Protease-activated receptor 1 (PAR-1), the main thrombin receptor on vascular cells, plays a key role in platelet activation. We examined the range of PAR-1 expression on platelets, obtained twice, 1 week apart, from 100 healthy subjects and found a 2-fold interindividual variation in receptor numbers (95% CI = 858-1700). Because PAR-1 density was stable with time (r2 = 76%,P < .001), we sought a genetic explanation for the observed variability. To validate this approach, we also analyzed the α2β1 genotype according to receptor density and platelet mRNA expression data. We found that the number of PAR-1 receptors on the platelet surface is associated with the intervening sequence IVSn−14 A/T intronic variation. The number of receptors was also found to govern the platelet response to the SFLLRN agonist, in terms of aggregation and P-selectin expression. The T allele (allelic frequency, 0.14) can be considered as an allele with decreased expression, because it was associated with lower PAR-1 expression on the platelet surface and with a lower response to SFLLRN. The IVSn−14 A/T intronic variation may therefore be clinically relevant.
Introduction
Platelets can be activated by a variety of physiologic agonists, such as collagen, adenosine diphosphate (ADP), and thrombin through interactions with specific membrane receptors.1 Activation results in platelet adhesion to extracellular matrix components and subsequent platelet aggregation. Thrombin, a key enzyme in blood coagulation, activates human platelets via two 7-transmembrane G-protein–coupled protease-activated receptors (PAR-1 and PAR-4) and activation of either is sufficient to trigger platelet secretion and aggregation.2-4 Thrombin at picomolar concentrations activates platelet PAR-1 by cleaving its amino-terminal end.5,6 The unmasked amino-terminus sequence acts as a tethered ligand, resulting in a rapid response that can be reproduced by a hexapeptide corresponding to the amino terminus (SFLLRN).5,7 PAR-4 can mediate platelet activation but only in the presence of nanomolar thrombin concentrations.3 PAR-1 density on resting platelets has been evaluated in a small number of subjects, by using radioiodinated monoclonal antibodies (mAbs).8,9 According to Brass et al,8 the PAR-1 number is approximately 1500/platelet. We have previously reported variable platelet responsiveness to SFLLRN in a study of 100 healthy volunteers.10 The response was stable over time in a given individual, pointing to genetic control. The PAR-1 gene is about 27-kilobase (kb) long and comprises 2 exons separated by a large intron (∼22 kb).11,12 Two polymorphisms have been identified in the 5′ regulatory region: a C>T transition 1426 base pair (bp) upstream of the transcription start site (−1426 C/T) and a 13-bp repeat (I) of the preceding −506 5′-CGGCCGCGGGAAG-3′ sequence (−506 I/D).13 A third polymorphism, an A>T transversion, has been located in the intervening sequence (IVS), 14 nucleotides upstream of the exon 2 start site (IVSn−14 A/T).13
Most of the genes encoding the platelet receptors have been sequenced, and polymorphisms have been described in coding and regulating regions. The consequences of these polymorphisms for platelet functions and their involvement in predispositions to excessive bleeding or thrombus formation begin to be determined.14-17 For instance, marked differences in platelet surface α2β1-integrin density are found among healthy individuals,18 and correlate with the degree of ex vivo adhesion to collagen.19-21 Associations between receptor density and polymorphisms within theα2 gene have also been reported, such as the silent exonic dimorphism 807 C/T.22,23 This polymorphism is in linkage disequilibrium with other polymorphisms that determine the density of platelet α2β1 receptors; high receptor density is associated with an increased risk of thrombosis, and low receptor density is associated with excessive bleeding (for reviews, see Furihata et al24 and Kunicki25).
The purpose of this study was to determine the impact ofPAR-1 gene polymorphisms on the expression and function of PAR-1 receptors on platelets from healthy individuals. We used quantitative flow cytometry to evaluate receptor density and real-time reverse transcription–polymerase chain reaction (RT-PCR) to quantify mRNA. To validate this approach, we also analyzed the α2β1 genotype according to receptor density and platelet mRNA expression data. Our data show that the differences in the platelet response to SFLLRN among healthy individuals are correlated to the variability of PAR-1 density on the cell surface and that this difference is, at least in part, due to the inheritance of one of the previously described PAR-1 polymorphisms.
Patients, materials, and methods
Subjects
One hundred unrelated healthy white male volunteers aged 18 to 35 years (mean, 24.3 ± 3.7 years) were recruited by public announcement from February to October 2001 and were investigated in the Clinical Investigation Center of Hôpital Européen Georges Pompidou (Paris). The volunteers were nonsmokers, had unremarkable personal and familial medical histories, and denied taking any medication for at least 10 days before blood collection. A physical examination and routine laboratory tests, including white blood cell and platelet counts, mean platelet volume, plasma fibrinogen, C-reactive protein (CRP), and von Willebrand factor plasma levels were performed before inclusion. Blood was obtained from all the volunteers on day 0 (visit 1) and day 7 (visit 2). A third blood sample was obtained from a subset of 20 volunteers between February and March 2002. All the subjects gave their written informed consent, and the study protocol was approved by the local ethics committee.
Sample preparation
Venous blood was collected between 8:00 and 10:00am, after an overnight fast, in tubes containing either 0.105 M sodium citrate (1:9 vol/vol; BD Vacutainer; Becton Dickinson, Le Pont-de-Claix, France) or 50 μg/mL lepirudine (Refludan, Hoechst, Paris, France), using a 19-gauge needle and no tourniquet. The first 2 mL blood was discarded. Platelet-rich plasma (PRP) was obtained by centrifugation at 150g for 10 minutes at room temperature. Autologous platelet-poor plasma (PPP), obtained by further centrifugation at 2300g for 15 minutes, was used to adjust the PRP platelet count to 250 × 109/L. PPP was stored at −80°C until analysis.
