SCID) can be immunologically classified by the absence or presence of T, B, and natural killer (NK) cells. About 30% of T−B−NK+ SCID patients carry mutations in the recombination activating genes (RAG). Some T−B−NK+ SCID patients withoutRAG gene mutations are sensitive to ionizing radiation, and several of these radiosensitive (RS) SCID patients were recently shown to have large deletions or truncation mutations in theArtemis gene, implying a role for Artemis in DNA double-strand break (dsb) repair. We identified 5 RS-SCID patients without RAG gene mutations, 4 of them withArtemis gene mutations. One patient had a large genomic deletion, but the other 3 patients carried simple missense mutations in conserved amino acid residues in the SNM1 homology domain of the Artemis protein. Extrachromosomal V(D)J recombination assays showed normal and precise signal joint formation, but inefficient coding joint formation in fibroblasts of these patients, which could be complemented by the wild-type Artemis gene. The cells containing the missense mutations in the SNM1 homology domain had the same recombination phenotype as the cells with the large deletion, indicating that these amino acid residues are indispensable for Artemis function. Immunogenotyping and immunophenotyping of bone marrow samples of 2 RS-SCID patients showed the absence of complete VH-JH gene rearrangements and consequently a complete B-cell differentiation arrest at the pre–B-cell receptor checkpoint—that is, at the transition from CyIgμ−pre-B-I cells to CyIgμ+ pre-B-II cells. The completeness of this arrest illustrates the importance of Artemis at this stage of lymphoid differentiation.
Introduction
Severe combined immunodeficiency disease (SCID) is clinically characterized by opportunistic infections, protracted diarrhea, and failure to thrive.1 Patients generally die within the first year of life unless treated with bone marrow (BM) transplantation. Other treatment options include enzyme substitution in case of adenosine deaminase–deficient SCID and gene therapy in case of common γ chain–deficient X-linked SCID.2
Although SCID consists of a heterogeneous group of diseases, it is immunologically characterized by the absence or dysfunctioning of T lymphocytes. SCID can be subdivided based on the additional presence or absence of B lymphocytes and natural killer (NK) cells in peripheral blood (PB). A number of T−B−NK+ SCID patients were shown to have mutations in the recombination activating genes (RAG1 and RAG2).3 However, not all patients with T−B−NK+ SCID have mutations in the RAG genes. Subsequently, it was shown that fibroblasts of several T−B−NK+SCID patients without RAG gene mutations were sensitive to ionizing radiation and thus likely to have a defect in DNA double-strand break (dsb) repair.4
During the process of immunoglobulin (Ig) and T-cell receptor (TCR) gene recombination in differentiating lymphocytes, the RAG proteins introduce DNA dsb's, which are repaired via nonhomologous end joining (NHEJ).5 NHEJ requires active DNA-dependent protein kinase (DNA-PK), which consists of Ku70, Ku80, and the catalytic subunit (DNA-PKcs). The DNA-PK protein complex functions as a DNA damage sensor, with Ku70 and Ku80 forming a heterodimer that binds to DNA ends, while DNA-PKcs has serine and threonine protein kinase activity.6 In the final phase, the 2 broken DNA ends are ligated by the DNA ligase IV–XRCC4 complex.7,8However, it was shown that a number of radiosensitive (RS) T−B−NK+ SCID patients without mutations in the RAG genes did not suffer from mutations in any of these 5 DNA dsb repair components.4 Recently, a new gene involved in the pathogenesis of RS-SCID was identified and namedArtemis.9,10 The Artemis protein can be phosphorylated by the DNA-PK complex, and the Artemis-DNA-PK complex is able to open hairpin-sealed coding ends and to process 5′ and 3′ overhangs.11 Several different large deletions and truncation mutations have been identified as disease-causingArtemis mutations.9 12
We studied 5 RS-SCID patients (from 4 families) without RAGgene mutations and found that 4 of them had mutations in theArtemis gene. Immunophenotyping and immunogenotyping of BM samples from 2 RS-SCID patients with Artemis gene mutations provided additional information about the role of Artemis during precursor B-cell differentiation.
Patients, materials, and methods
Cell samples
We received PB and/or BM samples and/or fibroblasts from 32 T−B−NK+ SCID patients. These patients were analyzed for the presence of mutations in theRAG genes.13 Fibroblasts from 13 T−B−NK+ SCID patients without mutations in the RAG genes were analyzed for sensitivity to ionizing radiation.
BM samples from T−B−NK+ SCID patients were subjected to Ficoll-Paque (density, 1.077 g/mL) (Pharmacia, Uppsala, Sweden) density centrifugation. The recovered mononuclear cells (MCs) were frozen and stored in liquid nitrogen and thawed later for flow cytometric immunophenotyping studies. Granulocytes were used for DNA extraction.
All cell samples were obtained according to the informed consent guidelines of the medical ethics committees of the Erasmus MC/University Medical Center Rotterdam and the Leiden University Medical Center.
