Relapse of B-lineage acute lymphoblastic leukemia (B-ALL) after allogeneic hematopoietic stem cell transplantation (HSCT) commonly results from the failure of a graft-versus-leukemia (GVL) effect to eradicate minimal residual disease. Augmenting the GVL effect by the adoptive transfer of donor-derived B-ALL–specific T-cell clones is a conceptually attractive strategy to decrease relapse rates without exacerbating graft-versus-host disease (GVHD). Toward this end, we investigated whether a genetic engineering approach could render CD8+ cytotoxic T lymphocytes (CTLs) specific for tumor cells that express the B-cell lineage cell surface molecule CD19. This was accomplished by the genetic modification of CTLs to express a chimeric immunoreceptor composed of a CD19-specific single-chain immunoglobulin extracellular targeting domain fused to a CD3-ζ intracellular signaling domain. CD19-redirected CTL clones display potent CD19-specific lytic activity and chimeric immunoreceptor-regulated cytokine production and proliferation. Because B-ALL cells can evade T-cell/natural killer- cell recognition by down-regulation of cell surface accessory molecules that participate in the formation of a functional immunologic synapse, we compared the CD19-specific effector function of genetically modified CD8+ CTLs toward CD19+ cells with disparate levels of intercellular adhesion molecule 1 (ICAM-1), leukocyte function-associated antigen 1 (LFA-1), and LFA-3. We observed that recognition of B-lineage tumor lines by CD19-specific CTLs was not impaired by low levels of ICAM-1, LFA-1, and LFA-3 cell surface expression, a functional attribute that is likely a consequence of our high-affinity CD19-specific chimeric immunoreceptor. Furthermore, the CD19-specific CTLs could lyse primary B-ALL blasts. These preclinical observations form the basis for implementing clinical trials using donor-derived CD19-specific T-cell clones to treat or prevent relapse of B-ALL after allogeneic HSCT.
Introduction
The ability of allogeneic hematopoietic stem cell transplantation (HSCT) to eradicate hematologic neoplasia is dependent not only on the cytotoxic effects of high-dose chemotherapy and chemoradiation therapy, but the subsequent immune-mediated destruction of neoplastic cells via effector cells derived from the donor hematopoietic stem cell (HSC) graft.1 The potency of the graft-versus-leukemia (GVL) effect varies widely depending on the type of leukemia. For B-lineage acute lymphoblastic leukemia (B-ALL), the GVL effect is modest and consequently disease relapse after transplantation is a major contributor to treatment failure. Although intensifying cytotoxic conditioning regimens has diminished relapse rates, the benefit of this approach to improve disease-free survival is frequently mitigated by the increased regimen-related toxicities.2-4
Alternately, the problem of B-ALL posttransplantation relapse can be approached by therapeutic strategies to augment the GVL effect. The adoptive transfer of donor leukocytes following HSC engraftment can induce durable remissions particularly in patients with chronic myelogenous leukemia (CML). However, donor lymphocyte infusions (DLIs) achieve remission rates of less than 10% in patients with B-ALL and are associated with a high incidence and severity of graft-versus-host disease (GVHD) morbidity and mortality.5,6 Although the genetic modification of donor lymphocytes to introduce suicide genes may improve the safety profile of DLIs by rendering transferred lymphocytes susceptible to in vivo ablation, the low potency and high association with GVHD continue to be major obstacles to achieving clinically robust anti-B-ALL GVL augmentation by this approach.7-9
Adoptive transfer of donor-derived T cells having antigen specificity restricted to the leukemic clone itself or to target antigens selectively expressed by host hematopoietic lineage cells has the potential to selectively augment GVL without exacerbating GVHD. Minor histocompatibility antigens (mHags) encoded by polymorphic genes selectively expressed in recipient leukemic cells/hematopoietic-derived cells can serve as target antigens for donor T cells and mediate selective tumor eradication.10-15 Significant challenges face the widespread implementation of adoptive therapy with donor mHag-specific T cells; these include the molecular identification of a panel of mHags with restricted hematopoietic expression, the delineation of immunogenic epitopes for a large number of HLA alleles, and the technologies for reliably isolating T cells with desired specificities from donors.16,17 Additionally, tumor escape mechanisms such as antigen-loss variants, the immune-mediated selection of leukemic clones that have down-regulated restricting HLA alleles, or critical adhesion/costimulatory molecules will need to be addressed, particularly for B-ALL.18
The genetic modification of T cells to introduce antigen receptors that are capable of recognizing leukemic cells is a strategy that can overcome the requirement of isolating leukemia/mHag-specific T cells endogenous to the donor. Moreover, chimeric immunoreceptors that use antibody-derived single-chain variable domains (scFvs) bind to native cell surface epitopes and bypass the requirement for antigen processing and presentation by HLA molecules and are therefore universal.19,20 We have constructed a chimeric immunoreceptor having a CD19-specific scFv extracellular targeting domain that activates T cells via its cytoplasmic CD3-ζ signaling domain. CD19 is an attractive target for immunotherapy because the vast majority of B-ALLs uniformly express CD19, whereas expression is absent on nonhematopoietic cells, as well as myeloid, erythroid, and T cells, and bone marrow stem cells.21-23 Importantly, the clonogenic progenitor cell of B-ALL is also CD19+ based on in vitro progenitor assays.24-28
Here we demonstrate that primary human CD8+ cytotoxic T lymphocyte (CTL) clones expressing our CD19-specific chimeric immunoreceptor specifically recognize and lyse CD19+leukemia/lymphoma cells and primary B-ALL blasts and are activated for chimeric immunoreceptor-regulated cytokine production and proliferation. Additionally, we find that the high efficiency of tumor cell killing/CTL activation achieved via our chimeric immunoreceptor is independent of disparities in the expression levels of adhesion molecules on leukemic cells that participate in the formation of a productive immunologic synapse, an escape mechanism relevant to B-ALL.