Platelet aggregation studies
Aggregation studies were performed within 2 hours after blood collection. A 280-μL aliquot of PRP was incubated for 3 minutes at 37°C and was then stirred at 1100 rpm for 2 minutes before adding 20 μL of one of the following agonists: collagen Horm (a 95% type I/5% type III mixture, Nycomed, Munich, Germany; 1 μg/mL), SFLLRN peptide (Serbio, Gennevilliers, France), or sodium chloride (spontaneous aggregation test). Platelet aggregation was recorded for 5 minutes by using a photometric method derived from the Born technique, on a 4-channel aggregometer (Regulest, Amneville, France). Results were expressed as the maximum percent increase in light transmission over that of the platelet suspension, relative to that of autologous PPP (arbitrarily 100%). The collagen lag time, that is, the interval between collagen addition and the onset of aggregation, was also recorded. SFLLRN aggregation was performed in the presence of 100 μM amastatin (Sigma-Aldrich, Saint-Quentin Fallavier, France), an aminopeptidase M inhibitor, to ensure the stability of the synthetic peptide in plasma.10 26 At visit 1, the tests were performed with 7, 10, and 15 μM SFLLRN (final concentration) to determine the range of SFLLRN concentrations causing irreversible aggregation. At the second visit, the precise SFLLRN concentration inducing biphasic aggregation was determined by varying by 1-μM intervals. In all samples the rate of spontaneous aggregation was lower than 5%.
Quantitative flow cytometry
Platelet receptors on resting and activated platelets were quantified in PRP by means of quantitative flow cytometry with a calibrator kit (Platelet Calibrator, Biocytex, Marseille, France), according to the manufacturer's instructions. The kit includes a mixture of 4 calibration beads coated with increasing concentrations of mouse IgGs (360, 8600, 29 000, and 90 000 molecules for the batch used throughout the study). Platelets were stained, in a no-wash indirect immunofluorescence technique, with the following mouse IgG1 mAbs (Immunotech, Marseille, France): anti–P-selectin (CD62p, clone CLB-Thromb/6), anti-α2β1 (CD49b, clone Gi9), and anti–PAR-1 WEDE15 and SPAN12. WEDE15 recognizes amino acids 51-64 of the PAR-1 N-terminus and is directed against both cleaved and uncleaved receptors. Anti–PAR-1 SPAN12 is directed against uncleaved receptors because it recognizes amino acids 35-46 (NATLDPR/SFLLR), where the virgule indicates the putative thrombin cleavage site. Combined use of the 2 mAbs theoretically yields the uncleaved receptor level. All mAbs were used at saturating conditions (10 μg/mL final concentration), as determined in preliminary experiments with concentrations ranging from 1 to 20 μg/mL. A negative isotypic control IgG1 was included in each series. The staining reagent was a polyclonal antimouse IgG-fluorescein isothiocyanate (FITC) antibody.
To assess the platelet response to agonists, PRP samples were incubated (15 minutes, 37°C) in static conditions with Horm collagen at 100 μg/mL (final concentration), 100 μM thrombin receptor-activating peptide (TRAP), or 10 μM U46619 (Calbiochem, VWR International, Meudon, France). To avoid platelet aggregation,27,28 PRP was first incubated for 3 minutes at 37°C with 4 μg/mL eptifibatide (Schering-Plough, Levallois-Perret, France), a cyclic heptapeptide inhibitor of the platelet fibrinogen receptor αIIbβ3.27 In preliminary experiments, we verified that no platelet aggregates were observed in the presence of 4 μg/mL eptifibatide, whatever the agonist used.
A calibration curve was constructed for each sample series, and a negative isotype control was run with each PRP sample. Five thousand events were acquired on a FACScan flow cytometer (Becton Dickinson), and data were analyzed using CellQuest software (Becton Dickinson). Receptor numbers were derived from the calibration curve, after subtracting the negative isotype control value (Figure1). The coefficient of variation of the method was below 10%. Experiments were performed within 3 hours after blood collection.
Flow cytometric analysis of PAR-1 density on platelets.
PRP (diluted 1:8, 20 μL) was incubated with 10 μg/mL (final concentration) WEDE15 anti–PAR-1 IgG1 mouse mAb or an irrelevant mouse IgG1 mAb (isotypic control) according to the manufacturer's instructions. After 15 minutes of incubation in the dark, an FITC-labeled antimouse antibody was added (20 μL). After a further 15 minutes in the dark, the mixture was diluted with 2 mL of the kit diluent. In parallel, 40 μL of calibration beads were stained with 20 μL FITC-labeled antimouse antibody, then diluted. Sample and calibration tubes were stored for 30 minutes at room temperature in the dark before flow cytometric analysis of 5000 events (FACScan flow cytometer; Becton Dickinson). (A) Histogram representing calibration bead fluorescence. The mean fluorescence intensity (M) of each peak was reported on a calibration curve. One representative example is shown (log-log scale, r2 = 0.998). The coefficient of variation of 100 curve slopes was 1.2%. (B) Typical histogram of PAR-1 labeling with WEDE15 mAb (solid line) and an isotypic control (dotted line). The number of specific PAR-1 sites was calculated after subtracting the negative isotypic control value with a calculation template (Biocytex).
Flow cytometric analysis of PAR-1 density on platelets.
PRP (diluted 1:8, 20 μL) was incubated with 10 μg/mL (final concentration) WEDE15 anti–PAR-1 IgG1 mouse mAb or an irrelevant mouse IgG1 mAb (isotypic control) according to the manufacturer's instructions. After 15 minutes of incubation in the dark, an FITC-labeled antimouse antibody was added (20 μL). After a further 15 minutes in the dark, the mixture was diluted with 2 mL of the kit diluent. In parallel, 40 μL of calibration beads were stained with 20 μL FITC-labeled antimouse antibody, then diluted. Sample and calibration tubes were stored for 30 minutes at room temperature in the dark before flow cytometric analysis of 5000 events (FACScan flow cytometer; Becton Dickinson). (A) Histogram representing calibration bead fluorescence. The mean fluorescence intensity (M) of each peak was reported on a calibration curve. One representative example is shown (log-log scale, r2 = 0.998). The coefficient of variation of 100 curve slopes was 1.2%. (B) Typical histogram of PAR-1 labeling with WEDE15 mAb (solid line) and an isotypic control (dotted line). The number of specific PAR-1 sites was calculated after subtracting the negative isotypic control value with a calculation template (Biocytex).