Sensitivity of fibroblasts to ionizing radiation
Primary skin fibroblasts in exponential growth were trypsinized, and 500 to 2000 cells (5 000 to 80 000 cells for the highest doses) were seeded into 10-cm plastic dishes (2 dishes per dose, 3 for nonirradiated control) and irradiated at room temperature at a dose of approximately 2.7 Gy/min (200 kV, 4.0 mA). After 12 to 14 days, the cells were rinsed with 0.9% NaCl and stained with 0.25% methylene blue for survival assessment. At least 3 independent survival experiments were performed for each sample.
Messenger RNA and DNA isolation and cDNA reaction
DNA was extracted from granulocytes or fibroblasts using the QIAamp Blood kit (Qiagen, Chatsworth, CA).14 Total RNA was isolated from BMMCs or fibroblasts using the GenElute Mammalian RNA kit (Sigma-Aldrich, St Louis, MO). Complementary DNA was prepared from mRNA as described before, using random hexamers and Superscript reverse transcriptase (Life Technologies, Paisley, United Kingdom).15
PCR amplification and analysis of Ig gene rearrangements
Polymerase chain reaction (PCR) was performed as described previously.15 In each 100-μL PCR reaction, 0.1 to 1 μg cDNA, 10 to 12.5 pmol of 5′ and 3′ oligonucleotides, and 1 unit of AmpliTaq gold polymerase (Applied Biosystems, Foster City, CA) were used. Oligonucleotides for amplification of theArtemis and Ig heavy chain (IGH) gene rearrangements (VH-JH and DH-JH) were published before.10,16,17 PCR conditions were 7 minutes at 95°C, followed by 45 seconds at 94°C, 90 seconds at 55°C to 60°C, 2 minutes at 72°C for 40 cycles, followed by a final extension step (7 minutes at 72°C). DNA from BMMCs was analyzed for rearrangements of DH1, DH2, DH3, DH4, DH5, DH6, DH7, VH1/7, VH2, VH3, VH4, VH5, and VH6 to JH.13
Fluorescent sequencing reaction and analysis
PCR products of Artemis were purified using QIAquick PCR purification kit (Qiagen). Cloned IGH gene rearrangements were isolated via GenElute Plasmid MiniPrep Kit (Sigma-Aldrich). A 2 to 9 μL template was sequenced with 5 μL BigDye terminator mix (Applied Biosystems) using 3.3 to 6.6 pmol sequencing primers. All sequencing was performed as described before16 and run on an ABI Prism 377 fluorescent sequencer (Applied Biosystems).
Western blot analysis of proteins involved in DNA dsb repair
Protein samples from fibroblasts were separated on an 11% polyacrylamide gel and analyzed by Western blotting. The expression of Mre11 (rabbit polyclonal antibody [Ab] no. 2244),18 XRCC4 (rabbit polyclonal Ab NIH14), Ku70 (goat polyclonal Ab C19, Santa Cruz Biotechnology, Santa Cruz, CA) and Ku80 (goat polyclonal Ab C20, Santa Cruz Biotechnology), DNA-PKcs (rabbit polyclonal Ab no. 2129), ligase IV (goat polyclonal Ab T20, Santa Cruz Biotechnology), ATM (gift from Dr S. Jackson, Wellcome Trust and Cancer Research, Cambridge, United Kingdom), and NBS1 (p95NBS1, Calbiochem, San Diego, CA) proteins was analyzed.
Analysis of NHEJ via transfection of linearized DNA constructs
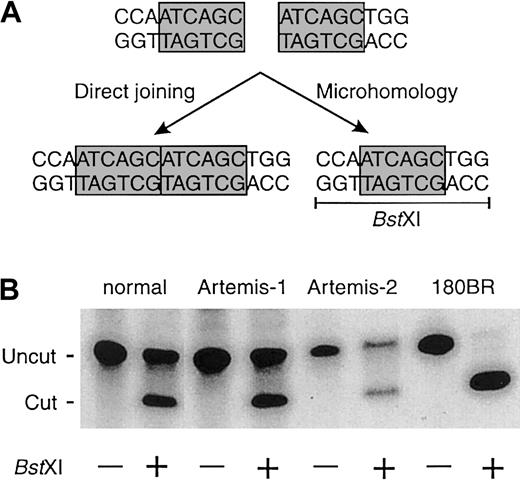
Linearized DNA constructs with homologous ends (ATCAGC sequence) were transfected into fibroblasts of RS-SCID patients and fibroblasts of a patient with a mutation in DNA ligase IV (180BR) as described before.19 Newly formed junctions were PCR amplified, and the relative use of a particular microhomology was assayed by digestion with the restriction enzyme BstXI (Figure2).19
Extrachromosomal VDJ recombination assay
For analysis of signal and coding joint formation in the different RS-SCID fibroblasts with Artemis mutations, 2 μgRAG1 and 2 μg RAG2 expression vectors together with 1 μg of the recombination substrate pGG49 or pGG51,20 each containing 2 recombination signal sequence (RSS) elements, were transfected into fibroblasts using Fugene Transfection Reagent (Roche Diagnostics, Indianapolis, IN) as described before.13 Transfected cells were cultured for 2 days at 37°C and 5% CO2 before isolation of extrachromosomal DNA. Upon V(D)J recombination, the sequence between the RSS elements is deleted, after which coding (pGG51) or signal joint (pGG49) formation can be detected by PCR analysis of the newly formed junctions (Figure 3). DNA recovered from these transfection experiments was resuspended in 20 μL H2O, of which 1 μL was used for PCR analysis. First, DNA was predigested with ClaI to reduce the signal derived from nonrecombined V(D)J substrate. To amplify the coding or signal joints, a nested PCR of 2 × 25 cycles was performed. The first round of PCR was performed with oligonucleotides NV09F and DG147 (Table1) in a total volume of 50 μL. Subsequently, 1 μL of this PCR reaction was used as a template for the second round of PCR using oligonucleotides NV08F and FM30, of which FM30 was radioactively labeled with γ32P–adenosine triphosphate (ATP). The PCR conditions were the same as described before.13 PCR fragments were separated on a 6% polyacrylamide gel, and products were visualized by phosphorimaging.13
Artemis complementation studies
To determine whether the recombination defect could be complemented, either wild-type (wt) Artemis or a mutant Artemis expression construct was cotransfected with the RAG1 and RAG2 expression vectors and the coding joint recombination substrate pGG51 into fibroblasts of the RS-SCID patients with Artemis gene mutations. The wtArtemis expression construct was made by performing a PCR with the DG238 and DG239 oligonucleotides (Table 1) on Hela cDNA. This 2.1-kb PCR product was digested with XhoI andMluI and cloned into the XhoI and MluI sites of plasmid pMS127B.21 The point mutations of patients Artemis-2 and Artemis-3.1 were cloned by performing this same PCR on cDNA generated from the fibroblasts of these patients. This 2.1-kb fragment was digested with NsiI and EcoRV, after which the 500-bp fragment containing the point mutations was exchanged for the NsiI-EcoRV part of the wt construct.
Flow cytometric analysis of BM samples from RS-SCID patients withArtemis gene mutations
Fifty microliter aliquots of thawed BMMCs (10 × 106 cells per milliliter) were incubated for 10 minutes at room temperature with combinations of optimally titrated monoclonal antibody (MAb): 50 μL fluorescein isothiocyanate (FITC)–conjugated MAb, 50 μL phycoerythrin (PE)–conjugated MAb, 50 μL peridinin chlorophyll protein (PerCP) cyanin (CY) 5.5–conjugated MAb, and 50 μL allophycocyanin (APC)–conjugated MAb were used to detect membrane-bound antigens. After incubation, the cells were washed and further processed depending on the type of quadruple labeling.22 23
Quadruple labelings for membrane-bound antigens were directly analyzed by flow cytometry using FACSCalibur (Becton Dickinson, San Jose, CA). For quadruple labelings involving intracellular staining of cytoplasmic (Cy) CD79a, CyIgμ, CyVpreB,24 and intranuclear staining of terminal deoxynucleotidyl transferase (TdT), we first performed the membrane labelings, followed by permeabilization of the BM cells using IntraPrep Permeabilization Reagent (Immunotech, Marseille, France) and subsequent intracellular staining.25 26
Results
Patient characteristics, Western blot analysis, and disease-causing Artemis gene mutations
We analyzed material from 32 T−B−NK+ SCID patients for the presence of RAG gene mutations, both at the genomic and at the transcriptional level. We could not detect mutations in theRAG genes in 23 patients (72%). From 13 T−B−NK+ SCID patients withoutRAG mutations, fibroblasts were available and analyzed for sensitivity to ionizing radiation in a clonogenic survival assay.
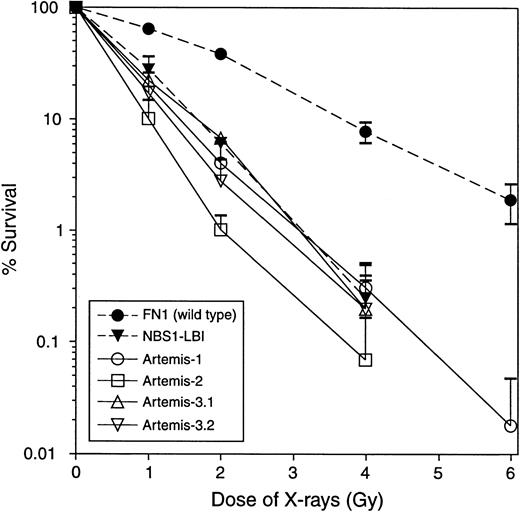
Fibroblasts from 5 T−B−NK+ SCID patients from 4 families showed a 2-fold to 3-fold increased sensitivity toward X-rays, compared with wild-type (wt) primary fibroblasts (FN1). The degree of the observed X-ray sensitivity was similar to fibroblasts derived from a patient with Nijmegen breakage syndrome (NBS1-LBI) (Figure 1). Western blot analysis of Mre11, XRCC4, Ku70, Ku80, DNA-PKcs, and ligase IV proteins derived from fibroblasts from the 5 RS-SCID patients showed normal expression (data not shown). Finally, genomic sequencing of Artemis exons (including splice sites) revealed mutations in 4 of the 5 RS-SCID patients. The clinical characteristics of the 4 patients with Artemis gene mutations are summarized in Table 2.