Materials and methods
Expression plasmid
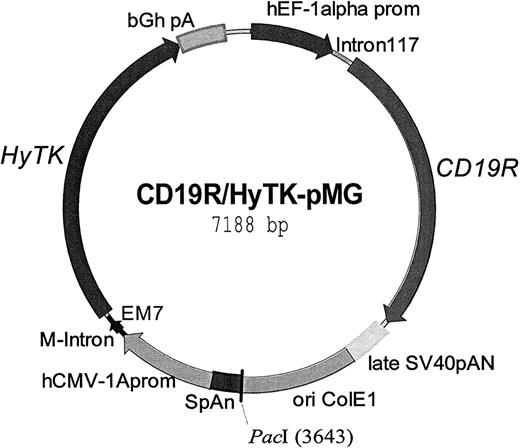
The CD19-specific chimeric immunoreceptor (designated CD19R) was assembled by polymerase chain reaction (PCR) using splicing by overlap extension (SOE).29 The scFv was constructed de novo using the human granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor α chain leader sequence and VL and VH regions derived from a CD19-specific murine IgG1 monoclonal antibody (mAb) designated FMC63,30 linked with the peptide GSTSGSGKPGSGEGSTKG. To generate the scFvFc, this motif was fused in-frame to residues 232-460 (using Kabat numbering system31) of the human IgG4 heavy chain corresponding to the Fc and hinge regions. The hinge was mutated from amino acids CPSC to CPPC to enhance stability of the dimerized IgG4 heavy chain.32 These domains were then fused in-frame to residues 372-393 of human CD4 transmembrane region (AIDS Research and Reference Reagen Program, catalog no. 157), which was, in turn, fused in-frame to residues 31-142 of the human cytoplasmic CD3-ζ chain, obtained by reverse transcription PCR from Jurkat cells,33to generate the scFvFcζ chimeric immunoreceptor. CD19R was ligated into the pMG expression plasmid (InvivoGen, San Diego, CA), that had been previously modified to delete the PacI restriction enzyme site at position 307 leaving a remainingPacI site for the purposes of linearization, under control of the hybrid human elongation factor-1α (hEF-1α) promoter. The cDNA encoding the bifunctional hygromycin phosphotransferase-thymidine kinase (HyTK) fusion selection/suicide enzyme34 was placed under the control of the cytomegalovirus (CMV) enhancer/promoter (hCMV-1A) to generate the plasmid CD19R/HyTK-pMG (Figure1). Correct assembly of CD19R and HyTK genes was verified from DNA sequence analyses. The plasmid was purified using Qiagen (Valencia, CA) endotoxin-free reagents and used for electroporation of peripheral blood mononuclear cells (PBMCs) after being digesting with PacI, precipitated, washed, and resuspended at 2 μg/μL in sterile water.
Schematic of the expression plasmid CD19R/HyTK-pMG.
This DNA vector was used to genetically modify primary human cytolytic T cells to coexpress the chimeric immunoreceptor specific for CD19, termed CD19R, and the bifunctional gene HyTK. Regulatory elements include the promoters hEFl-1 and hCMV-1A driving the expression of CD19R and HyTK genes, respectively. The polyadenylation signals from SV40 and bovine growth hormone are included after the termination codons of CD19Rand HyTK, respectively. The synthetic promoter EM7 drives the prokaryotic expression of hygromycin to select for bacterial drug resistance. The plasmid was linearized at the uniquePacI site (at base pair position 3643) prior to use in electroporation.
Schematic of the expression plasmid CD19R/HyTK-pMG.
This DNA vector was used to genetically modify primary human cytolytic T cells to coexpress the chimeric immunoreceptor specific for CD19, termed CD19R, and the bifunctional gene HyTK. Regulatory elements include the promoters hEFl-1 and hCMV-1A driving the expression of CD19R and HyTK genes, respectively. The polyadenylation signals from SV40 and bovine growth hormone are included after the termination codons of CD19Rand HyTK, respectively. The synthetic promoter EM7 drives the prokaryotic expression of hygromycin to select for bacterial drug resistance. The plasmid was linearized at the uniquePacI site (at base pair position 3643) prior to use in electroporation.
Cells
Primary human T cells from PBMCs, isolated by density gradient centrifugation over Ficoll-Paque-Plus (Pharmacia Biotech, Pistacaway, NJ), were maintained in tissue culture using 14-day stimulation cycles as previously described.35 Briefly, on day 0 of each cycle, 105 T cells were mixed with 50 × 106PBMCs irradiated at 3500 cGy, 10 × 106 lymphoblastoid cells (LCLs) irradiated at 8000 cGy, and 30 ng/mL OKT3 (Ortho Biotech, Raritan, NJ) in RPMI 1640 (Biowhittaker, Walkersville, MD) supplemented with 2 mM l-glutamine (Irvine Scientific, Santa Ana, CA), 25 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; Irvine Scientific), 100 U/mL penicillin, 0.1 mg/mL streptomycin (Irvine Scientific), and 10% heat-inactivated defined fetal calf serum (FCS; Hyclone, Logan, UT). Beginning on day 1 of each stimulation cycle recombinant human interleukin 2 (rhIL-2; Proleukin, Chiron, Emeryville, CA) at either 25 U/mL (T-cell “bulk” culture) or 50 U/mL (T-cell clone) was added every 48 hours. On day 5 of each stimulation cycle hygromycin B (InvivoGen) at 0.2 mg/mL was added. T cells were cloned on day 0 of a cycle in 96-well plates by limiting dilution using a combined mixture of irradiated PBMCs and LCLs in media containing OKT3 at 30 ng/mL and rhIL-2 at 50 U/mL. Hygromycin B at 0.2 mg/mL was added 5 days after plating. Twenty-one days after plating, the cells from wells that demonstrated growth were expanded. The human CD19+ leukemia lines RS4,36SUP-B15,37 JM1, and the Daudi38 Burkitt lymphoma line, and the CD19−K562 erythroleukemia line were obtained from the American Type Culture Collection (Manassas, VA). CD19+ LCLs were established from human Epstein-Barr virus (EBV)–infected PBMCs cultured in the presence of cyclosporin.39 These lines were grown using the RPMI 1640 media supplemented as described. The adherent CD19−Be-2 neuroblastoma cell line (kindly provided by Dr Maurer, Children's Hospital Los Angeles, Los Angeles, CA) was grown in Dulbecco modified Eagle medium high glucose (Irvine Scientific) supplemented with 2 mM l-glutamine, 25 mM HEPES, 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 10% heat-inactivated FCS. The primary B-ALL blasts were obtained from discarded material after obtaining informed consent from the individuals.