Platelet RNA extraction and reverse transcription
Four milliliters PRP (1 × 109 platelets) was pelleted by centrifugation for 15 minutes at 2300g and immediately stored at −80°C. Platelet was extracted with the RNeasy kit (Qiagen, Courtabœuf, France) according to the manufacturer's instructions, and eluted in 30 μL Rnase-free water. Eight microliters RNA solution was immediately used for cDNA synthesis. The remaining RNA was stored at −80°C.
Reverse transcription was performed in a final volume of 20 μL containing 1 × RT-PCR buffer (3 mM MgCl2, 75 mM KCl, 50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 8.3; Invitrogen, Cergy-Pontoise, France), 500 μM each deoxyribonucleoside triphosphate (dNTP; Amersham Pharmacia Biotech, Orsay, France), 10 U RNasin ribonuclease inhibitor (Promega, Charbonnières, France), 10 mM dithiothreitol (Invitrogen), 100 U Superscript II Rnase H− reverse transcriptase (Invitrogen), 1.5 mM random hexanucleotides (Amersham Pharmacia Biotech), and 8 μL mRNA. The samples were incubated at 20°C for 10 minutes then at 42°C for 30 minutes; reverse transcriptase was inactivated by heating for 5 minutes at 99°C, then cooling for 5 minutes at 5°C. The cDNA was stored at −20°C.
Real-time RT-PCR
Theoretical basis.
Quantitative values are obtained from the threshold cycle number (Ct value) at which the increase in fluorescent signal associated with exponential growth of PCR products begins to be detected by the laser detector of the ABI Prism 7700 Sequence Detection System (Applera, Courtabœuf, France), using the Applera analysis software according to the supplier's instructions.
The precise amount of total RNA added to each reaction mix (based on optical density) and its quality (ie, lack of extensive degradation) are both difficult to assess.29 We therefore quantified transcripts of the RPLP0 gene (also known as36B4) encoding human acidic ribosomal phosphoprotein P0 as an endogenous RNA control, and each test result was normalized to the sample RPLP0 content. The relative gene expression level was also normalized to an arbitrarily selected reference sample (R). Final results, expressed as n-fold differences in target gene expression relative to the RPLP0 gene and the reference sample R, and designated Ntarget, were determined as follows:
Ntarget = 2((Ct RPLP0 sample − Ct target gene sample) − (Ct RPLP0 reference sample R − Ct target gene reference sample R))
Each data point was determined in duplicate, and data with a Ct coefficient of variation of more than 2% were determined again.
Primers and PCR consumables.
Primers for PAR-1, α2, and RPLP0 were chosen with the assistance of the computer program Oligo 5.0 (National Biosciences, Plymouth, MN). We conducted BLASTN searches against the database for expressed sequence tags (dbEST), high throughput genomic sequences (htgs), and nr (the nonredundant set of GenBank, EMBL, and DDJB database sequences, respectively) to confirm the total gene specificity of the nucleotide sequences chosen for the primers and the absence of DNA polymorphisms. The primer nucleotide sequences are shown in Table 1. To avoid amplification of contaminating gDNA, 1 of the 2 primers was placed at the junction between 2 exons or in a different exon.
PCR amplification.
All PCRs were performed using an ABI Prism 7700 Sequence Detection System (Applera), according to the manufacturer's instructions, and the SYBR Green PCR Core Reagents kit (Applera). For each PCR run, a master mixture was prepared on ice with 1 × Sybr Green buffer, 200 μM deoxyadenosine triphosphate (dATP), deoxycytidine triphosphate (dCTP), and deoxyguanosine trhiphosphate (dGTP), 400 μM deoxyuridine triphosphate (dUTP), 1.25 U AmpliTaq Gold DNA polymerase, 5 mM MgCl2, and 200 nM each primer (Invitrogen). Ten microliters of each appropriately diluted reverse transcription sample was added to 40 μL of the PCR master mix. The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 minutes, and 50 cycles at 95°C for 15 seconds and 65°C for 1 minute.
Analysis of the exon 1/exon 2 junction sequences in platelet PAR-1 cDNA
We amplified the platelet cDNA fragment flanking the region at the junction of the 2 PAR-1 exons by using a reverse primer 5′-ACA ATG GGG CCG CGG CGG-3′ and a forward primer 5′-CCG GAG GCA TCT TCT GAG ATG-3′ corresponding to the exon 1 and exon 2 sequences, respectively. PCR products were then purified using the Pre-Sequencing Kit (Amersham Pharmacia Biotech) and directly sequenced with an ABI Prism 3700 (Applera) and DNA Sequencing Analysis 3.6 NT software (Applera).
Genotyping of polymorphisms
gDNA was isolated from peripheral blood leukocytes by using the Qiamp Maxi Kit (Qiagen) according to the manufacturer's instructions and was stored at 4°C until analysis. Polymorphisms were determined by means of genomic PCR. All primers were from Genset (Proligo, Paris, France).
−506 I/D PAR-1 polymorphism.
A fragment encompassing the polymorphic site at position −506 was amplified (Table 2). To detect the 13-bp insertion, the PCR products were electrophoresed in 3% agarose gel and stained with ethidium bromide.
−1426 PAR-1 C/T polymorphism and IVSn−14 A/T transition.
To detect the polymorphism in the 5′ regulatory region, a DNA fragment encompassing the polymorphic site at position −1426 was amplified. The intronic polymorphism was genotyped after amplification of the DNA fragment flanking 14 nucleotides upstream of the exon 2 start site, using appropriate primers (Table 2). PCR products were then purified and purification was followed by a cycle sequencing reaction with the same primers and sequencing on an ABI Prism 3700 (Applera) with DNA Sequencing Analysis 3.6 NT software (Applera).
α2-Integrin 807 C/T polymorphism.
We used allele-specific restriction digestion to detect the substitution of cytosine by thymidine at position 807 of the gene encoding integrin α2 (GP Ia). After amplification with primers flanking position 807 of theα2-integrin gene (Table 2), PCR products were digested with HinfI restriction enzyme (New England Biolabs, Ozyme, Saint-Quentin en Yvelines, France), which is specific for the 807 C allele. The resulting fragments were analyzed on 6% polyacrylamide gel with ethidium bromide staining.
β3-integrin 1565 C/T polymorphism (PlA1/PlA2).