Clonogenic survival assay of fibroblasts after ionizing radiation.
Fibroblasts from 5 T−B−NK+ SCID patients from 4 families without RAG mutations were radiosensitive. Data from the 4 patients with Artemis gene mutations are shown. NBS indicates Nijmegen breakage syndrome. Each survival curve represents the mean of at least 3 independent experiments. Error bars represent standard errors of the mean.
Clonogenic survival assay of fibroblasts after ionizing radiation.
Fibroblasts from 5 T−B−NK+ SCID patients from 4 families without RAG mutations were radiosensitive. Data from the 4 patients with Artemis gene mutations are shown. NBS indicates Nijmegen breakage syndrome. Each survival curve represents the mean of at least 3 independent experiments. Error bars represent standard errors of the mean.
Patient Artemis-1 showed a homozygous deletion of exons 10, 11, and 12 of the Artemis gene. At the mRNA level, exon 9 was coupled to exon 13, resulting in a frameshift and premature stop at codon 269 in the SNM1 homology domain (numbering according to Moshous et al).10
Patient Artemis-2 showed a homozygous G>T mutation at position 47 in exon 5 of the Artemis gene. This resulted in mutation of a glycine to valine residue at position 111 in the SNM1 homology domain of the Artemis protein. We did not identify this point mutation in 18 healthy controls. Furthermore, glycine at position 111 is highly conserved between yeast PSO2, mouse SNM1, human SNM1A, and human SNM1B proteins and therefore is likely to represent an essential amino acid (aa).27
Patients Artemis-3.1 and -3.2 showed a homozygous G>A mutation at position 42 in exon 6 of the Artemis gene. This resulted in mutation of a glycine to glutamic acid residue at position 128 in the SNM1 homology domain of the Artemis protein. We did not identify this point mutation in 14 healthy controls. Furthermore, glycine at position 128 is highly conserved between yeast PSO2, mouse SNM1, human SNM1A, and human SNM1B proteins and therefore likely to represent an essential aa.27 Because the parents of patients Artemis-3.1 and -3.2 had a nonconsanguineous relationship, we investigated whether the point mutation was really homozygous. We excluded a large deletion (containing exon 6) of one allele by Southern blot analysis of theArtemis gene using a PCR-based exon 6 probe (data not shown). We did not find a lower level of hybridization (normalized to Igκ J5) in the patients relative to a healthy control, indicating that these patients are homozygous for the point mutation.
In addition to the full-length transcripts, one of the Artemis-3 patients showed alternatively spliced transcripts in which exon 4 was spliced to exon 6. Sequencing analysis of the alternatively spliced transcripts showed the presence of the disease-causing G>A mutation in exon 6, fully consistent with the homozygous mutation. Such alternatively spliced Artemis transcripts were not found in patient Artemis-2. No mutations were present in the donor and acceptor splice sites of any Artemis exon in the 4 studied patients.
Additional nondisease-causing mutations in the Artemisgene
The entire Artemis gene was sequenced in all 4 RS-SCID patients with Artemis mutations to exclude the presence of additional mutations. We identified a total of 4 additional alterations compared with the published sequence. Two base changes were present in all 4 RS-SCID patients analyzed and were therefore likely to represent polymorphisms or sequencing errors in the published sequence. The first alteration concerned a G>T change at position 522 in exon 14 of theArtemis gene. This alteration resulted in a change from a valine to leucine residue at position 553 outside the SNM1 homology domain of the Artemis protein. The second alteration concerned a C>T change at position 653 in exon 14 of the Artemis gene, which did not result in an aa substitution.
We identified another silent alteration at position 106 in exon 8 of the Artemis gene (a T>C change) in patients Artemis-3.1 and -3.2. Finally, patient Artemis-1 with the large deletion carried a homozygous A>G change at position 50 in exon 9 of theArtemis gene. This alteration resulted in a change from the basic aa histidine to the basic aa arginine at position 236 in the SNM1 homology domain. We did not identify this point mutation in 12 healthy controls.
Dsb repair
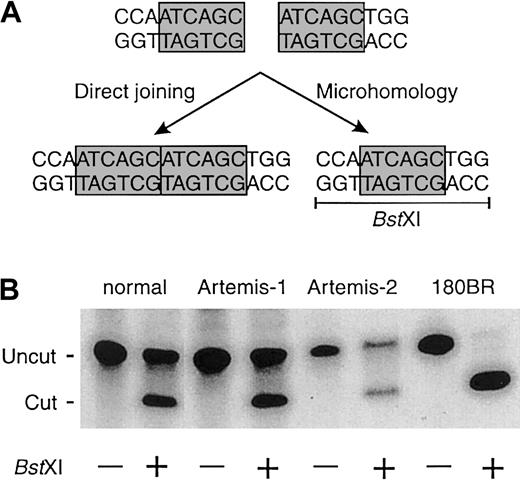
Mutation of Ku80, DNA-PKcs, XRCC4, or DNA ligase IV leads to a shift from precise joining to microhomology-directed joining in mammalian cells.19 Therefore, we transfected linearized DNA constructs into fibroblasts of all 4 patients withArtemis gene mutations to analyze the use of direct joining and microhomology pathways. The Artemis-mutated cells showed equal usage of the direct joining and microhomology pathways (Figure2), suggesting that Artemis is not involved in the ligation process itself.