Genetic modification
T-cell genetic modification was carried out as previously described.40 Briefly, 3 days after activation with OKT3 the PBMCs were resuspended in hypotonic electroporation buffer (Eppendorf, Brinkmann Instruments, Westbury, NY) at 20 × 106 cells/mL. Then, 25 μg of the linearized plasmid DNA CD19R/HyTK-pMG together with 400 μL cell suspension were added to sterile 0.2-cm electroporation cuvettes and subjected to a single electrical pulse of 250 V/40 μs delivered by the Multiporator electroporation device (Eppendorf).
Flow cytometry
The cell surface phenotype of the genetically modified T cells and tumor lines was determined by staining with the following: fluorescein isothiocyanate (FITC)–conjugated or phycoerythrin (PE)–conjugated mAbs specific for T-cell receptor α/β (TCRαβ), CD2, CD3ε, CD4, CD8, CD11a, CD16, CD18, CD28, CD54, CD57, and CD58 (BD Biosciences, San Jose, CA). Surface expression of the CD19-specific immunoreceptor was detected using the F(ab′)2 fragment of FITC-conjugated goat antibody specific for human IgG Fc fragment (Immunotech, Marseilles, France). Briefly, 105 to 106 washed cells were resuspended in 100 μL Hanks balanced salt solution (HBSS) supplemented with 2% FCS, 0.2 mg/mL NaN3 staining buffer, and 0.5 to 2 μL of the stock antibody preparation. Following a 60-minute incubation on ice, the cells were washed twice and resuspended in staining buffer containing propidium iodide (PI) at 1 μg/mL. Dead cells were excluded from analysis on uptake of PI. Intracellular staining with PE-conjugated mAb specific for perforin (BD Biosciences) was undertaken on T cells that had been fixed and permeabolized using the Pharmingen (BD Biosciences) system. Cells were analyzed on a FACSCalibur cytometer (BD Immunocytometry Systems, San Jose, CA). CellQuest software (BD Immunocytometry Systems) was used to calculate the percentage of cells and mean fluorescent intensity (MFI) within a given region of a histogram.
Western blot
Expression of the CD19R was determined by Western blot using a mAb specific for CD3-ζ to detect the chimeric immunoreceptor. Whole cell lysates of transfected T-cell clones were prepared by lysis of 106 washed cells in 1 mL RIPA buffer (phosphate-buffered saline [PBS], 1% NP40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing 1 tablet/10 mL Complete Protease Inhibitor Cocktail (Boehringer Mannheim, Penzberg, Federal Republic of Germany). After a 60-minute incubation on ice, aliquots of centrifuged whole cell lysate supernatant were harvested and boiled in an equal volume of loading buffer under reducing conditions and then subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on precast 12% acrylamide gels (Bio-Rad Laboratories, Hercules, CA). Following transfer to nitrocellulose, membranes were blocked for 2 hours in Blotto solution containing 0.07 g/mL nonfat dried milk. Membranes were washed in T-TBS (0.05% Tween 20 in Tris [tris(hydroxymethyl)aminomethane]–buffered saline, pH 8.0) and then incubated with primary mouse anti–human CD3-ζ mAb 8D3 (Pharmingen, San Diego, CA) at a concentration of 1 μg/mL for 2 hours. Prior to developing, the membranes were washed 4 times in T-TBS and then incubated for 1 hour with a 1:500 dilution of alkaline phosphatase–conjugated goat antibody specific for murine IgG. After rinsing in T-TBS the membranes were developed with 30 mL AKP solution (Promega, Madison, WI) per the manufacturer's instructions.
Chromium release assay
The cytolytic activity of genetically modified CD8+T-cell clones was determined in a 4-hour chromium release assay (CRA) using 51Cr-labeled RS4, Daudi, JM-1, SupB-15, and erythroleukemia K562 target cells. The T-cell effectors were harvested 12 to 14 days following stimulation with OKT3, washed, and plated in V-bottom microtiter plates (Costar, Cambridge, MA) at 37°C for 4 hours in triplicate at 2.5 × 105, 1.25 × 105, 0.25 × 105, and 0.05 × 105/well with 5 × 103 target cells. After centrifugation and incubation, 100-μL aliquots of cell-free supernatant were harvested and counted and the percent specific cytolysis was calculated from the release of 51Cr as follows:
Control wells contained target cells incubated in media. The maximal 51Cr was determined by measuring the51Cr content released by target cells lysed with 2% SDS. Data are reported as an average ± SD.
Cytokine production
Duplicate wells containing 106 cells of a genetically modified CD8+ T-cell clone were coincubated with 106 stimulator cells irradiated to 8000 cGy in 2 mL culture media for 72 hours with rhIL-2 at 5 U/mL. Cell-free supernatants were harvested and assayed for cytokine content by enzyme-linked immunosorbent assay (ELISA; R & D Systems, Minneapolis, MN) and the concentration was extrapolated from a standard curve.
T-cell proliferation
Cells (105) of a CD19-specific CD8+T-cell clone were cocultured with 2 × 106 irradiated PBMCs, with and without a panel of 105 irradiated stimulator cells. Parallel culture conditions were established without the addition of genetically modified T cells. T cells were also plated without PBMCs and stimulator cells. Then, rhIL-2 at 5 U/mL was added every 48 hours. The viable cells, based on exclusion with trypan blue, were enumerated at day 7 and day 14. On day 7 of the culture, 2 × 106 irradiated PBMCs were re-added with and without the irradiated stimulator cells at a 1:1 ratio with the T cells. The assay was repeated 3 times. Fold expansion was calculated by dividing the average cell counts by the input T-cell number of 105. The proliferative activity was also measured by means of a [3H]thymidine incorporation assay. T cells (105) were cocultured with 106 irradiated PBMCs with and without irradiated 105 Daudi and LCL stimulator cells. Then, rhIL-2 at 5 U/mL was added at the beginning of the assay and again at 48 hours. On the fourth day of culture [3H]thymidine (ICN, Costa Mesa, CA) was added at a concentration of 50 μCi/mL (7.4 MBq). The following day the incorporation of [3H]thymidine into T cells was measured by seeding the culture wells into replicates of 6 wells on 96-well plates and harvesting the cells on glass fiber filters with a PHD Harvester (Brandel, Gaithersburg, MD), and then radioactivity was measured with a liquid scintillation LS 6500 counter (Beckman Coulter, Fullerton, CA). Data are reported as an average ± SD.