We used restriction fragment length polymorphism (RFLP) analysis to detect the substitution of cytosine by thymidine at position 1565 of exon 2 of the β3-integrin gene, which is responsible for the PlA2 polymorphism.10
Genotyping was repeated in ambiguous cases, and all 100 subjects were successfully genotyped.
Statistical analysis
Data are shown as means ± SEM, except for skewed variables, which are expressed as medians. The distribution of continuous variables was estimated by using the Schapiro-Wilk test. Skewed variables were log-transformed before analysis. Individual subjects' values obtained at visits 1 and 2 (1 week apart) were compared using a concordance test.30 When values of a given parameter obtained at the 2 visits were concordant (defined as r2 > 50%), the mean of the 2 measurements was used for subsequent analyses. The χ2 test was used to compare the observed allele and genotype frequencies with the Hardy-Weinberg equilibrium prediction. A paired t test was used to estimate changes in receptor numbers before and after platelet activation. An univariate linear regression was used to compare phenotypic results. The association between genotype and phenotype was tested using analysis of variance (ANOVA). Genotype-phenotype or phenotype-phenotype associations were further tested after adjustment for other parameters (white blood cell count, fibrinogen plasma level, mean platelet volume, CRP, von Willebrand factor plasma level, PlA1/PlA2 polymorphism), by using multiple linear regression model.
Statistical tests were performed using the STATA 7.0 software package (Stata, College Station, TX) and differences with P < .05 were considered statistically significant.
Results
Platelet PAR-1 quantification
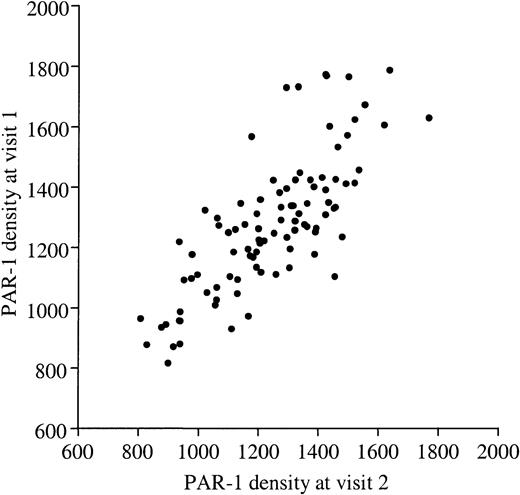
The number of PAR-1 receptors on platelets was determined from a calibration curve, after subtracting nonspecific binding. A typical example is given in Figure 1. As shown in Figure2, PAR-1 expression, as determined by mAb WEDE15 binding, ranged from 853 to 1713 copies/platelet (mean, 1262 ± 198 copies/platelet) among the 100 volunteers. Values obtained in each subject at the 2 visits (1 week apart) showed a high degree of concordance (r2 = 76%,P < .001; Figure 2). We also tested a subset of 63 donors with mAb SPAN12, which is specific for uncleaved PAR-1 receptors. The mean number of uncleaved PAR-1 receptors was 687 ± 154/platelet (range, 407-1114/platelet) and, again, the 2 measurements performed 1 week apart showed good concordance (r2 = 61%,P < .001). Thus, 45% of PAR-1 receptors on resting platelets were not recognized by SPAN12, that is, were cleaved. Such an observation has already been reported by others.31 To rule out the possibility of platelet activation during PRP preparation, we quantified the expression of P-selectin, a marker of α-granule exposure.32 33 P-selectin expression on resting platelets was below 1000 sites in most cases (mean, 502 ± 335 sites) and “cleaved” PAR-1 expression did not correlate with the P-selectin level. One possible explanation for artefactual PAR-1 cleavage is the generation of trace amounts of thrombin in citrated PRP. We thus randomly selected 20 of the 100 subjects and quantified total and uncleaved PAR-1 on resting platelets isolated from a third blood sample collected on hirudin or citrate as anticoagulant. The ratio of uncleaved PAR-1 to total PAR-1 was stable whatever the anticoagulant (data not shown), implying that PAR-1 was not cleaved by thrombin during PRP preparation.
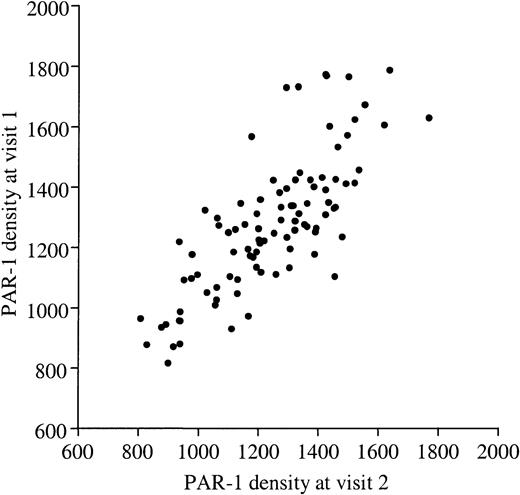
PAR-1 density on human platelets.
PAR-1 density was determined on resting platelets from 100 healthy volunteers by quantitative flow cytometry using mAb WEDE15 directed against both cleaved and intact PAR-1. The number of PAR-1 receptors at visit 1 (y-axis) is plotted against the number at visit 2 (x-axis), and shows significant concordance (r2 = 76%,P < .001). Values ranged from 804 to 1713 sites/platelet (mean, 1262 ± 198 sites/platelet).
PAR-1 density on human platelets.
PAR-1 density was determined on resting platelets from 100 healthy volunteers by quantitative flow cytometry using mAb WEDE15 directed against both cleaved and intact PAR-1. The number of PAR-1 receptors at visit 1 (y-axis) is plotted against the number at visit 2 (x-axis), and shows significant concordance (r2 = 76%,P < .001). Values ranged from 804 to 1713 sites/platelet (mean, 1262 ± 198 sites/platelet).
