Abstract
The BCL6 proto-oncogene encodes a transcriptional repressor whose expression is deregulated by chromosomal translocations in approximately 40% of diffuse large B-cell lymphomas (DLBCLs). The BCL6 regulatory sequences are also targeted by somatic hypermutation in germinal center (GC) B cells and in a fraction of all GC-derived lymphomas. However, the functional consequences of these mutations are unknown. Here we report that a subset of mutations specifically associated with DLBCL causes deregulated BCL6 transcription. These mutations affect 2 adjacent BCL6 binding sites located within the first noncoding exon of the gene, and they prevent BCL6 from binding its own promoter, thereby disrupting its negative autoregulatory circuit. These alterations were found in approximately 16% of DLBCLs devoid of chromosomal translocations involving the BCL6 locus, but they were not found in normal GC B cells. This study establishes a novel mechanism for BCL6 deregulation and reveals a broader involvement of this gene in DLBCL pathogenesis.
Introduction
The BCL6 proto-oncogene was originally identified because of its involvement in 3q27 chromosomal translocations associated with diffuse large B-cell lymphomas (DLBCLs).1-4BCL6 encodes a POZ/zinc finger (ZF) transcriptional repressor expressed in mature germinal center (GC) B cells and required for GC formation and for affinity maturation during T-cell–dependent antibody responses.5-10 The BCL6 protein acts as a potent transcriptional repressor of promoter sequences containing its specific DNA recognition motif.8,11 This function requires DNA binding through the ZF domain and 2 noncontiguous transrepression domains, including its N-terminal POZ motif and a second region located in the middle of the molecule. Because of the similarity between the DNA binding sites of BCL6 and the signal transducer and activator of transcription 6 (STAT6) transcription factor, BCL6 can modulate incoming interleukin-4 (IL-4) signaling by repressing STAT6-induced transcription from the immunoglobulin (Ig) germline ε promoter, thereby modulating isotype switching toward IgE in vitro and in vivo.12 Based on gene expression profiling of B cells that differentially express BCL6, it has also been proposed that BCL6 suppresses genes involved in lymphocyte activation, differentiation, cell cycle arrest, and apoptosis.13,14 Thus, BCL6 represents an important modulator of B-cell responses in the GC, and its transcriptional silencing is required for GC exit and, possibly, for differentiation toward plasma cells or memory B cells.15
Consistent with its essential role in the GC reaction, BCL6 expression is regulated by a number of signals important for GC development and differentiation. At the protein level, activation of the B-cell receptor induces mitogen-activated protein (MAP) kinase-mediated phosphorylation of BCL6, which, in turn, targets BCL6 for rapid degradation by the ubiquitin proteasome pathway.16 At the transcriptional level, a variety of stimuli, including CD40 receptor engagement and mitogenic stimulation, lead to the down-regulation of BCL6 RNA in cultured B cells.43 More recently, a novel mechanism has been identified that down-regulates BCL6 activity by inhibiting its transrepression function through p300-mediated acetylation.17
In tumors, chromosomal translocations affecting the BCL6 locus at band 3q27 represent the most common and specific genetic abnormality associated with DLBCL, accounting for more than 40% of the cases.1,18,19 These translocations juxtapose heterologous promoters from the partner chromosome with intact BCL6 coding sequences, leading to deregulated expression of BCL6 by a mechanism known as promoter substitution.20 21
In addition, the 5′ regulatory sequences of BCL6 can be altered by multiple somatic mutations in 30% to 40% of GC B cells.22-24 These mutations cluster within approximately 2 kb of the transcription initiation site at average frequencies of 1.2 × 10−3/bp in the mutated cells.22-24They are often biallelic and extremely heterogeneous, and they display features of the IgV-associated somatic hypermutation mechanism, including a preference for single base-pair substitutions, the predominance of transitions over transversions, and a preferential (RGYW) targeting motif.22-24 Consistent with their association with the physiologic GC reaction, BCL6 mutations are also found in a fraction of all B-cell tumors that carry mutated IgV sequences and display a GC or a post-GC phenotype, including B-cell chronic lymphocytic leukemia (B-CLL), Burkitt lymphoma (BL), follicular lymphoma (FL), DLBCL, and multiple myeloma (MM).25-27 The structural features of these mutations, as well as their distribution and the specificity for GC B cells, strongly suggest that BCL6 mutations and IgV mutations are derived by the same mechanism. Nonetheless, the biologic role of BCL6 mutations in normal and malignant B-cell development remains unclear.
In the present study, we investigated the functional consequences of BCL6 mutations by performing a structure–function analysis on 55 mutant alleles (a total of 371 mutations) derived from normal GC lymphocytes or from various lymphoma subtypes. We identified a subset of mutations, located within the first noncoding exon of BCL6, that disrupts the mechanism of negative autoregulation normally involved in controlling BCL6 expression levels. These mutations were found only in DLBCL cases, suggesting that they might have been selected during lymphomagenesis for their ability to deregulate BCL6 expression.