Results
Genetically modified T-cell clones express the CD19-specific chimeric immunoreceptor
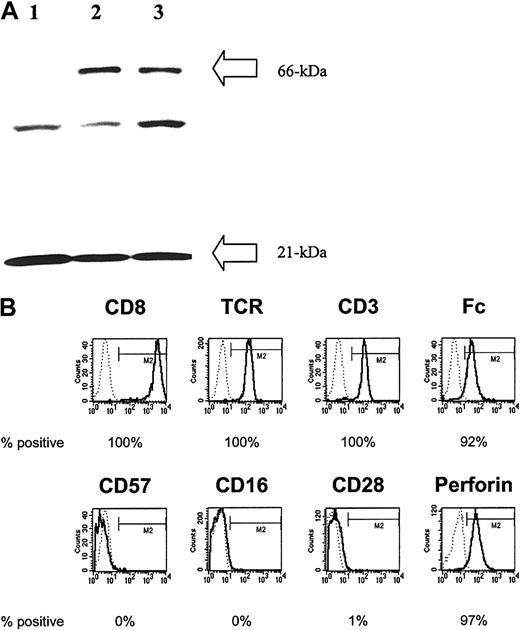
To generate CD19-specific T cells that coexpress CD19Rand HyTK genes, PBMCs were activated with OKT3 mAb specific for CD3 and the naked DNA linearized plasmid CD19R/HyTk-pMG was introduced by electrotransfer. Four weeks after electroporation, an outgrowth of cells in the presence of cytocidal concentrations of hygromycin was observed. Culture systems were developed to retrieve clones of stable transfectants by plating T cells in limiting dilution. This procedure has yielded drug-resistant clones in multiple separate electroporations of T cells and these clones can be expanded to 1010 in number over approximately 10 weeks with retention of both transgene expression and redirected CTL effector function. Although these culture conditions tend to favor the emergence of CD8+ CTL clones whose growth is supported by the addition of rhIL-2, CD4+ CTL clones can be identified. Genetically modified hygromycin-resistant CD8+ T-cell clones were evaluated for expression of the chimeric immunoreceptor by Western blot of reduced whole cell lysates probed using a mAb specific for CD3-ζ chain. This technique revealed that the genetically modified T cells express the introduced 66-kDa chimeric ζ chain in addition to the 21-kDa endogenous ζ chain (Figure2A). An additional band between 21 and 66 kDa was routinely observed in both unmodified and genetically modified T cells and is of uncertain significance. Flow cytometry using mAbs specific for CD8, CD4, TCRαβ, and F(ab′)2 fragments of goat polyclonal antibody specific for human Fc were used to validate that the majority of genetically modified hygromycin-resistant T-cell clones from healthy donors uniformly express CD8+CD4−TCRαβ+Fc+on the surface (Figure 2B). However, as is typical for CD8+T cells expanded ex vivo using the described techniques, these differentiated effector cells failed to express CD28, rendering these CTLs unable to receive costimulatory signals from the binding of CD28 to B7 molecules on tumor cells.
Genetically modified lymphocytes express the introduced CD19-specific chimeric immunoreceptor and are differentiated effector T cells.
(A) Western blot result from reduced whole T-cell lysates probed with an anti–CD3-ζ mAb. The unmodified T-cell line (lane 1) and representative genetically modified T-cell clones (lanes 2 and 3) each display a 21-kDa band consistent with the wild-type ζ chain. In addition, the 2 hygromycin-resistant T-cell clones demonstrate a second band of approximately 66 kDa consistent with the introduced chimeric ζ chain. (B) Phenotype of a CD19-specific T-cell clone using flow cytometry. Binding of specific mAbs (bold lines) is relative to isotype control (dotted lines), and the percentage of cells in the M2 gate is indicated.
Genetically modified lymphocytes express the introduced CD19-specific chimeric immunoreceptor and are differentiated effector T cells.
(A) Western blot result from reduced whole T-cell lysates probed with an anti–CD3-ζ mAb. The unmodified T-cell line (lane 1) and representative genetically modified T-cell clones (lanes 2 and 3) each display a 21-kDa band consistent with the wild-type ζ chain. In addition, the 2 hygromycin-resistant T-cell clones demonstrate a second band of approximately 66 kDa consistent with the introduced chimeric ζ chain. (B) Phenotype of a CD19-specific T-cell clone using flow cytometry. Binding of specific mAbs (bold lines) is relative to isotype control (dotted lines), and the percentage of cells in the M2 gate is indicated.
CD8+ T cells induce GVL cytotoxic effects primarily through perforin-mediated cytotoxicity by releasing granule contents after target cell recognition.41 Therefore, we investigated the perforin content in the genetically modified CTLs using flow cytometry and found that the CD8+ CD19-specific T cells uniformly express the cytotoxic effector protein perforin (Figure 2B). Because human natural killer (NK) cells, expressing CD57 or CD16 or both, can also exhibit cytotoxicity, we used the lack of surface expression of these markers to distinguish the genetically modified T-cell clones from NK/T cells.
CD19+ tumor lines express variable levels of T-cell adhesion molecules
A panel of CD19+ B-ALL and lymphoma lines was assembled to investigate the role of adhesion molecules in the effector function of CD19-specific T cells. The adhesion molecules were selected based on an ability to form stable conjugates between T cells and target cells, thereby contributing to the assembly of the immune synapse. For example, the binding of intercellular adhesion molecule-1 (ICAM-1, CD54) with lymphocyte function-associated antigen-1 (LFA-1, CD11a/18) and CD2 with LFA-3 (CD58) can facilitate T-cell coactivation on docking of the TCR with cognate antigen.42-44 The relative cell surface expression of these adhesion molecules on the tumor lines was demonstrated by flow cytometry using a panel of adhesion molecule-specific mAbs. Even though CD19-specific CD8+ T cells expressed relatively high levels all of the accessory molecules tested, the pattern of expression of these molecules varied on the CD19+ tumor lines, both in percentage of cells expressing adhesion molecules and the density of adhesion molecules detected on the surface (Figure3). The flow cytometry data revealed that the RS4 and SUP-B15 cells expressed the lowest levels of CD11a and CD18, whereas Daudi and JM1 cells expressed the lowest levels of CD58 and the expression of CD54 was reduced on SUP-B15 cells. Furthermore, there was variation between the tumor lines in the density of adhesion molecule expression as measured by MFI. For example, there was approximately a 10-fold range in the MFI associated with CD54 and CD58 on tumor lines that uniformly express these determinants. However, whereas the leukemia and lymphoma cell lines, RS4, Sup-B15, Daudi, and JM-1 varied in the relative expression and intensity of anti-ICAM-1, LFA-1, and LFA-3 staining, the EBV-transformed LCLs uniformly expressed the highest levels of these adhesion molecules.