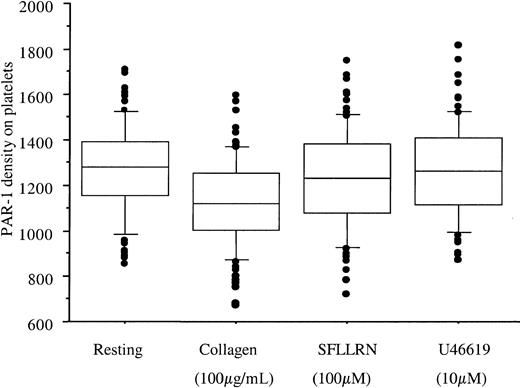
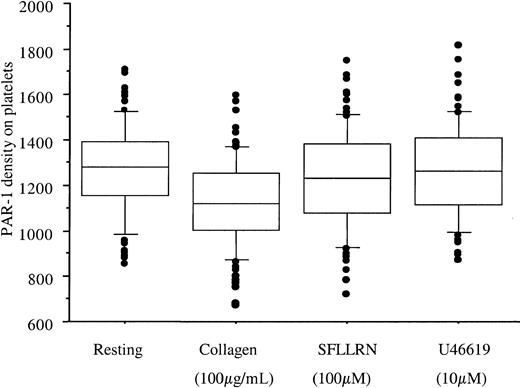
PAR-1 density was also determined after ex vivo platelet stimulation with collagen, SFLLRN, and the thromboxane A2 receptor agonist U46619. U46619 did not modify PAR-1 expression, whereas SFLLRN and collagen induced a small but statistically significant decrease in total PAR-1 receptor expression (mean decrease of 27 and 133 sites, respectively, P < .001; Figure3). The number of PAR-1 copies after platelet activation was also measured on the second blood sample. As with resting platelets, the 2 values showed good concordance (r2 > 68% whatever the agonist,P < .001).
PAR-1 expression after platelet activation.
Boxes represent PAR-1 density quantified by flow cytometry at rest and on PRP stimulation for 15 minutes at 37°C by collagen 100 μg/mL, SFLLRN 100 μM, or U46619 10 μM (final concentrations). To avoid platelet aggregation, PRP was first incubated with 4 μg/mL eptifibatide. Because the response was stable at a 1-week interval, for each subject the mean value of the 2 visits is used. Collagen and SFLLRN induced a small but significant decrease in receptor numbers, from 1262 ± 198 sites at rest to 1125 ± 192 and 1231 ± 215 sites after stimulation by collagen and SFLLRN, respectively. U46619 had no effect (1264 ± 203 sites). Boxes represent the median values with 25th and 75th percentiles, and the bar chart shows 90th and 10th percentiles.
PAR-1 expression after platelet activation.
Boxes represent PAR-1 density quantified by flow cytometry at rest and on PRP stimulation for 15 minutes at 37°C by collagen 100 μg/mL, SFLLRN 100 μM, or U46619 10 μM (final concentrations). To avoid platelet aggregation, PRP was first incubated with 4 μg/mL eptifibatide. Because the response was stable at a 1-week interval, for each subject the mean value of the 2 visits is used. Collagen and SFLLRN induced a small but significant decrease in receptor numbers, from 1262 ± 198 sites at rest to 1125 ± 192 and 1231 ± 215 sites after stimulation by collagen and SFLLRN, respectively. U46619 had no effect (1264 ± 203 sites). Boxes represent the median values with 25th and 75th percentiles, and the bar chart shows 90th and 10th percentiles.
Platelet aggregation and secretion responses to SFLLRN
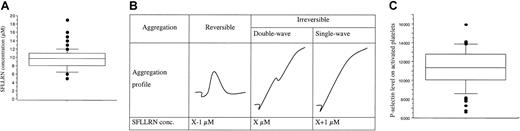
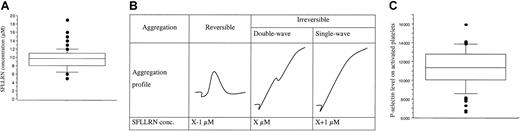
SFLLRN is specific for PAR-1 and does not therefore bind other thrombin receptors on platelets. The median SFLLRN concentration inducing a double-wave aggregation profile (Figure4A inset) was 9.75 μM (range, 5-19 μM; Figure 4A). These results confirmed the interindividual variability of platelet sensibility to SFLLRN.10 The SFLLRN concentration causing double-wave aggregation on sample 2 was always close to the SFLLRN concentration determined on sample 1, suggesting that the platelet aggregation response to SFLLRN is stable with time in a given subject. We also observed interindividual differences in the platelet secretory response to SFLLRN. Indeed, P-selectin density on platelets activated by 100 μM SFLLRN ranged from 6613 to 15 933 sites (11 223 ± 1934 sites; Figure 4B) and this response was stable at a 1-week interval (r2 = 50%, P < .001). We therefore used the mean value of the 2 visits in each subject for subsequent analyses.
Platelet responses to SFLLRN.
(A) The box plot represents the distribution of SFLLRN concentrations inducing a double wave of aggregation (B). Values ranged from 5 to 19 μM, with a median of 9.75 μM (center horizontal bar). Errors bars indicate 90th and 10th percentiles, respectively. Panel B shows determination of SFLLRN concentration inducing double-wave aggregation profile. At visit 1, the tests were performed with 7, 10, and 15 μM SFLLRN to determine the range of SFLLRN concentrations inducing an irreversible single wave of aggregation. At the second visit, the precise SFLLRN concentration inducing an irreversible double-wave aggregation profile (X value) was determined. (C) Distribution of P-selectin levels, as quantified by flow cytometry on SFLLRN-activated platelets (100 μM) preincubated with eptifibatide to avoid aggregation. The number of P-selectin sites ranged from 6613 to 15 933 sites/platelet (11 223 ± 1934 sites/platelet). Because the response was stable at a 1-week interval (r2 = 50%,P < .01), the mean value of the 2 visits is used for each subject. Box plot displays median value with 25th and 75th percentiles and the bar chart shows 90th and 10th percentiles.
Platelet responses to SFLLRN.