Materials and methods
Cell lines, stable transfections, and sorting of tonsillar GC B cells
All B-cell lines used were cultured in Iscove modified Dulbecco medium (IMDM) supplemented with 10% fetal calf serum (FCS), 100 U/mL penicillin, 100 U/mL streptomycin, and 2 mM L-glutamine. The Epstein-Barr virus (EBV)–transformed EREB cell line required the addition of 1 μM β-estradiol to the culture medium. To generate the stable EREB-MT and EREB-MT-BCL6 cell lines, EREB cells were transfected by electroporation with 20 μg pMEP4 (Invitrogen, Carlsbad, CA) or pMEP4-HA-BCL-6 and were selected in hygromycin (150 μg/mL). For the induction of exogenous BCL6 expression in the EREB-MT-HA-BCL6 stable line, 2 μM CdCl2 was added to the culture medium, and cells were harvested at various time points as indicated in Figure 5B. The 293T fibroblast cell line was maintained in Dulbecco minimum essential medium (DMEM) containing 10% FCS, penicillin/streptomycin, and 2 mM L-glutamine. The protocol for isolation of tonsillar GC B lymphocytes is reported in detail in Pasqualucci et al.23
DNA extraction and polymerase chain reaction amplification of the BCL6 promoter region
Genomic DNA from sorted GC B cells or lymphoma cases (n = 46; 32 DLBCL, 5 B-CLL, 4 FL, and 5 BL) was extracted according to standard methods. For amplification of the approximately 2.7-kb BCL6 genomic region cloned into the pLA/S5 reporter plasmids, the Expand High Fidelity PCR System (Roche Diagnostics, Indianapolis, IN) and the following primers were used: 5′-AGGAGAGACACACTTCAGC-3′ (sense) and 5′-ATCGTAGAAGGGAACACAAC-3′ (antisense). For mutational analysis of the BCL6 exon 1 sequences, a 596-bp genomic fragment spanning this region was amplified through polymerase chain reaction (PCR) for 30 cycles using the primers E1.15 and E1.20C described in Migliazza et al, in 1995.25
Cloning and sequence analysis
Purified amplicons were sequenced directly from both strands, as described.23 The 2.7-kb BCL6 amplicons were digested withNdeI and SpeI, gel purified using the QIAquick gel extraction kit (Qiagen, Valencia, CA), and cloned into the corresponding sites of the pLA/S5 vector (see “Plasmid constructs”) using the Rapid Ligation Kit (Roche); approximately 100 recombinant clones from GC B cells and 6 to 10 clones from each tumor case were analyzed to identify individual alleles. Full-length sequencing of the 2.7-kb BCL6 promoter region was then performed on selected clones to confirm the mutations observed by direct sequencing and to detect additional changes, possibly representing subclonal variants or DNA polymerase-introduced errors. The BCL6 5′ noncoding sequence has been deposited in GenBank under accession number AY189709 (note that the numbering has been corrected (+1 bp) compared with that reported in our previous publications23,25 27).
Plasmid constructs
The original BCL6 luciferase reporter plasmid (pLA/S14wt) was constructed in multiple steps by inserting an approximately 14-kbAvrII/NcoI genomic fragment of the native BCL6 promoter (spanning the 5′ flanking sequences, exon 1, intron 1, and exon 2) into the multiple cloning site of the pGL3 vector (Promega, Madison, WI). This construct displayed comparable basal activity to a deletion mutant lacking the approximately 7.7-kbSpeI/Bsu36I fragment at the 3′ end of intron 1 (pLA/S5wt; Figure 1A). The latter was therefore used as a backbone to derive all the mutant reporter constructs by exchanging the wild-type 2776-bp NdeI–SpeI region of the BCL6 5′ sequences with the corresponding fragment PCR amplified from genomic DNA of GC B cells and lymphoma cases. Four of these constructs (Ly1A, 93-611A, 93-611B, and 93-2889A) were further dissected by swapping—in different combinations—mutant and wild-type fragments corresponding to the 5′ flanking, first exon and first intron of BCL6, as defined by the restriction sites NdeI (−694), PmlI (−64),SexAI (+276), and SpeI (+2082). Single point mutants were generated by site-directed mutagenesis using the pLA/S5wt plasmid as a target and the Transformer Site-Directed Mutagenesis Kit (BD Biosciences Clontech, Palo Alto, CA). The expression vector pMT2T-HA-BCL6 and its deletion mutants pMT2T-HA-BCL6-ZF and pMT2T-HA-BCL6-ΔZF have been described previously.8 The metallothionein-inducible expression vector pMEP4-HA-BCL6 was constructed by cloning a 2.4-kb BCL6 cDNA fragment, hemagglutinin (HA)–tagged, into the BamHI site of pMEP4 (Invitrogen).
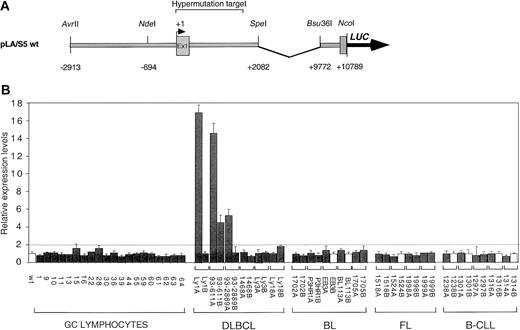
Abnormal expression of DLBCL-derived mutant BCL6 alleles.
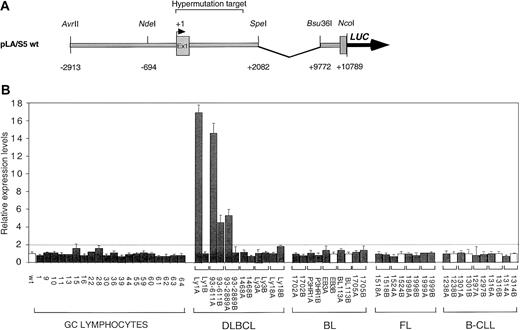
(A) Schematic diagram of the pLA/S5 wild-type reporter containing the BCL6 5′ noncoding region. Mutant constructs were generated by exchanging the 2.7-kb NdeI/SpeI fragment with the corresponding region of mutant BCL6 alleles derived from GC B cells or various lymphoma types. (B) Ly1 cells were transfected by electroporation with equimolar amounts of the indicated pLA/S5 reporter constructs and the pRL-TK plasmid as an internal control for transfection efficiency. In each tumor case, identified by a bracket, both alleles were tested (solid bars indicate mutated alleles; white bars, wild-type alleles). After 48 hours, cells were harvested and the luciferase activities were measured. One representative experiment of 3 to 8 independent transfections performed in duplicate with similar results is shown. Average luciferase activity (± SD) is expressed as relative increases with respect to the wild-type construct (set as 1), after normalization for Renilla activity; differences in activity were defined as significant when greater than 2-fold (dotted line). The DLBCL-derived alleles Ly1A, 93-611A, 93-611B, and 93-2889A were found overexpressed (4- to 18-fold), whereas no significant changes were observed in transfectants deriving from normal GC cells or other lymphoma types. DLBCL indicates diffuse large B-cell lymphoma; FL, follicular lymphoma; BL, Burkitt lymphoma; B-CLL, B-cell chronic lymphocytic leukemia.
Abnormal expression of DLBCL-derived mutant BCL6 alleles.