B-ALL and lymphoma CD19+ tumor lines differ in relative expression of T-cell adhesion molecules.
Cell surface expression of molecules that participate in activation of CD19-specific T cells assessed by flow cytometry on a panel of CD19+B-ALL and lymphoma lines (A) and a genetically modified T cell (B). Relative expression of the adhesion molecules and CD19 (solid lines) within the gate M2 is reported as a percentage and was determined based on binding of the specific mAbs compared with isotype-matched controls (dotted lines). The MFI for the cells in gate M2 reflects the relative density of adhesion molecules expressed on the cell surface. Dead cells were excluded on uptake of PI.
B-ALL and lymphoma CD19+ tumor lines differ in relative expression of T-cell adhesion molecules.
Cell surface expression of molecules that participate in activation of CD19-specific T cells assessed by flow cytometry on a panel of CD19+B-ALL and lymphoma lines (A) and a genetically modified T cell (B). Relative expression of the adhesion molecules and CD19 (solid lines) within the gate M2 is reported as a percentage and was determined based on binding of the specific mAbs compared with isotype-matched controls (dotted lines). The MFI for the cells in gate M2 reflects the relative density of adhesion molecules expressed on the cell surface. Dead cells were excluded on uptake of PI.
CD19+ tumor lines are lysed equivalently by CD19-specific T cells
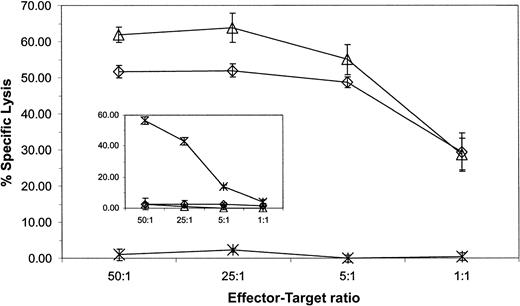
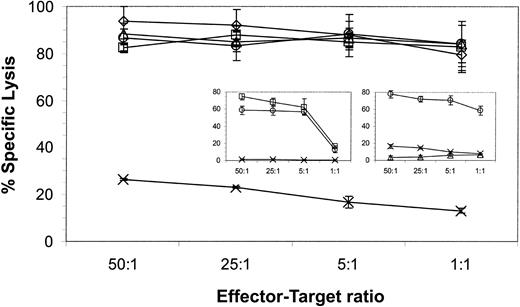
We investigated whether the altered expression of adhesion molecules on CD19+ tumor lines correlated with a susceptibility to be lysed by CD19-specific CD8+ T cells. These B-ALL and lymphoma lines expressed approximately equivalent levels of CD19 determinant; thus, differences in lysis could not be explained by variation of antigen expression (Figure 3). The lines were used as targets in a 4-hour CRA and the analysis revealed that the ability of a genetically modified T-cell clone to lyse the CD19+ lymphoma and leukemia lines was independent of the relative expression of LFA-1, LFA-3, and ICAM-1. Furthermore, both CD8+ and CD4+ genetically modified CTL clones efficiently killed HLA-mismatched human CD19+ leukemia and lymphoma cells because maximal lysis was achieved with an effector-target ratio of 5:1 (Figure 4). The genetically modified T cells preserve their cytolytic activity after extensive in vitro culture because the results from the 4-hour CRA presented are from a CD19-specific T-cell clone that has been cultured in vitro for approximately 100 days. The specificity of the genetically modified T cells for CD19 was demonstrated by the relative lack of lysis of the K562 CD19− cell line (Figure 4) and lack of lysis of the CD19−Be-2 neuroblastoma line (Figure7). In addition, CD8+ T cells genetically modified to be CD20 specific (M.C.J., L.J.N.C., A. M. Wu, S.J.F., A.R., manuscript submitted) fail to lyse the CD20−CD19+ JM1 leukemia line, but can lyse CD20+CD19+ Daudi cells, indicating that the specificity of activation is mediated via the specificity of the chimeric immunoreceptor's scFv (Figure 4 insert right).
CD19-specific CTL clone, but not CD20-specific CTL-clone, lyses CD19+ B-ALL and lymphoma targets.
CD19-specific effector cells from a CD8+ CTL clone and a CD19-specific CD4+ CTL clone (left insert ) and CD20-specific effector cells from a CD8+ CTL clone (right insert) were incubated with some or all of 51Cr-labeled CD19+ B-ALL and lymphoma lines: Daudi (○), SUP-B15 (⋄), JM1 (▵), RS4 (■), and CD19−target line K562 (×). Daudi cells, but not JM1 and K562 cells, express CD20 determinant. Lytic activity was calculated by measuring chromium release after 4 hours. Spontaneous release for each target was 10% or less. Average counts ± SD from triplicates are shown.
CD19-specific CTL clone, but not CD20-specific CTL-clone, lyses CD19+ B-ALL and lymphoma targets.
CD19-specific effector cells from a CD8+ CTL clone and a CD19-specific CD4+ CTL clone (left insert ) and CD20-specific effector cells from a CD8+ CTL clone (right insert) were incubated with some or all of 51Cr-labeled CD19+ B-ALL and lymphoma lines: Daudi (○), SUP-B15 (⋄), JM1 (▵), RS4 (■), and CD19−target line K562 (×). Daudi cells, but not JM1 and K562 cells, express CD20 determinant. Lytic activity was calculated by measuring chromium release after 4 hours. Spontaneous release for each target was 10% or less. Average counts ± SD from triplicates are shown.