Fibroblasts of a healthy control and RS-SCID patients with mutations in the Artemis gene show equal usage of direct joining and microhomology when rejoining linearized DNA constructs.
(A) Linearized DNA constructs with homologous ends (ATCAGC) can be rejoined via direct joining or via microhomology. Joining via microhomology results in the generation of a BstXI restriction site (CCAN6TGG). (B) In contrast to normal fibroblasts and fibroblasts from patients with an Artemismutation, fibroblasts with a mutation in DNA ligase IV (180BR) show complete absence of direct joining and full usage of the microhomology pathway.
Fibroblasts of a healthy control and RS-SCID patients with mutations in the Artemis gene show equal usage of direct joining and microhomology when rejoining linearized DNA constructs.
(A) Linearized DNA constructs with homologous ends (ATCAGC) can be rejoined via direct joining or via microhomology. Joining via microhomology results in the generation of a BstXI restriction site (CCAN6TGG). (B) In contrast to normal fibroblasts and fibroblasts from patients with an Artemismutation, fibroblasts with a mutation in DNA ligase IV (180BR) show complete absence of direct joining and full usage of the microhomology pathway.
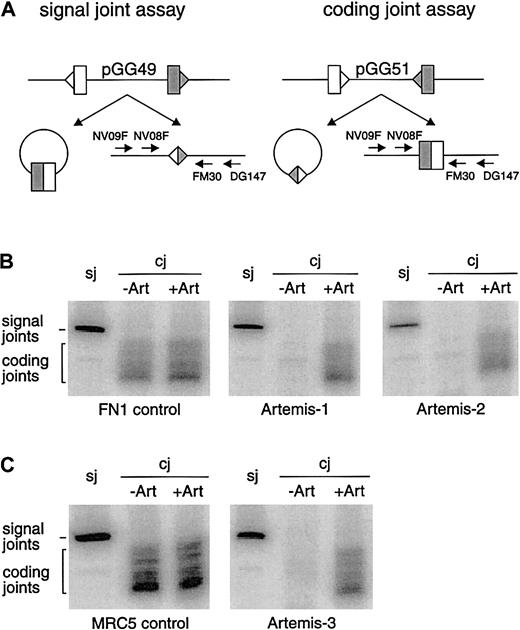
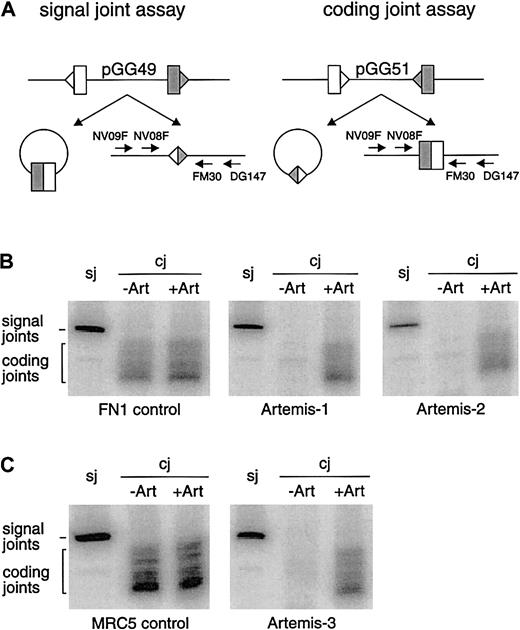
V(D)J recombination also involves dsb repair by NHEJ. We analyzed coding and signal joint formation in fibroblasts of all 4 patients withArtemis gene mutations. In all cases we found normal levels of precise signal joint formation but absence of coding joint formation (Figure 3).
V(D)J recombination assay in RS-SCID fibroblasts withArtemis gene mutations.
(A) Schematic representation of the signal joint assay (sj) and the coding joint assay (cj) in which the recombination substrates pGG49 and pGG51 were used, respectively. Recombination signal sequences are depicted as triangles and the flanking coding sequence as rectangles. The signal joints are retained in the recombination substrate when pGG49 is used, while coding joints are retained in the recombination substrate when pGG51 is used. Signal joints and coding joints were detected with nested PCR analysis using a 32P-labeled primer (see “Materials and methods”). (B) Primary fibroblasts of Artemis-1 and Artemis-2 transfected with RAG1 andRAG2 expression constructs and the recombination substrate formed signal joints but no coding joints. FN1 control primary fibroblasts can form both signal joints and coding joints. Defective coding joint formation in Artemis-1 and Artemis-2 could be complemented by cotransfection of the wt Artemis expression construct (+Art). (C) SV40-immortalized fibroblasts of Artemis-3 were also able to form signal joints but not coding joints. Like in Artemis-1 and Artemis-2, coding joint formation occurred upon cotransfection of the wt Artemis expression construct. SV40-immortalized MCR5 fibroblasts were used as control.