(A) Schematic diagram of the pLA/S5 wild-type reporter containing the BCL6 5′ noncoding region. Mutant constructs were generated by exchanging the 2.7-kb NdeI/SpeI fragment with the corresponding region of mutant BCL6 alleles derived from GC B cells or various lymphoma types. (B) Ly1 cells were transfected by electroporation with equimolar amounts of the indicated pLA/S5 reporter constructs and the pRL-TK plasmid as an internal control for transfection efficiency. In each tumor case, identified by a bracket, both alleles were tested (solid bars indicate mutated alleles; white bars, wild-type alleles). After 48 hours, cells were harvested and the luciferase activities were measured. One representative experiment of 3 to 8 independent transfections performed in duplicate with similar results is shown. Average luciferase activity (± SD) is expressed as relative increases with respect to the wild-type construct (set as 1), after normalization for Renilla activity; differences in activity were defined as significant when greater than 2-fold (dotted line). The DLBCL-derived alleles Ly1A, 93-611A, 93-611B, and 93-2889A were found overexpressed (4- to 18-fold), whereas no significant changes were observed in transfectants deriving from normal GC cells or other lymphoma types. DLBCL indicates diffuse large B-cell lymphoma; FL, follicular lymphoma; BL, Burkitt lymphoma; B-CLL, B-cell chronic lymphocytic leukemia.
Transient transfections and reporter gene assays
The B-cell lymphoma lines Ly1, MUTU I, and MUTU III (1.5 × 107/sample) were transfected by electroporation at 250 V and 960 μF using a Bio-Rad Gene Pulser apparatus (Bio-Rad Laboratories, Hercules, CA). Equimolar amounts (2 pmol) of wild-type and mutant reporter plasmids were used in each experiment, together with 0.5 pmol plasmid expressing Renilla luciferase (pRL-TK) as a control. Transient transfections into 293T cells were performed by the calcium-phosphate precipitation method,28 using 0.1 pmol of the various reporter constructs and increasing amounts (0.01, 0.02, 0.04, 0.1 pmol) of the effector plasmids (pMT2T-HA-BCL6, pMT2T-HA-BCL6-ZF, and pMT2T-HA-BCL6-ΔZF). The total amount of transfected DNA was kept constant in each experiment by adding pMT2T vector sequences to a final amount of 10 μg. The pRL-TK vector (0.15 μg) was also cotransfected as an internal control for transfection efficiency. Luciferase activities were measured 48 hours after transfection using the Dual-Luciferase Reporter Assay Kit (Promega), according to the manufacturer's protocol.
Electrophoretic mobility shift assay
Chromatin immunoprecipitation assay
The detailed protocol for formaldehyde cross-linking and chromatin immunoprecipitation (ChIP) has been published.29Briefly, samples (each obtained from 1-2.5 × 107 B cells) were immunoprecipitated overnight at 4°C with 2 μg anti-BCL6 polyclonal antibody N3 (Santa Cruz Biotechnology, Santa Cruz, CA). In each experiment, one sample with no antibody and one sample with an irrelevant antibody (anti-IgG) were included as negative controls for nonspecific binding. Immune complexes were recovered by adding 30 μL blocked salmon sperm DNA/Protein A agarose beads (50% slurry; Upstate Biotechnology, Lake Placid, NY) and incubating the samples for 2 hours at 4°C. Following extensive washing and reverse cross-linking, DNA was purified by phenol-chloroform extraction, ethanol precipitated, and resuspended in 30 μL TE (100 μL for the total input sample, representing 0.5% of the soluble chromatin before immunoprecipitation). Fragments containing the BCL6 exon 1 binding sites were detected by PCR amplification using 2.5 μL of the above DNA preparations as template and 2 different oligonucleotide pairs—5′-ACGCTCTGCTTATGAGGA-3′ (sense) and 5′-CGGCAGCAACAGCAATAA-3′ (antisense) for the B1 fragment (position +39 to +300) and 5′-GGGTTCTTAGAAGTGGTG-3′ (sense) and 5′-CAAAGCATTTGGCAAGAG-3′ (antisense) for the B2 fragment (+227 to +421). Two additional primer sets were designed to amplify control region A, located approximately 1 kb upstream of the transcription initiation site (5′-ACTAGGACCCACAATGAA-3′ and 5′-CGTTTCAAGATCGTTGTA-3′), and control region D, corresponding to the coding exon 5 (5′-GTGTGCCACAGCAATATC-3′ and 5′-TGGCAGTCAGATTTCTGG-3′). Following 32 cycles of amplification, PCR products were resolved on 2% agarose gels and visualized by ethidium bromide staining. The same amplicons were subsequently gel purified and sequenced directly and after cloning into pGEM-T vector (Promega) as described.23
Northern blot, Western blot, and reverse transcription–polymerase chain reaction analysis
Total RNA was prepared from various B-cell lines and lymphoma cases using the Trizol reagent (Invitrogen). Northern blot and Western blot analyses for BCL6 expression were performed as described.6 For cDNA synthesis, 2 μg RNA was reverse transcribed using oligo(dT)18 primer and SuperScript II reverse transcriptase (Invitrogen). The resultant first-strand DNA was PCR amplified using primers that annealed to BCL6 exons 1 and 2 and was directly sequenced.
Results
Deregulated expression of DLBCL-associated mutant alleles
To investigate the consequences of BCL6 mutations on transcriptional regulation of the BCL6 gene, we compared the transcriptional activity of mutant BCL6 alleles derived from normal cells (ie, sorted GC B cells) and lymphoma cells by using a transient transfection/reporter gene assay in 2 lymphoma lines (Ly1 and MUTU I) permissive for BCL6 expression. Initial studies showed that a luciferase reporter gene under the control of the native BCL6 promoter (approximately 14-kb AvrII/NcoI genomic fragment spanning the 5′ flanking sequences, exon 1, intron 1, and exon 2) had activity comparable to that of a deletion mutant lacking approximately 7.7 kb at the 3′ end of intron 1 (data not shown). Therefore, we used this shorter plasmid (Figure 1A) as a backbone to generate a series of mutant reporter constructs in which the wild-type 2.7-kb NdeI–SpeI fragment was substituted with variant BCL6 sequences amplified from normal GC cells (20 mutated alleles) or from various tumor cases (6 DLBCL, 5 BL, 4 FL, and 5 B-CLL). For each tumor sample, both alleles, designated A and B, were tested. Although mutations were biallelically distributed in 15 cases, the remaining cases displayed one mutated and one wild-type allele (a detailed description of the mutations present in each construct is available in Supplemental data). The basal expression level of the corresponding plasmids (pLA/S5 series) was then evaluated in the transfection/reporter gene assay in Ly1 and MUTUI.