CD19-specific T cells produce cytokines in response to stimulation with CD19+ tumor cells
The ability of T cells to modulate cytokine secretion in response to antigenic stimulation is important for an effective immune response.45 46 To test whether the genetically modified T-cell clones are activated via the chimeric immunoreceptor for regulated CD19-specific cytokine secretion in response to human CD19+ leukemia and lymphoma cells, the CD19-specific T-cell clones were cocultured with the panel of tumor cells. Conditioned supernatants from these cultures were tested for production of the cytokines GM-CSF, tumor necrosis factor α (TNF-α), and interferon γ (IFN-γ). The results for a CD8+ CD19-specific cytolytic T-cell clone are shown in Figure5. The genetically modified T-cell clone produced cytokines in response to each of the CD19+leukemia and lymphoma stimulator cells, but not by a CD19−stimulator cell (K562), nor by media alone. The T-cell production of IFN-γ in response to each CD19+ tumor line was approximately equivalent, whereas RS4 and SUP-B15 cells were able to stimulate the highest levels of TNF-α, and Daudi cells and LCLs stimulated the highest levels of GM-CSF. Thus, the CD19-specific T cells could be activated to secrete Tc1 cytokines despite alterations in the expression of adhesion molecules on the tumor cells.
CD19-specific CD8+ T cells produce cytokines in response to CD19+ B-ALL and lymphoma stimulator cells.
T cells (106) from a CD8+ CTL clone were cocultured with mitomycin-treated CD19+ B-ALL and lymphoma lines SUP-B15, LCL, RS4, Daudi, or the CD19− line K562. After 7 days, secretion of the cytokines GM-CSF (A), TNF-α (B), and IFN-γ (C) were quantified from conditioned supernatant by an ELISA.
CD19-specific CD8+ T cells produce cytokines in response to CD19+ B-ALL and lymphoma stimulator cells.
T cells (106) from a CD8+ CTL clone were cocultured with mitomycin-treated CD19+ B-ALL and lymphoma lines SUP-B15, LCL, RS4, Daudi, or the CD19− line K562. After 7 days, secretion of the cytokines GM-CSF (A), TNF-α (B), and IFN-γ (C) were quantified from conditioned supernatant by an ELISA.
CD19-specific T cells proliferate in response CD19+tumor lines
A proliferative response of CD19-specific CTLs to CD19+ tumor targets indicates that the chimeric immunoreceptor is competent to deliver a stimulatory response for growth and establishes that the docking of genetically modified T cells with CD19+ tumor cells does not lead to T-cell degradation. Figure 6 demonstrates that a CD8+ CD19-specific T-cell clone could exhibit up to an 8-fold expansion in numbers over a 2-week period when cocultured with CD19+ leukemia and lymphoma stimulators. The proliferation of genetically modified T cells in response to the leukemia SUP-B15 and lymphoma Daudi lines was superior to using LCLs as stimulators, despite the decreased expression of adhesion molecules relative to LCLs. Furthermore, the proliferation of the genetically modified T-cell clones in response to Daudi stimulators was similar to the cell growth achieved using OKT3, PBMCs, and LCLs to activate the CTL—culture conditions that are similar to those used for the in vitro expansion of T cells. The proliferation of CD19-specific T cells was dependent on the expression of CD19 on tumor cells because tissue culture conditions lacking CD19+ tumor cells resulted in only minimal T-cell expansion. Similar results were obtained using an assay that measured the incorporation of [3H]thymidine by proliferating T cells (Figure 6 insert). Each of the culture conditions included low-dose rhIL-2 added at 5 U/mL; however, the use of this cytokine alone was insufficient to sustain the survival of the CD19-specific CD8+ T cells, as demonstrated both by the failure of the T cells to proliferate within 14 days when cultured with rhIL-2 in the absence of CD19+ tumor cells, and by the correspondingly near-background incorporation of [3H]thymidine by the T cells. The addition of PBMCs, containing normal CD19+ B cells, along with low-dose rhIL-2, could maintain the survival of the genetically modified T cells, but this condition resulted in only a 2-fold expansion and was associated with background levels of [3H]thymidine incorporation.
CD19-specific T cells proliferate in response to stimulation by CD19+ B-ALL and lymphoma cells.
T cells (1 × 105) from a CD19-specific CD8+CTL clone were cocultured with 2 × 106 irradiated PBMCs. Some cultures received 1 × 105 irradiated CD19+ or CD19− stimulator cells. The 2 × 106 irradiated PBMCs and irradiated stimulator cells, at a 1:1 ratio with the T cells, were re-added after 7 days. One culture condition was the T-cell clone incubated without PBMCs and stimulator cells. Each culture condition was supplemented with 5 U/mL rhIL-2 every 48 hours. The cells were counted after 14 days and nonviable cells excluded by trypan blue staining. The average cell counts and SDs from 3 repeats are presented. In the insert, 105 T cells from a CD19-specific CD8+ CTL clone were cocultured as above. Methyl-[3H]thymidine was added after 96 hours and the incorporated counts were measured 18 hours later. The background incorporation of [3H]thymidine by Daudi cells and a LCL was 827 ± 358 and 909 ± 493, respectively. The average cell counts and SDs from triplicates are presented.
CD19-specific T cells proliferate in response to stimulation by CD19+ B-ALL and lymphoma cells.
T cells (1 × 105) from a CD19-specific CD8+CTL clone were cocultured with 2 × 106 irradiated PBMCs. Some cultures received 1 × 105 irradiated CD19+ or CD19− stimulator cells. The 2 × 106 irradiated PBMCs and irradiated stimulator cells, at a 1:1 ratio with the T cells, were re-added after 7 days. One culture condition was the T-cell clone incubated without PBMCs and stimulator cells. Each culture condition was supplemented with 5 U/mL rhIL-2 every 48 hours. The cells were counted after 14 days and nonviable cells excluded by trypan blue staining. The average cell counts and SDs from 3 repeats are presented. In the insert, 105 T cells from a CD19-specific CD8+ CTL clone were cocultured as above. Methyl-[3H]thymidine was added after 96 hours and the incorporated counts were measured 18 hours later. The background incorporation of [3H]thymidine by Daudi cells and a LCL was 827 ± 358 and 909 ± 493, respectively. The average cell counts and SDs from triplicates are presented.