(A) The box plot represents the distribution of SFLLRN concentrations inducing a double wave of aggregation (B). Values ranged from 5 to 19 μM, with a median of 9.75 μM (center horizontal bar). Errors bars indicate 90th and 10th percentiles, respectively. Panel B shows determination of SFLLRN concentration inducing double-wave aggregation profile. At visit 1, the tests were performed with 7, 10, and 15 μM SFLLRN to determine the range of SFLLRN concentrations inducing an irreversible single wave of aggregation. At the second visit, the precise SFLLRN concentration inducing an irreversible double-wave aggregation profile (X value) was determined. (C) Distribution of P-selectin levels, as quantified by flow cytometry on SFLLRN-activated platelets (100 μM) preincubated with eptifibatide to avoid aggregation. The number of P-selectin sites ranged from 6613 to 15 933 sites/platelet (11 223 ± 1934 sites/platelet). Because the response was stable at a 1-week interval (r2 = 50%,P < .01), the mean value of the 2 visits is used for each subject. Box plot displays median value with 25th and 75th percentiles and the bar chart shows 90th and 10th percentiles.
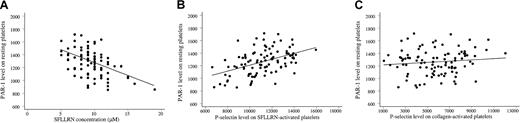
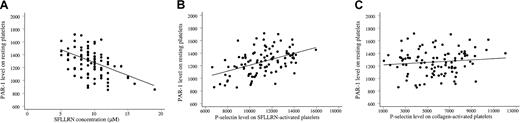
Although the transduction mechanisms underlying platelet activation are complex and multiple, the platelet response to SFLLRN correlated with PAR-1 expression; the SFLLRN concentration causing double-wave aggregation correlated negatively with the number of PAR-1 sites on the platelet surface (r2 = 23%, P < .001; Figure 5A). This correlation remained significant after adjustment for the PlA1/PlA2genotype, the white blood cell count, the fibrinogen level, platelet mean volume, and CRP and von Willebrand factor levels (P < .001). Platelet secretion (P-selectin expression) also correlated with PAR-1 density (r2 = 30%,P < .001; Figure 5B). Adjustment for other blood parameters did not influence the correlation. Importantly, P-selectin expression did not correlate with PAR-1 density when platelets were activated with collagen (P = .28; Figure 5C). Thus, response capacity to SFLLRN activation of platelets from healthy volunteers was associated with platelet surface PAR-1 density.
PAR-1 density influences the platelet response to SFLLRN.
(A) The SFLLRN concentration causing a double wave of aggregation (x-axis) correlated negatively with PAR-1 density (ordinate;r2 = 23%, P < .001). The mean value of the 2 determinations 1 week apart is plotted for each variable. (B) Platelet secretion in response to 100 μM SFLLRN (P-selectin expression, x-axis) correlated with PAR-1 density (y-axis;r2 = 30%, P < .001). The mean value of the 2 determinations 1 week apart is plotted for each variable. (C) Platelet secretion in response to 100 μg/mL collagen stimulation (P-selectin expression, x-axis) did not correlate with PAR-1 density (y-axis; P = .28).
PAR-1 density influences the platelet response to SFLLRN.
(A) The SFLLRN concentration causing a double wave of aggregation (x-axis) correlated negatively with PAR-1 density (ordinate;r2 = 23%, P < .001). The mean value of the 2 determinations 1 week apart is plotted for each variable. (B) Platelet secretion in response to 100 μM SFLLRN (P-selectin expression, x-axis) correlated with PAR-1 density (y-axis;r2 = 30%, P < .001). The mean value of the 2 determinations 1 week apart is plotted for each variable. (C) Platelet secretion in response to 100 μg/mL collagen stimulation (P-selectin expression, x-axis) did not correlate with PAR-1 density (y-axis; P = .28).
Genotype-phenotype relationships
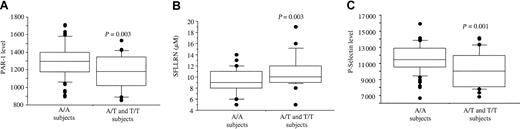
To determine whether the platelet PAR-1 phenotype was associated with genetic variations, we genotyped the volunteers for 3 previously described PAR-1 polymorphisms, namely −1426 C/T, −506 I/D, and IVSn−14 A/T. The respective allelic frequencies of −1426T, −506I, and IVSn−14T were 0.12, 0.27, and 0.14, and the distribution of heterozygotes was close to that predicted by Hardy-Weinberg equilibrium. These allelic frequencies were similar to those previously obtained in 1214 healthy subjects.13 The −506 I/D and −1426 C/T polymorphisms were not associated with the platelet PAR-1 phenotype, whereas the intronic polymorphism IVSn−14 A/T was significantly associated with the platelet PAR-1 expression level (P = .003; Figure 6A). Homozygous carriers of the IVSn−14A allele (n = 74) had a significantly higher PAR-1 level (1297 ± 186) than heterozygous carriers (1182 ± 199, n = 24, P = .013). The 2 subjects homozygous for the IVSn−14T allele had 857 and 1022 PAR-1 sites. The presence of an IVSn−14T allele was also associated with decreased platelet sensitivity to SFLLRN (Figure 6B). Indeed, biphasic aggregation occurred with 9.1 ± 2.1 μM SFLLRN in AA subjects and 10.6 ± 2.5 μM in AT subjects (P = .005). The highest SFLLRN concentration required for biphasic aggregation (19 μM) was found in 1 of the 2 TT homozygotes (subject H1), who had only 857 PAR-1 sites/platelet. The IVSn−14 A/T polymorphism was also associated with the maximal platelet secretory response to 100 μM SFLLRN (Figure 6C). Indeed, higher P-selectin expression was observed in carriers of 2 A alleles (11 548 ± 1735 sites/platelet) than in carriers of a single T allele (10 530 ± 2078 sites/platelet, P = .011). The TT homozygous subjects H1 and H2 had, respectively, 6778 and 10 211 P-selectin sites per platelet after SFLLRN stimulation.
Platelet PAR-1 expression and function according to the PAR-1 IVSn−14 A/T polymorphism.