Results showed that all the mutant alleles derived from GC cells (n = 20) had a transcriptional activity comparable to that of the wild-type allele, arbitrarily set as 1 (Figure 1B). Similarly, mutant alleles from BL (n = 8), FL (n = 7), and B-CLL (n = 6) cases were also indistinguishable from the wild-type allele in this assay. However, 4 of 12 (33%) DLBCL-associated alleles were significantly and reproducibly overexpressed (4- to 18-fold) in both transfected cell lines (see Figure 1B for representative data with the Ly1 cell line). Of the 4 deregulated alleles, 2 were derived from 2 distinct DLBCL cases (Ly1 and 93-2889), whereas the remaining 2 were derived from a single case (93-611). Interestingly, deregulation was not observed when the same reporters were transfected in a cell line (MUTU III) that lacks BCL6 expression (not shown), suggesting the involvement of the BCL6 protein itself in the observed effect (see below). These results indicate that some mutations associated with DLBCL can transcriptionally deregulate the BCL6 gene.
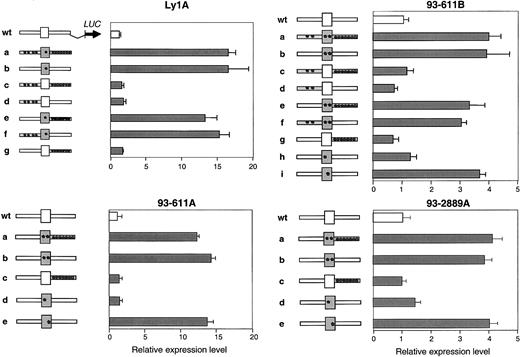
DLBCL-associated deregulating mutations cluster within exon 1 of BCL6
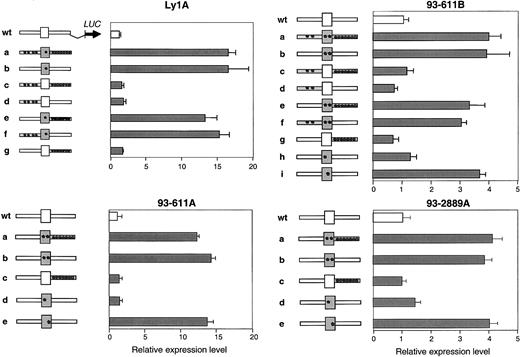
Each of the 4 BCL6 alleles displaying deregulated activity in the transient transfection assay contains multiple base-pair substitutions scattered throughout the first noncoding exon, the first intron, and, in 2 cases, the 5′ flanking sequences (Supplemental data). To identify the mutation(s) responsible for DLBCL-associated deregulation, we generated a series of reporter constructs in which wild-type and mutant fragments corresponding to the 5′ flanking, first exon, and first intron of each mutated allele were swapped in all possible combinations. The activity of these constructs was then tested using the same transient transfection/reporter gene assay in Ly1 cells. Results in Figure 2 document that, in all 4 alleles, the mutated sequences responsible for transcriptional deregulation were located in exon 1 (see construct B in each panel). Notably, this region only contains a single mutation in the deregulated allele Ly1A (T257C, indicated by an asterisk) or 2 distinct changes in deregulated alleles 93-611A, 93-611B, and 93-2889A. To ascertain which of these changes is responsible for transcriptional deregulation, each was incorporated into the pLA/S5 wt reporter construct by site-directed mutagenesis, and their effect on transcriptional activation was tested in the same assay. Remarkably, a single nucleotide substitution from each deregulated allele (T257C in Ly1A, A260G in 93-611A, T231G in 93-611B, and T234C in 93-2889A) recapitulated the abnormal behavior displayed by the original mutant. These results demonstrate that, within multiply mutated BCL6 alleles, individual mutations—all located in the first exon—were responsible for the observed deregulation.
Mapping the deregulating mutations.
Transient transfection assays were performed as described in Figure 1, using the reporter constructs shown on the left of each panel. Grey fragments correspond to the mutated 5′ flanking, exon 1 (box), and intron 1 region of the original allele (construct “a”), which were swapped in all possible combinations to map the deregulating mutation(s). Asterisks indicate the presence of mutations (note that 2 distinct nucleotide changes were present in the exon 1 sequences of allele 93-611A, 93-611B, and 93-2889A). Bars represent the activities of the resultant constructs (mean ± SD), obtained from 2 independent experiments performed in duplicate. In all 4 cases, a single mutation located in the BCL6 first noncoding exon recapitulates the activity displayed by the original construct.
Mapping the deregulating mutations.
Transient transfection assays were performed as described in Figure 1, using the reporter constructs shown on the left of each panel. Grey fragments correspond to the mutated 5′ flanking, exon 1 (box), and intron 1 region of the original allele (construct “a”), which were swapped in all possible combinations to map the deregulating mutation(s). Asterisks indicate the presence of mutations (note that 2 distinct nucleotide changes were present in the exon 1 sequences of allele 93-611A, 93-611B, and 93-2889A). Bars represent the activities of the resultant constructs (mean ± SD), obtained from 2 independent experiments performed in duplicate. In all 4 cases, a single mutation located in the BCL6 first noncoding exon recapitulates the activity displayed by the original construct.