CD19-specific T cells recognize CD19+ primary B-ALL cells
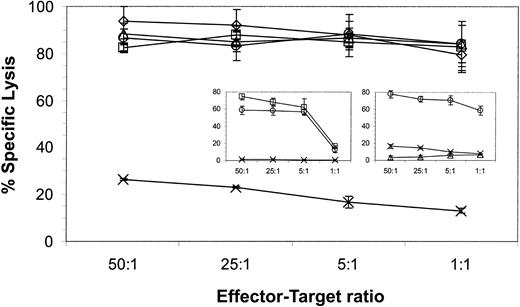
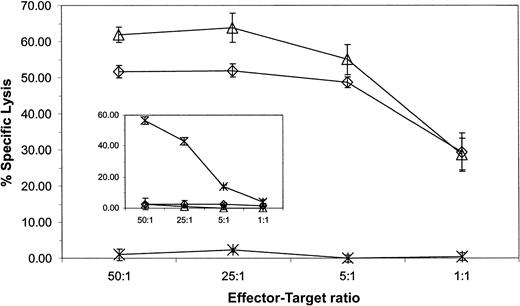
The ability of CD19-specific T cells to recognize B-ALL and lymphoma lines suggested that these genetically modified T cells might also recognize primary B-ALL cells. Cryopreserved blasts were used as targets in a 4-hour CRA to test this hypothesis. Figure7 demonstrates that a CD19-specific CTL clone could lyse CD19+ blasts almost to the same extent as the CD19+ JM1 leukemic line. A CTL clone genetically modified to express a chimeric immunoreceptor specific for a neuroblastoma-specific antigen failed to recognize the blasts, but did lyse the CD19−Be-2 neuroblastoma line (Figure 7 insert). These data indicate that the CD19-specific immunoreceptor is sufficient to trigger killing of primary CD19+ leukemia cells.
CD19-specific CTL clone lyses CD19+ primary B-ALL.
CD19-specific effector cells from a CD8+ CTL clone and a neuroblastoma-specific CD8+ CTL clone (insert) were incubated with 51Cr-labeled CD19+CD10+ primary B-ALL blasts (⋄), JM1 CD19+ B-ALL line (▵) and CD19−Be2 neuroblastoma line (×). Lytic activity was calculated by measuring chromium release after 4 hours. Spontaneous release for each target was 15% or less. Average counts ± SD from triplicates are shown. The percentage, as measured by flow cytometry, of CD19+CD10+ blasts was 84% and of CD19+CD10− B cells was 4%, in the primary sample.
CD19-specific CTL clone lyses CD19+ primary B-ALL.
CD19-specific effector cells from a CD8+ CTL clone and a neuroblastoma-specific CD8+ CTL clone (insert) were incubated with 51Cr-labeled CD19+CD10+ primary B-ALL blasts (⋄), JM1 CD19+ B-ALL line (▵) and CD19−Be2 neuroblastoma line (×). Lytic activity was calculated by measuring chromium release after 4 hours. Spontaneous release for each target was 15% or less. Average counts ± SD from triplicates are shown. The percentage, as measured by flow cytometry, of CD19+CD10+ blasts was 84% and of CD19+CD10− B cells was 4%, in the primary sample.
Discussion
Our data demonstrate that CD19 can serve as a target for primary human CTL clones with specificity redirected via a CD19-specific chimeric immunoreceptor. CD19+ lymphoma and leukemic lines, as well as primary B-ALL cells, are efficiently killed by clonal populations of these engineered effector cells although the chimeric immunoreceptor also regulates CD19-specific cytokine secretion and proliferation. We observed that the efficiency of CTL activation for these effector functions by B-ALL and lymphoma cells was not impaired despite the low-level expression of ICAM-1, LFA-1, and LFA-3 accessory molecules on target cells. This result is likely attributable to the high affinity of the chimeric immunoreceptor's scFv for CD19 as a means to overcome the impaired avidity of the T-cell immunologic synapse with targets expressing low levels of ICAM-1 or LFA-1 or LFA-3. Down-regulation of these accessory molecules is common in B-ALL and is likely to be an immunologic escape mechanism used by leukemic cells to avoid recognition by T cells and NK cells.47-52
CD19 represents a ubiquitous target moiety for B-lineage leukemias and lymphomas. Targeting CD19 with mAb-based therapeutics is the subject of clinical investigation and has not, to date, revealed a high incidence of selection for CD19− escape variants.53-55The engineering of bispecific antibody constructs with dual specificity for CD19 and TCR complex CD3 is under development as a strategy to redirect T-cell specificity to CD19.56 Targeting CD19 with genetically modified T cells has potential advantages over anti-CD19×CD3 bispecific antibodies, because the engineered T cells themselves can recycle the chimeric immunoreceptor by stable expression and are not dependent on diffusion-limited colocalization of antibody into tissue compartments to have their redirected CD19 specificity.
Because nonmalignant CD19+ B cells will be subject to recognition by redirected CTLs, the persistence of the adoptively transferred CD19-specific CTLs has the potential to exacerbate/prolong the B-cell immunodeficiency associated with allogeneic HSCT. However, the in vivo persistence of the genetically modified T cells can be limited by using CD19-specific CD8+ CTLs that are dependent on exogenous rhIL-2 for survival, or by coexpression of a suicide gene, such as HSV-TK. In patients who are predicted to relapse with B-ALL after allogeneic HSCT, the clinical sequelae of temporary B-cell lymphopenia may be an acceptable side effect of CD19-directed immunotherapy, especially because prolonged ablation of normal CD20+ B cells in patients receiving rituximab therapy does not appear to result in clinically significant complications attributable to depleted numbers of normal B cells.57
Although the safety of the patient in initial adoptive immunotherapy trials may be enhanced through the coexpression of a suicide gene along with the CD19-specific immunoreceptor in T-cell clones, the expression of the HyTK gene may limit the in vivo survival of the genetically modified T cells, as a result of an immune response directed against the transgenes. To reduce the immunogenicity of genetically modified T cells, our laboratory is exploring the use of flow cytometry operating in compliance with current good manufacturing practices (cGMPs) to sort on T cells displaying an introduced chimeric immunoreceptor that have not undergone hygromycin drug selection, humanizing the CD19-specific scFvs, and use of the human Fas-based suicide switch.58
Although cloned genetically modified CTLs represent a homogeneous high-potency cell product for clinical experimentation that maximizes safety due to dependency on exogenous rhIL-2, the optimal cell product will have to account for the potential adverse impact of immunogenicity of expressed transgenes, the lack of costimulatory ligands on B-ALL, and deficits in homing of ex vivo propagated T cells to all tissue compartments harboring leukemic blasts. Under development is the further engineering of CTLs to express engineered costimulatory receptors to enhance their function and prevent activation-induced cell death,59 and the genetic modification of clones to express selectin/chemokine receptor pairs for specified tissue homing.60-62 Our naked DNA T-cell transfection procedure provides for ample flexibility to engineer polycistronic vectors and is a cost-effective manufacturing practice to produce clinical grade material. Finally, as Brenner et al have suggested, future studies will also use T cells that are selected on the basis of the clone's endogenous αβ TCR specificity, to generate genetically modified T cells that are capable of recognizing 2 defined target antigens.63 Our laboratory is currently generating T-cell clones that are both CMV reactive and CD19 specific to help restore a CMV immune response as well as augment the antitumor GVL effect after allogeneic T cell-depleted HSCT for B-ALL.