(A) Distribution of PAR-1 levels according to the genotype. Left, A/A homozygotes (74 subjects). Right, A/T heterozygotes (24 subjects) and T/T homozygotes (2 subjects). A/A homozygotes had a significantly higher PAR-1 level (1297 ± 186 sites) than carriers of at least one T allele (A/T and T/T subjects, 1164 ± 203 sites). (B) Relation between PAR-1 receptor numbers and aggregation responses to SFLLRN according to the genotype. PAR-1 numbers correlate negatively with the platelet response to SFLLRN. Double-wave aggregation occurred with 9.1 ± 2.1 μM SFLLRN in A/A subjects (left) and with 10.9 ± 3.0 μM SFLLRN in A/T and T/T subjects. (C) Platelet secretory response to SFLLRN according to the genotype. On SFLLRN (100 μM) platelet stimulation, higher P-selectin expression was observed in carriers of 2 A alleles (11 548 ± 1735 sites) than in A/T and T/T subjects (10 160 ± 2117 sites). Boxes represent the median values with 25th and 75th percentiles and the bar chart shows 90th and 10th percentiles.
Platelet PAR-1 expression and function according to the PAR-1 IVSn−14 A/T polymorphism.
(A) Distribution of PAR-1 levels according to the genotype. Left, A/A homozygotes (74 subjects). Right, A/T heterozygotes (24 subjects) and T/T homozygotes (2 subjects). A/A homozygotes had a significantly higher PAR-1 level (1297 ± 186 sites) than carriers of at least one T allele (A/T and T/T subjects, 1164 ± 203 sites). (B) Relation between PAR-1 receptor numbers and aggregation responses to SFLLRN according to the genotype. PAR-1 numbers correlate negatively with the platelet response to SFLLRN. Double-wave aggregation occurred with 9.1 ± 2.1 μM SFLLRN in A/A subjects (left) and with 10.9 ± 3.0 μM SFLLRN in A/T and T/T subjects. (C) Platelet secretory response to SFLLRN according to the genotype. On SFLLRN (100 μM) platelet stimulation, higher P-selectin expression was observed in carriers of 2 A alleles (11 548 ± 1735 sites) than in A/T and T/T subjects (10 160 ± 2117 sites). Boxes represent the median values with 25th and 75th percentiles and the bar chart shows 90th and 10th percentiles.
The relationship between the IVSn−14 A/T polymorphism and the platelet PAR-1 phenotype might be explained by an effect on intron splicing. Indeed, the sequence variation is located in the intervening sequence (IVS), 14 nucleotides upstream of the exon 2 start site, which may affect the recognition of the splicing site. Interestingly, we found no sequence differences in the region of the junction of the 2 exons on platelet cDNA between 5 AA homozygotes, 3 AT heterozygotes, and the 2 TT homozygotes.
PAR-1 expression might also be regulated at the transcriptional level. We used real-time RT-PCR to quantify gene expression at the mRNA level. The NPAR-1 value, calculated as described in “Patients, materials, and methods,” was determined in a subset of 75 subjects in whom platelet mRNA isolation and reverse transcription yielded enough material for a reliable quantification (Ct values below 30). NPAR-1 values ranged from 0.03 to 3.61 (1.02 ± 0.74) for PAR-1, but did not correlate either with the PAR-1 receptor number or with the intronic polymorphism IVSn−14 A/T.
Platelet α2β1 characterization
To validate our approach, we quantified α2-integrin, the density of which on the platelet surface is known to be associated with the 807 C/T polymorphism. We confirmed the strong variability of α2-integrin expression on resting platelets from healthy subjects. Density ranged from 1607 to 5631 receptors/platelet (mean, 3587 ± 833 receptors/platelet) and remained stable in a given individual at a 1-week interval (r2 = 77%,P < .001). No correlation was found between α2-receptor expression and either maximal platelet aggregation or P-selectin expression induced by collagen. However, the aggregation lag time (mean, 62 ± 16 seconds; range, 35-120 seconds), which did not vary between the 2 samples 1 week apart (r2 = 55%, P < .001), correlated with α2β1-receptor density (r2 = 8%, P = .004).
We also genotyped the volunteers for the 807 C/T polymorphism in theα2 gene. We found an 807 T allele frequency of 0.44, in agreement with the normal range for a white population.25 As expected, the 807 C/T polymorphism was associated with platelet α2-integrin density. Subjects with the C/C genotype (n = 31) had low α2 expression (2810 ± 756 receptors) relative to C/T carriers (n = 50, 3747 ± 541 receptors) and T/T homozygotes (n = 19, 4434 ± 449 receptors, P < .001). Moreover, platelet aggregation started within 58 ± 16 seconds in the carriers of two 807 T alleles, compared to 68 ± 16 seconds in the carriers of two 807 C alleles (P = .056). The 807 C/T polymorphism is in linkage disequilibrium with another polymorphism that affects a regulatory domain in the α2 gene promoter. It is thus highly likely that this polymorphism influences the level of gene expression, and, probably, the amount of mRNA produced. However, platelet α2 mRNA expression evaluated after real-time RT-PCR (n = 84, Nα2 = 2.12 ± 2.71; range, 0.02-13) did not differ according to the polymorphism nor to the platelet α2-integrin density.
Discussion
PAR-1, the main thrombin receptor on vascular cells, plays a key role in platelet activation. Here, we studied the variability of the platelet PAR-1 phenotype in 100 healthy male volunteers, to avoid fluctuating hormonal influences on platelet functions. The number of PAR-1 molecules on the platelet surface was measured by using a new quantitative flow cytometry method with the WEDE15 antibody, which recognizes both intact and cleaved PAR-1. The mean receptor number per platelet (1200 copies) was similar to the reported number of moderate-affinity 125I-thrombin–binding sites on platelets, and to the number obtained by radioimmunoassay in a small number of subjects.8,9 34 We observed 2-fold variability of PAR-1 levels, in keeping with the expression range (1000-1800 sites) reported in the literature. PAR-1 levels were stable on 2 occasions 1 week apart (ie, when most platelets had been renewed) and, in a subset of 20 subjects, over a period of 6 months (data not shown). This strongly suggests that genetic factors are involved in the control of platelet PAR-1 expression.
We also quantified PAR-1 expression after platelet activation because previous data suggest that activation of U46619 induces expression of surface-connecting system PAR-1 receptors, whereas SFLLRN induces a strong decrease of PAR-1.31 We did not find such a wide change in PAR-1 expression on platelet activation; the number of receptors fell only slightly on SFLLRN and collagen activation and did not change significantly after U46619 activation. This implies that only small numbers of PAR-1 receptors are internalized in platelets, in contrast to nucleated cells such as endothelial cells and megakaryocytes.35-37 Noteworthy, αIIbβ3 inhibitors, used here during flow cytometric study of isolated platelets, interferes with the events subsequent to fibrinogen binding to activated αIIbβ3 (“outside-in” signaling),38 which may be involved in the modification of the level of surface platelet receptors.