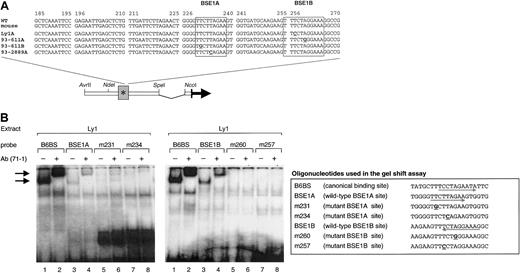
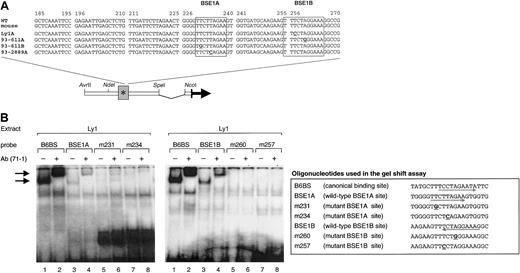
Deregulating mutations cluster within 2 BCL6 binding sites
The 4 deregulating mutations are clustered in 2 sequence motifs within exon 1 of BCL6. Significantly, both motifs share extensive homology with the preferred DNA-binding sequence of BCL6 polypeptides (Figure 3A, boxed areas; active mutations are in bold and underlined), suggesting that the BCL6 protein may bind and regulate its own promoter region. To test this hypothesis, we examined whether the 2 potential binding sites within exon 1 (called BSE1A and BSE1B) could bind BCL6 in an electrophoretic mobility shift assay (EMSA). Double-stranded oligonucleotide probes corresponding to the wild-type BSE1A (+225 + 244 from the minor BCL6 promoter) and BSE1B (+249 + 268) sequences were incubated with nuclear extracts from the BCL6-expressing cell line Ly1. Figure 3B shows that both probes were associated with a DNA-binding protein complex similar in migration to the one formed by B6BS, a probe representing the canonical BCL6 binding site8 (lanes 1 and 3 in each gel). Supershift analysis using antisera against the N-terminus of BCL-6 (N71-1) confirmed that these complexes contained BCL6 (lanes 2 and 4). The difference in intensities between the BSE1A- and BSE1B-associated bands may reflect differences in the abundance of these complexes or distinct binding affinities of the BSE1A (8 of 9 matches with the core domain of the canonical BCL6 binding site) and BSE1B (100% identity in the 9-bp core) sequences. Notably, oligonucleotide probes carrying the tumor-derived deregulating mutations (m231, m234, m257, and m260) failed to bind BCL6 (lanes 5-8). In addition, it may be relevant that alleles bearing mutations of the high-affinity BSE1B site (ie, Ly1A and 93-611A) were significantly more deregulated than those harboring mutations of the low-affinity BSE1A site (93-611B and 93-2889A) (Figure 1A-B). Thus, the BCL6 protein can bind the exon 1 sequences in vitro and this binding is abrogated by the DLBCL-associated mutations.
In vitro binding of BCL6 to the BSE1 motifs.
(A) Sequence of the BCL6 exon 1 region in which the 4 deregulating mutations (in bold, underlined) were mapped. The corresponding murine sequence is also aligned to show the complete sequence identity between the 2 species. Boxed areas represent the 2 BCL6 binding motifs (BSE1A and BSE1B). Nonderegulating mutations are shown in gray. (B) BCL6 deregulating mutations abolish binding of BCL6 to its exon 1 sequences in vitro. EMSA was performed on Ly1 nuclear extracts using as probes the indicated oligonucleotides. The B6BS probe containing the BCL6 canonical binding site was used as a control.8 Arrows point to BCL6-containing complexes. Supershift analysis was performed using an anti-BCL6 (N71-1) antiserum that recognizes the N-terminus of the BCL6 protein.
In vitro binding of BCL6 to the BSE1 motifs.
(A) Sequence of the BCL6 exon 1 region in which the 4 deregulating mutations (in bold, underlined) were mapped. The corresponding murine sequence is also aligned to show the complete sequence identity between the 2 species. Boxed areas represent the 2 BCL6 binding motifs (BSE1A and BSE1B). Nonderegulating mutations are shown in gray. (B) BCL6 deregulating mutations abolish binding of BCL6 to its exon 1 sequences in vitro. EMSA was performed on Ly1 nuclear extracts using as probes the indicated oligonucleotides. The B6BS probe containing the BCL6 canonical binding site was used as a control.8 Arrows point to BCL6-containing complexes. Supershift analysis was performed using an anti-BCL6 (N71-1) antiserum that recognizes the N-terminus of the BCL6 protein.
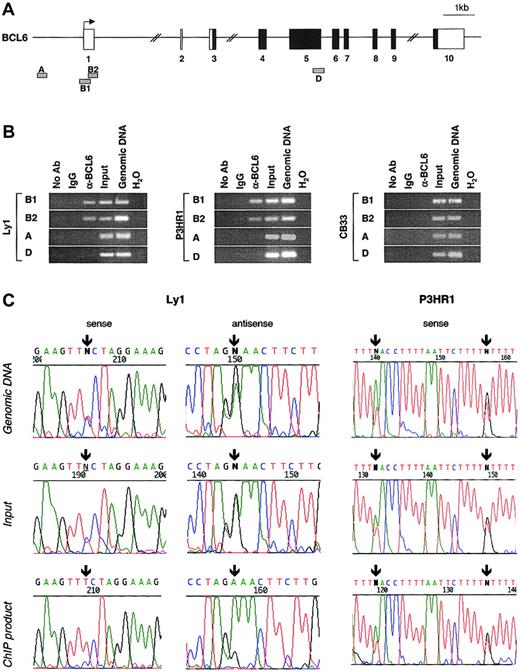
To confirm that endogenous BCL6 can bind to its native promoter in vivo, we performed ChIP assays using an anti-BCL6 polyclonal antibody (N3) in 2 BCL6-positive cell lines. Ly1 cells carry a mutation in the BSE1B site of allele A, which confers deregulation in the reporter assay (Figure 2) and inactivates BCL6 binding in the EMSA (Figure 3B). The P3HR1 line was used as a control because it contains mutations in the BCL6 exon 1/intron 1 boundary (172G>C, 380T>A, and 397T>G) located outside the BSE1A and BSE1B motifs and not associated with deregulation. Four different oligonucleotide pairs were designed to amplify genomic fragments corresponding to the relevant exon 1 sequences (products B1 and B2) or to 2 distal control regions (products A and D) located approximately 1 kb upstream and more than 10 kb downstream of exon 1, respectively (Figure4A). The results show that BCL6 binds specifically to regions B1 and B2, but not to the control regions A and D, in both BCL6-expressing cell lines (Figure 4B); no product was obtained using an irrelevant antibody (IgG) or a control line (CB33) that did not express BCL6 (Figure 4B, right panel). Thus, the exon 1 sequence containing the BSE1A and BSE1B sites represents a specific target for BCL6 binding in vivo.
BCL6 binds to its exon 1 sequence in vivo (ChIP assay), and the binding is abrogated by BCL6 deregulating mutations.