These preclinical data lay the groundwork for clinical trials to determine the safety, feasibility, and efficacy of using donor-derived CD19-specific CTL clones after allogeneic HSCT. Using cGMP-compliant methodologies currently used in Food and Drug Administration–authorized clinical trials at City of Hope National Medical Center for isolating and expanding plasmid vector-modified CTL clones, we will be initiating a clinical trial in which patients experiencing a molecular B-ALL relapse will be treated with escalating cell doses of donor-derived CD19-specific CTL clones selected on the basis of expressing an endogenous αβ TCR devoid of GVHD reactivity. These early feasibility and safety studies will provide the basis for subsequent trials involving second-generation T-cell products with enhanced antileukemic potency and capacity for persistence and homing.
Prepublished online as Blood First Edition Paper, October 10, 2002; DOI 10.1182/blood-2002-07-1989.
Supported by CA30206, CA33572, Leukemia and Lymphoma Society, Altschul and Abe and Estelle Sanders foundations, Zagoria Foundation, Deutsche Forschungsgemeinschaft (To208/1-1) and the Cancer Research Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Laurence J. N. Cooper, Division of Molecular Medicine Beckman Research Institute, Division of Pediatric Hematology/Oncology, City of Hope National Medical Center, 1500 E Duarte Rd, Duarte, CA 91010; e-mail: lcooper@coh.org.



![Fig. 6. CD19-specific T cells proliferate in response to stimulation by CD19+ B-ALL and lymphoma cells. / T cells (1 × 105) from a CD19-specific CD8+CTL clone were cocultured with 2 × 106 irradiated PBMCs. Some cultures received 1 × 105 irradiated CD19+ or CD19− stimulator cells. The 2 × 106 irradiated PBMCs and irradiated stimulator cells, at a 1:1 ratio with the T cells, were re-added after 7 days. One culture condition was the T-cell clone incubated without PBMCs and stimulator cells. Each culture condition was supplemented with 5 U/mL rhIL-2 every 48 hours. The cells were counted after 14 days and nonviable cells excluded by trypan blue staining. The average cell counts and SDs from 3 repeats are presented. In the insert, 105 T cells from a CD19-specific CD8+ CTL clone were cocultured as above. Methyl-[3H]thymidine was added after 96 hours and the incorporated counts were measured 18 hours later. The background incorporation of [3H]thymidine by Daudi cells and a LCL was 827 ± 358 and 909 ± 493, respectively. The average cell counts and SDs from triplicates are presented.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/4/10.1182_blood-2002-07-1989/3/m_h80433830006.jpeg?Expires=1765930375&Signature=2AzBbbaW6TDF1y0thVZLk7y~wSvKSpzr8BP8CRe8HsIgGSj6VKMi9-rVGsYoA3rx4HuwOhJbntsfbbs5g~C1c5~Xp4eew8QA~HKqSiz9iSmTbzpIjkJTxXYee5HMIBR00I8-kLYSHmUedWlIQ1IIgRO92MVsQsyWxSxDoOZSF9cAlx2lsLi7QOfWbBONLZQyvjmvjzEH693RprJe7fVtRV5u6XpfmLV9JkzqDgYmscvGh1Y2ykzniyc8DKdfziF9~CS5f6NITzYiTkBDvHGPGyKAU-gGyRuT1l1y9eiguxGQiGihhYWOgx8yXPot73tO2FA05QaOS5eMDHBUxSazug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





![Fig. 6. CD19-specific T cells proliferate in response to stimulation by CD19+ B-ALL and lymphoma cells. / T cells (1 × 105) from a CD19-specific CD8+CTL clone were cocultured with 2 × 106 irradiated PBMCs. Some cultures received 1 × 105 irradiated CD19+ or CD19− stimulator cells. The 2 × 106 irradiated PBMCs and irradiated stimulator cells, at a 1:1 ratio with the T cells, were re-added after 7 days. One culture condition was the T-cell clone incubated without PBMCs and stimulator cells. Each culture condition was supplemented with 5 U/mL rhIL-2 every 48 hours. The cells were counted after 14 days and nonviable cells excluded by trypan blue staining. The average cell counts and SDs from 3 repeats are presented. In the insert, 105 T cells from a CD19-specific CD8+ CTL clone were cocultured as above. Methyl-[3H]thymidine was added after 96 hours and the incorporated counts were measured 18 hours later. The background incorporation of [3H]thymidine by Daudi cells and a LCL was 827 ± 358 and 909 ± 493, respectively. The average cell counts and SDs from triplicates are presented.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/4/10.1182_blood-2002-07-1989/3/m_h80433830006.jpeg?Expires=1766066901&Signature=twa9ZxqZjXx2gzHwWhIEjq8Sy7Xk-xyEIJDHWdbWX6bhkAjeARIq8wMp1UbGeHHzIA6pl49nNo-ItFeMfOyv2rJc-~m-FSVTFbe~jtDqBl51Y7fmYYfdl32sIGuFJvrhA5tXpgovnRiABOcv9lC7l1RoLnCwQMAhAnwOLzLpoIfy7HlUelkcINGvY1cguYub2eBOjkpniP4eKr94YDDr9AAMJkHlUsH0GrYnthnV0d-D5~okw84O6mjyovRuJzhD~SbZAXexuboyCVuTsmTdRfPXa9P2NJ5P5cxaSPsOr6MjE5j1Um2EDPbO6q-A65QdOsaMXbcb4MCsh9PN1EwQfg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)