V(D)J recombination assay in RS-SCID fibroblasts withArtemis gene mutations.
(A) Schematic representation of the signal joint assay (sj) and the coding joint assay (cj) in which the recombination substrates pGG49 and pGG51 were used, respectively. Recombination signal sequences are depicted as triangles and the flanking coding sequence as rectangles. The signal joints are retained in the recombination substrate when pGG49 is used, while coding joints are retained in the recombination substrate when pGG51 is used. Signal joints and coding joints were detected with nested PCR analysis using a 32P-labeled primer (see “Materials and methods”). (B) Primary fibroblasts of Artemis-1 and Artemis-2 transfected with RAG1 andRAG2 expression constructs and the recombination substrate formed signal joints but no coding joints. FN1 control primary fibroblasts can form both signal joints and coding joints. Defective coding joint formation in Artemis-1 and Artemis-2 could be complemented by cotransfection of the wt Artemis expression construct (+Art). (C) SV40-immortalized fibroblasts of Artemis-3 were also able to form signal joints but not coding joints. Like in Artemis-1 and Artemis-2, coding joint formation occurred upon cotransfection of the wt Artemis expression construct. SV40-immortalized MCR5 fibroblasts were used as control.
Complementation of the coding joint formation defect by cotransfection of wt Artemis constructs
The genomic deletion in patient Artemis-1, resulting in a premature stop codon, was most probably the disease-causing defect. However, we considered the possibility that the missense mutations in patients Artemis-2, -3.1, and -3.2 could represent polymorphisms. Therefore, we cotransfected the wt Artemis expression vector together with the RAG1 and RAG2 expression vectors and the recombination substrates into fibroblasts of the RS-SCID patients and analyzed the signal and coding joint formation. As shown in Figure 3, this resulted in complementation of the coding joint formation defect, thereby proving the disease-causing effect of the missense mutations in the Artemis gene. Furthermore, transfection of an Artemis expression vector containing the mutation of Artemis-3 could not complement coding joint formation in Artemis-2 fibroblasts and vice versa (data not shown).
Ig gene rearrangements in BMMCs