(A) Schematic representation of the human BCL6 locus; the 4 genomic fragments amplified for analysis are approximately positioned below the map (B1 and B2, test region; A and D, control regions). (B) Ethidium bromide–stained agarose gels of PCR products A to D obtained from 2 BCL6-expressing cell lines (Ly1 and P3HR1) and a control line that lacks BCL6 expression (CB33). After formaldehyde cross-linking, chromatin was immunoprecipitated using the anti-BCL6 antibody N3 or an irrelevant antibody (IgG) as control, and PCR reactions were performed on ChIP products, total chromatin before immunoprecipitation (input), and genomic DNA as a positive control for PCR. One sample was also processed with no antibody to serve as a negative control. (C) BCL6 binds to the wild-type allele but not to the mutated allele in the Ly1 cell line. Direct sequencing of PCR products obtained from genomic DNA, total input, and immunoprecipitated chromatin (ChIP) in the Ly1 cell line and in P3HR1 as a control. Arrows indicate the position of the mutations (Ly1, 257T>C; P3HR1, 380T>A and 397T>G).
BCL6 binds to its exon 1 sequence in vivo (ChIP assay), and the binding is abrogated by BCL6 deregulating mutations.
(A) Schematic representation of the human BCL6 locus; the 4 genomic fragments amplified for analysis are approximately positioned below the map (B1 and B2, test region; A and D, control regions). (B) Ethidium bromide–stained agarose gels of PCR products A to D obtained from 2 BCL6-expressing cell lines (Ly1 and P3HR1) and a control line that lacks BCL6 expression (CB33). After formaldehyde cross-linking, chromatin was immunoprecipitated using the anti-BCL6 antibody N3 or an irrelevant antibody (IgG) as control, and PCR reactions were performed on ChIP products, total chromatin before immunoprecipitation (input), and genomic DNA as a positive control for PCR. One sample was also processed with no antibody to serve as a negative control. (C) BCL6 binds to the wild-type allele but not to the mutated allele in the Ly1 cell line. Direct sequencing of PCR products obtained from genomic DNA, total input, and immunoprecipitated chromatin (ChIP) in the Ly1 cell line and in P3HR1 as a control. Arrows indicate the position of the mutations (Ly1, 257T>C; P3HR1, 380T>A and 397T>G).
Based on the EMSA results showing that BCL6 binds to the wild-type, but not mutant, exon 1 sequences, we investigated whether this differential binding can also be detected in vivo by examining the allelic representation of chromatin immunoprecipitates obtained from Ly1 cells (and P3HR1 cells as a control) using the anti-BCL6 antibody. Because all mutations were heterozygous in these lines, the presence of each individual allele could be uniquely identified by PCR sequencing. Figure 4C shows that both alleles are present in the genomic DNA and in the chromatin solution before immunoprecipitation (input), from both Ly1 and P3HR1 (top and middle panels). However, we consistently observed preferential amplification of the wild-type allele in the ChIP product obtained from Ly1 but not from P3HR1 cells (bottom panels). This predominance of the unmutated allele B in the immunoprecipitated chromatin from Ly1 was confirmed by cloning and sequencing the corresponding PCR amplicon, which revealed wild-type sequences in 83% of the clones as opposed to the approximately 50% expected and observed in the control genomic DNA and in the total input (not shown). Taken together, these results demonstrate that the BCL6 exon 1 sequence is a direct physiologic target for BCL6 binding in vivo and that this binding is impaired by the so-called deregulating mutations associated with DLBCL.
BCL6 exon 1 binding sites mediate a negative autoregulatory circuit
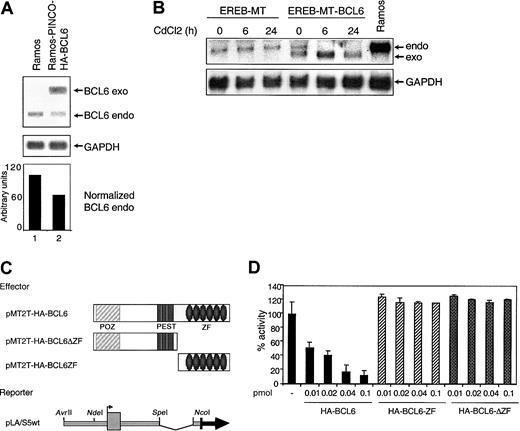
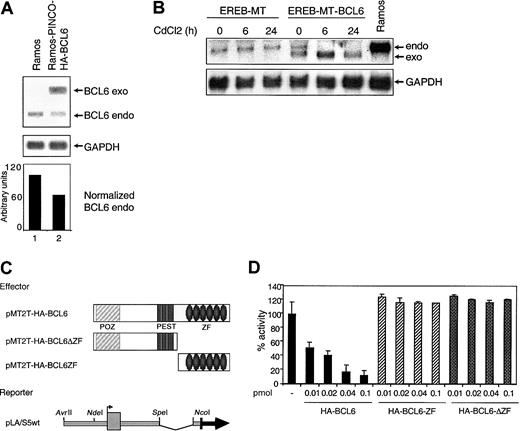
Given that BCL6 is a transcriptional repressor, these data suggested the existence of an autoregulatory circuit in which BCL6 binds its own promoter and inhibits its own transcription. To examine whether BCL6 genes are subjected to this autoregulatory circuit in vivo, we first analyzed endogenous BCL6 expression levels in a BL cell line, Ramos, which had been engineered to constitutively express exogenous BCL6 under the control of a retroviral vector (PINCO). The results shown in Figure 5A indicate that even modest levels of exogenous BCL6 expression (Ramos cells do not tolerate higher levels) lead to a proportional reduction (approximately 40%) in endogenous BCL6 levels.
The BCL6 promoter is a target for BCL6-mediated transcriptional repression.