The in vivo PAR-1 cleavage by thrombin is complex because it is modulated by other thrombin-binding sites on platelets (PAR-4 and GP Ib).3,4,39,40 Thus, to determine whether platelet activation via PAR-1 is dependent on the number of receptors, we used the specific PAR-1–activating peptide SFLLRN, which allows platelets to be studied in plasma, their natural environment. SFLLRN is widely used to monitor antithrombotic therapy with αIIbβ3 antagonists.41 We determined the minimal SFLLRN concentration inducing biphasic aggregation, and α-granule secretion (resulting in P-selectin expression) on SFLLRN activation. P-selectin is present in platelet α granules and rapidly translocates to the cell surface after activation.32 P-selectin is involved both in rolling adhesion of platelets to activated endothelial cells and in activated platelet interactions with neutrophils and monocytes.42,43We observed strong interindividual variability in the SFLLRN response, which remained stable at a 1-week interval in a given subject. Moreover, both SFLLRN-induced aggregation and P-selectin expression correlated strongly with the PAR-1 expression level on the platelet surface. Both platelet aggregation in response to SFLLRN and platelet secretion were dependent on the number of PAR-1 molecules expressed on the membrane, despite the complexity of the signal transduction pathway involved during PAR-1 activation.44 45 Other factors are probably involved in the platelet response to SFLLRN because the second TT homozygote was a high responder to SFLLRN, that is, the aggregation occurred at a 10-μM concentration, despite a low PAR-1 platelet density.
Analysis of the platelet PAR-1 genotype according to the PAR-1 phenotype showed that the intronic polymorphism IVSn−14 A/T was associated with PAR-1 expression level and the platelet response to SFLLRN. Subjects carrying the T allele (frequency 0.14) had a significant lower expression level. Activation of a cryptic splice site was ruled out by platelet cDNA analysis of both alleles (IVSn−14 A and IVSn−14 T), which were found to have the same sequence at the exon 1/exon 2 junction. This does not, however, exclude an effect on the rate of mRNA processing. The intronic polymorphism may also be in linkage disequilibrium with other unknown variations situated in regulatory regions of the PAR-1 gene. Polymorphisms in coding and noncoding regions that augment transcription efficiency and, thus, the quantity of protein synthesized, have been described for many genes.46-48 In an attempt to confirm that the IVSn−14 A/T polymorphism affects gene expression itself, we quantified platelet PAR-1 mRNA by using real-time RT-PCR and found no difference between carriers and noncarriers of the T allele. To validate our approach, we applied it to the collagen receptor α2β1, whose expression is influenced by gene polymorphisms such as the 807 C/T dimorphism. We found that receptor numbers were variable among individuals and stable with time, the numerical data being close to those previously reported.18,20 The receptor number correlated with the platelet response to collagen and with the 807 C/T polymorphism. The 807 C allele, which is associated with low platelet α2β1-receptor density, was recently shown to be in linkage disequilibrium with another polymorphism in the promoter (−52 C/T), leading to decreased binding of the nuclear transcription factors specificity protein (Sp)1 and Sp3 in megakaryocytic cell lines.49 Because the molecular mechanisms underlying α2β1-receptor expression are due to a modulation of α2 gene transcription, we quantified platelet α2 mRNA. Despite marked variability of platelet α2β1 levels among healthy subjects, no relation between the platelet α2 mRNA level and the genotype was found. Few previous studies have focused on platelet mRNA quantification.50,51Qualitative and semiquantitative studies have shown that platelets contain undegraded ribosomal RNA as well as intact mRNAs. The absence or quantitative alteration of αIIbβ3-integrin mRNA has been reported in Glanzmann thrombasthenia,52,53 but no relation between the mRNA and protein expression level has been reported. Seidl et al51 found no correlation between the number of platelet receptors (GP Ia, Ib, IIb, and IV) in healthy subjects and their respective mRNA abundance measured by quantitative RT-PCR. Circulating platelets have an average half-life of 8 days, whereas eukaryotic mRNA typically has a half-life of 6 to 24 hours54 and might be present in only a small fraction of circulating platelets. Thus, platelet mRNA levels might not be representative of the gene expression in megakaryocytes, that is, expression of genes such as PAR-1 and α2 may be induced early in megakaryocyte maturation, which may explain the lack of relationship between protein expression and platelet mRNA.
In conclusion, we report a relationship between the platelet PAR-1 phenotype and the IVSn−14 A/T polymorphism in the intron of thePAR-1 gene. The T allele can be considered as having decreased expression, because it is associated with lower PAR-1 expression on the platelet surface and with a weaker aggregation and secretion response to SFLLRN. Animal studies with PAR-1 antagonists suggest that lower PAR-1 availability might enhance the bleeding tendency.55 Thus, inherited differences in receptor density may be clinically relevant for individuals who are already at an increased risk of bleeding, such as those with mild hemophilia or von Willebrand disease, and surgical patients.
We thank Alvine Bissery for her help with the statistical analysis, and the nursing staff of the Clinical Investigation Center 9201-INSERM AP-HP of Hôpital Européen Georges Pompidou. We thank Véronique Remones for excellent technical assistance. We are grateful to Biocytex for generously providing the calibrator kits for flow cytometry.
Prepublished online as Blood First Edition Paper, October 24, 2002; DOI 10.1182/blood-2002-07-2149.
Supported in part by a grant from Programme Hospitalier de Recherche Clinique (Ministère chargé de la Santé, PHRC AOR01023, sponsor: INSERM) and the Claude Bernard Association. A.D. was supported by a grant from Assistance Publique-Hôpitaux de Paris.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Pascale Gaussem, INSERM U 428, Service d'Hématologie Biologique A, Hôpital Européen Georges Pompidou, 20 rue Leblanc, F-75908 Paris Cedex 15, France; e-mail:pascale.gaussem@egp.ap-hop-paris.fr.