To study the capacity of mutated Artemis proteins to perform DNA dsb repair during human precursor B-cell differentiation in vivo, we isolated DNA from BMMCs and investigated the occurrence of incomplete DH-JH and complete VH-JH gene rearrangements. BM samples were available from patients Artemis-1 and Artemis-2. In both patients, incomplete DH-JH gene rearrangements could be amplified by PCR, but no complete VH-JH gene rearrangements could be detected.
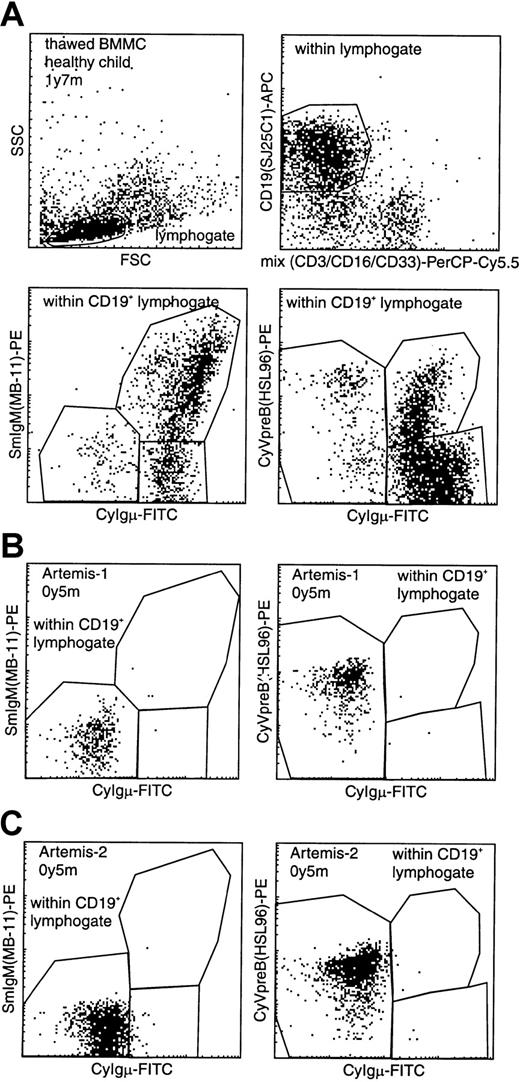
Flow cytometric analysis of precursor B cells in BM of RS-SCID patients with Artemis gene mutations
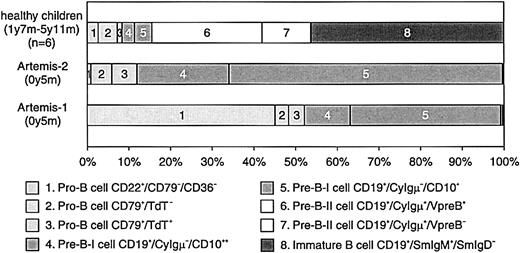
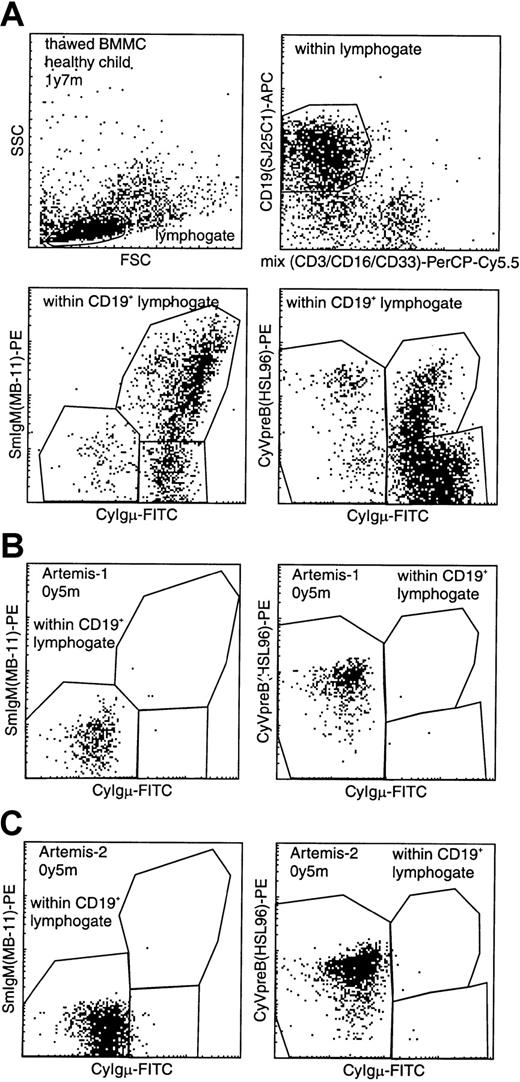
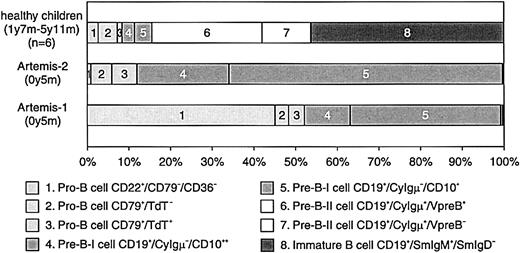
BM samples from 2 RS-SCID patients with Artemis gene mutations (Artemis-1 and -2) were available and analyzed as described before.22 23 The percentage of B cells within the lymphocyte gate differed between the 2 RS-SCID patients withArtemis gene mutations (Table3). In BM from healthy children, approximately 15% of the precursor B-cell compartment consisted of CyIgμ− precursor B cells, distributed over pro-B cells and pre-B-I cells with a pro-B/pre-B-I ratio of 1.3 ± 0.8, whereas in BM from both RS-SCID patients with Artemis gene mutations, 100% of the precursor B-cell compartment was located in the pro-B and pre-B-I cell stages (Figures 4and 5). Therefore, our results indicate that the differentiation arrest in the 2 RS-SCID patients with Artemis gene mutations resulted in a more than 6-fold relative accumulation of precursor B-cell subpopulations located before the transition from CyIgμ− pre-B-I cells to CyIgμ+ pre-B-II cells, with an inverted pro-B/pre-B-I ratio of 0.6 ± 0.7 in the 2 RS-SCID patients with Artemisgene mutations.
Flow cytometric analysis of CD19+lymphocytes in BM from a healthy control and 2 RS-SCID patients withArtemis gene mutations.
(A) Gating on CD19+ lymphocytes with exclusion of CD3+ T lymphocytes, CD16+ NK cells, and CD33+ myeloid cells, resulting in a purified “B-cell gate” (upper panels). Within this B-cell gate, expression of cytoplasmic VpreB (CyVpreB), cytoplasmic Igμ (CyIgμ), and surface membrane IgM (SmIgM) can be evaluated (lower panels), showing several subsets in normal BM with most cells in the most mature (Igμ+) precursor B-cell stages. (B-C) Patients Artemis-1 and -2 showed a complete arrest at the transition from CyIgμ− pre-B-I cells to CyIgμ+ pre-B-II cells. Although no CyIgμ expression was detected, virtually all CD19+ precursor B cells were positive for CyVpreB. This is fully in line with the expression of CyVpreB in early precursor B cells.
Flow cytometric analysis of CD19+lymphocytes in BM from a healthy control and 2 RS-SCID patients withArtemis gene mutations.