(A) Negative regulation of endogenous BCL6 by constitutive expression of exogenous BCL6 in Ramos cells. Northern blot analysis of endogenous (endo) and exogenous (exo) BCL6 expression in Ramos cells and in Ramos clones transduced with the PINCO-HA-BCL6 retroviral vector. The signal intensity ratio between endogenous BCL6 and GAPDH, quantitated by PhosphorImager analysis, is shown in the lower panel. (B) CdCl2-induced down-regulation of endogenous BCL6 gene expression in EREB cells stably transfected with an inducible BCL6 gene. Northern blot analysis of EREB cells transfected with MT (EREB-MT) or MT-BCL6 (EREB-MT-BCL6) plasmids. Cells were treated with CdCl2 as described in “Materials and methods” and were collected at the indicated intervals. The Ramos cell line was included as a control for size of the endogenous BCL6 transcripts. Filters were sequentially hybridized with a radiolabeled BCL6 cDNA probe and with GAPDH to control for amount of RNA loading. (C) Schematic representation of the plasmids used in transient cotransfection experiments. POZ indicates protein–protein interaction domain; and ZF, zinc finger DNA-binding domain. (D) The reporter construct indicated in panel C was transfected alone or in the presence of increasing amounts of various BCL6-expressing plasmids into 293T cells. Luciferase activities measured 48 hours after transfection revealed a strong and dose-dependent repression of the reporter gene when cotransfected with the wild-type BCL6-expressing plasmid (solid bars), but not with 2 deletion mutants that lack the DNA-binding (ΔZF) or the transrepression domain (ZF) (hatched bars). All experiments were performed in duplicate, and standard deviations are indicated.
The BCL6 promoter is a target for BCL6-mediated transcriptional repression.
(A) Negative regulation of endogenous BCL6 by constitutive expression of exogenous BCL6 in Ramos cells. Northern blot analysis of endogenous (endo) and exogenous (exo) BCL6 expression in Ramos cells and in Ramos clones transduced with the PINCO-HA-BCL6 retroviral vector. The signal intensity ratio between endogenous BCL6 and GAPDH, quantitated by PhosphorImager analysis, is shown in the lower panel. (B) CdCl2-induced down-regulation of endogenous BCL6 gene expression in EREB cells stably transfected with an inducible BCL6 gene. Northern blot analysis of EREB cells transfected with MT (EREB-MT) or MT-BCL6 (EREB-MT-BCL6) plasmids. Cells were treated with CdCl2 as described in “Materials and methods” and were collected at the indicated intervals. The Ramos cell line was included as a control for size of the endogenous BCL6 transcripts. Filters were sequentially hybridized with a radiolabeled BCL6 cDNA probe and with GAPDH to control for amount of RNA loading. (C) Schematic representation of the plasmids used in transient cotransfection experiments. POZ indicates protein–protein interaction domain; and ZF, zinc finger DNA-binding domain. (D) The reporter construct indicated in panel C was transfected alone or in the presence of increasing amounts of various BCL6-expressing plasmids into 293T cells. Luciferase activities measured 48 hours after transfection revealed a strong and dose-dependent repression of the reporter gene when cotransfected with the wild-type BCL6-expressing plasmid (solid bars), but not with 2 deletion mutants that lack the DNA-binding (ΔZF) or the transrepression domain (ZF) (hatched bars). All experiments were performed in duplicate, and standard deviations are indicated.
To exclude that the observed phenomenon may be related to cell culture selection or to clonal variability, we examined the temporal and quantitative responses of endogenous BCL6 transcription in a lymphoblastoid cell line, EREB, transfected with an inducibleBCL6 gene vector (pMEP4-HA-BCL6 [MT-BCL6]) or with an empty vector (pMEP4 [MT]) as control. In this system, transcripts from the exogenous BCL6 sequences are driven by the heavy metal-inducible metallothionein promoter30 and can be distinguished from endogenous transcripts because of their lower molecular weights. Stably transfected EREB-MT and EREB-MT-BCL6 cells were analyzed by Northern blot for BCL6 expression at different time points after the addition of CdCl2 to the cell culture medium. As indicated in Figure 5B, endogenous BCL6 transcripts can be clearly detected in the EREB-MT-BCL6 clone at time 0, when very low levels of exogenous BCL6 are present, possibly because of the leaky activity of the MT promoter (line 4). However, CdCl2-mediated induction of exogenous BCL6 caused a progressive decrease in endogenous BCL6 mRNA levels (lanes 5 and 6). Thus, the down-regulation of BCL6gene expression in the presence of exogenous BCL6 represents a specific and dose-dependent effect of increased BCL6 protein levels, fulfilling the criteria for an autoregulatory circuit.
Finally, we examined whether the autoregulatory circuit functions at the transcriptional level and whether it targets the BCL6 promoter region. To this end, the pLA/S5 reporter construct containing the wild-type BCL6 promoter was cotransfected with increasing amounts of a BCL6 expression plasmid (pMT2T-HA-BCL6) into 293T cells (Figure 5C). Luciferase activities revealed a strong and dose-dependent repression of the reporter gene in the presence of exogenous BCL6 (8% residual activity at 0.1 pmol input) (Figure 5D, solid bars). This effect was attributed to genuine BCL6-mediated transcriptional repression because it was not detected when the same reporter was cotransfected with BCL6 deletion mutants that lack either the DNA-binding (pMT2T-HA-BCL6-ΔZF) or the N-terminal transrepression (pMT2T-HA-BCL6-ZF) domain (Figure 5D, hatched bars). Taken together, these data document the existence of an autoregulatory circuit in which BCL6 modulates its own transcription by binding to its promoter sequences in native B cells.
BCL6 mutations disrupt the BCL6 autoregulatory mechanism
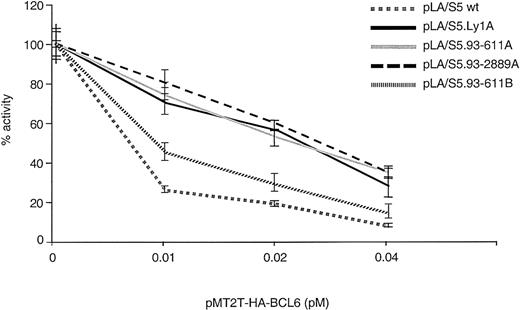
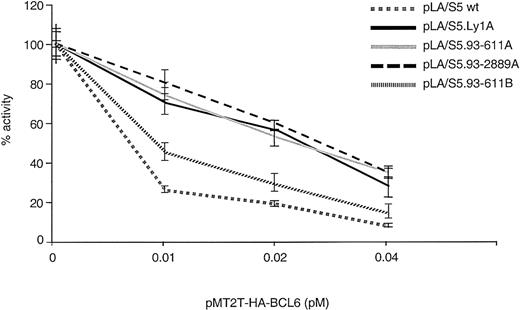
To study whether BCL6 autoregulation targets specifically the BSE1A and BSE1B motifs and is disrupted by the 4 DLBCL-associated mutations, we used the same transient transfection/reporter gene assay and tested the degree of resistance of the mutated alleles to this negative feedback loop. To this end, the responses of the 4 deregulated pLA/S5 single mutant reporters (Figure 2) were compared with those of the wild-type construct on cotransfection with increasing amounts of pMT2T-HA-BCL6 in 293T cells. Variable degrees of resistance to exogenous BCL6 repression were reproducibly observed in each of the 4 mutants (Figure 6). This abnormal response was especially evident in the Ly1A-, 93-611A-, and 93-2889A–derived transfectants and, to a lesser extent, in 93-611B. These data indicate that the DLBCL-associated mutations confer resistance to the autoregulatory feedback loop and also establish the BSE1 motifs as the in vivo targets for autoregulation.