(A) Gating on CD19+ lymphocytes with exclusion of CD3+ T lymphocytes, CD16+ NK cells, and CD33+ myeloid cells, resulting in a purified “B-cell gate” (upper panels). Within this B-cell gate, expression of cytoplasmic VpreB (CyVpreB), cytoplasmic Igμ (CyIgμ), and surface membrane IgM (SmIgM) can be evaluated (lower panels), showing several subsets in normal BM with most cells in the most mature (Igμ+) precursor B-cell stages. (B-C) Patients Artemis-1 and -2 showed a complete arrest at the transition from CyIgμ− pre-B-I cells to CyIgμ+ pre-B-II cells. Although no CyIgμ expression was detected, virtually all CD19+ precursor B cells were positive for CyVpreB. This is fully in line with the expression of CyVpreB in early precursor B cells.
Composition of the precursor B-cell compartment in healthy controls and in the 2 studied RS-SCID patients with Artemis gene mutations.
In contrast to healthy controls, the precursor B-cell compartment in the 2 RS-SCID patients with Artemis gene mutations showed a complete arrest at the transition of pre-B-I cells to pre-B-II cells. The relative distribution between pro-B and pre-B-I cells was different in the 2 patients.
Composition of the precursor B-cell compartment in healthy controls and in the 2 studied RS-SCID patients with Artemis gene mutations.
In contrast to healthy controls, the precursor B-cell compartment in the 2 RS-SCID patients with Artemis gene mutations showed a complete arrest at the transition of pre-B-I cells to pre-B-II cells. The relative distribution between pro-B and pre-B-I cells was different in the 2 patients.
Discussion
From 13 T−B−NK+ SCID patients without RAG mutations, fibroblasts were analyzed for sensitivity to ionizing radiation. We found 5 of these 13 T−B−NK+ SCID patients to be RS, 4 of which contained Artemis mutations, indicating thatArtemis mutation is the most common reason for RS-SCID. We did not find a founder effect in our patients. In accordance with the suggestion of Moshous et al10 that the Artemisgene might represent a hot spot for gene deletion, we identified a large genomic deletion in patient Artemis-1. So far, none of the reported Artemis mutations were simple missense mutations, but the RS-SCID phenotype in patients Artemis-2, -3.1, and -3.2 was proven to be caused by aa substitutions in the SNM1 homology domain of the Artemis protein, because the coding joint formation defect in the fibroblast recombination assay of these RS-SCID patients could be complemented by cotransfection of the wt Artemis expression vector (Figure 3). The 2 point mutations were in evolutionary highly conserved aa residues in the SNM1 homology domain, suggesting that this domain is required for catalytic activity, probably the nuclease activity that can open the DNA hairpins at the coding ends.
Increased microhomology use in a plasmid recircularization assay proved to be highly diagnostic of mutations in the DNA-PK or ligase IV–XRCC4 complex.19 However, our 4 RS-SCID patients with mutations in the Artemis gene showed normal direct joining (Figure 2). This suggests that the Artemis protein functions in a different step of the end joining process. These results are consistent with the recent biochemical evidence that Artemis may be required for hairpin opening and DNA end processing.11
We previously described detection of IGH gene rearrangements in human BMMCs as a parameter for in vivo RAG activity.13Now we have used detection of IGH gene rearrangements in BMMCs of Artemis-1 and Artemis-2 as a parameter for residual activity of the Artemis protein. The complete absence of normal coding joints in patient Artemis-1 is in line with the large genomic deletion, which results in a truncated Artemis protein with absence of part of the SNM1 homology domain. Patient Artemis-2 contained relatively high frequencies of precursor B cells (23% of BM lymphocytes, Table 3), but we could detect only incomplete DH-JHrearrangements and no VH-JH gene rearrangements.
Flow cytometric evaluation of the BM precursor B-cell compartment in patients Artemis-1 and -2 showed a complete arrest at the transition from CyIgμ− pre-B-I cells to CyIgμ+pre-B-II cells. The completeness of this B-cell differentiation arrest illustrates the importance of Artemis at this stage of lymphoid differentiation.
In conclusion, both deletions and missense mutations in theArtemis gene can cause RS-SCID with defective coding joint formation and lead to an early and complete B-cell differentiation block.
The authors thank Penny Jeggo for 180BR, Jean-Pierre de Villartay for communication of results before publication, Michael Lieber for the pGG49 and pGG51 constructs, Mauro Modesti for XRCC4 antisera, Steve Jackson for anti-ATM antiserum, R. E. E. van Lange for technical assistance, and H. Karasuyama for making available the MAb HSL96 directed against the human VpreB protein.
Prepublished online as Blood First Edition Paper, October 24, 2002; DOI 10.1182/blood-2002-01-0187.
Supported by the Revolving Fund 2000 of the University Hospital Rotterdam, the Dutch Cancer Society/Koningin Wilhelmina Fonds (grants EUR 98-1775 and EMCR 2002-2734), and the Netherlands Scientific Organization (NWO grant AGIKO 920-03-089).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
J. J. M. van Dongen, Department of Immunology, Erasmus MC/University Medical Center Rotterdam, Dr Molewaterplein 50, 3015 GE Rotterdam, The Netherlands; e-mail:vandongen@immu.fgg.eur.nl.