Mutations within the BCL6 exon 1 binding sites confer resistance to BCL6-mediated transrepression activity.
Responses of the 4 DLBCL-derived BSE1-mutant reporter constructs to BCL6-mediated transrepression activity were compared with those of a wild-type plasmid in cotransfection experiments in 293T cells, as described in Figure 5D.
Mutations within the BCL6 exon 1 binding sites confer resistance to BCL6-mediated transrepression activity.
Responses of the 4 DLBCL-derived BSE1-mutant reporter constructs to BCL6-mediated transrepression activity were compared with those of a wild-type plasmid in cotransfection experiments in 293T cells, as described in Figure 5D.
Deregulated expression of BCL6 in DLBCL cases carrying BSE1 mutations
The results obtained in the transient transfection experiments imply that deregulated alleles escape the negative autoregulatory circuit and are overexpressed, whereas the wild-type allele remains responsive and transcriptionally silent. Consistent with this hypothesis, Northern and Western blot analyses of DLBCL cell lines showed that the Ly1 cells, which carry a BCL6 allele mutated in the BSE1 site, express levels of BCL6 mRNA and protein higher than lines carrying either translocated, non–BSE1-mutated, or wild-type BCL6 alleles (see Figure 7A-B for representative results). Furthermore, an analysis of BCL6 allele-specific transcription by RT-PCR and sequencing revealed that only one allele—the one that carries the mutation in the BSE1 site—is transcribed in Ly1 cells (Figure 7C, top panel). In contrast, BCL6 is biallelically expressed in the BJAB (Figure 7C, bottom panel) and P3HR1 (not shown) lymphoma lines, each of which is heterozygous for nonderegulating mutations. The correlation between exon 1 mutations and deregulation could not be validated in primary tumors because the presence of contaminating normal cells makes it difficult to quantitatively analyze allelic expression in tumor cells by RT-PCR with accuracy. Nonetheless, these data demonstrate that the deregulating mutations alter the transcriptional regulation of endogenousBCL6 genes in native DLBCL cells.
Deregulated BCL6 expression in DLBCL cases carrying BSE1 mutations.
(A) Northern blot analysis of BCL6 expression in various DLBCL-derived cell lines. Equal amounts (12 μg) of total RNA were loaded on a formaldehyde-agarose gel, blotted, and sequentially hybridized with a full-length BCL6 cDNA probe and with GAPDH as control for the amount and integrity of the RNA. Quantitative analysis of the data was performed using PhosphorImager (bottom panel). For each sample, the presence of 3q27 chromosomal translocations [t(3q27)], mutations affecting the BSE1 sites [M (BSE1)], and mutations located outside the BSE1 sites [M (others)] are indicated. The lymphoblastoid cell line CB33, which does not express BCL6, was included as a negative control. (B) Western blot analysis of BCL6 protein expression in the same cell lines, using the anti-BCL6 (N3) antibody and an anti–β-tubulin antibody as control for protein loading. (C) Nucleotide sequencing of RT-PCR products generated from the mutated lymphoma cell lines Ly1 and BJAB. Only the allele A, identified by the 257T>C mutation in the BSE1 site, but not the allele B (containing a normal sequence in exon 1) is expressed in Ly1, whereas BCL6 is biallelically expressed in the control line BJAB, heterozygous for nonderegulating mutations. Ramos cells are shown as a control for unmutated exon 1 sequences.
Deregulated BCL6 expression in DLBCL cases carrying BSE1 mutations.
(A) Northern blot analysis of BCL6 expression in various DLBCL-derived cell lines. Equal amounts (12 μg) of total RNA were loaded on a formaldehyde-agarose gel, blotted, and sequentially hybridized with a full-length BCL6 cDNA probe and with GAPDH as control for the amount and integrity of the RNA. Quantitative analysis of the data was performed using PhosphorImager (bottom panel). For each sample, the presence of 3q27 chromosomal translocations [t(3q27)], mutations affecting the BSE1 sites [M (BSE1)], and mutations located outside the BSE1 sites [M (others)] are indicated. The lymphoblastoid cell line CB33, which does not express BCL6, was included as a negative control. (B) Western blot analysis of BCL6 protein expression in the same cell lines, using the anti-BCL6 (N3) antibody and an anti–β-tubulin antibody as control for protein loading. (C) Nucleotide sequencing of RT-PCR products generated from the mutated lymphoma cell lines Ly1 and BJAB. Only the allele A, identified by the 257T>C mutation in the BSE1 site, but not the allele B (containing a normal sequence in exon 1) is expressed in Ly1, whereas BCL6 is biallelically expressed in the control line BJAB, heterozygous for nonderegulating mutations. Ramos cells are shown as a control for unmutated exon 1 sequences.
Frequency of deregulating mutations in DLBCL
To determine the frequency of BSE1 mutations in DLBCL, we screened by PCR amplification and direct sequencing a panel of 32 DLBCL cases, all cytogenetically characterized, for the presence of mutations affecting the 2 BCL6 binding sites. Our survey revealed that 4 of 25 (16%) samples lacking 3q27 translocations (approximately 13% of all DLBCLs) carry alterations in these sequences; conversely, mutations within the BSE1 sites were not found in cases in which theBCL6 gene had been disrupted by 3q27 abnormalities. These results indicate that mutations affecting the BCL6 exon 1 binding sites are associated with a sizable fraction of DLBCL and appear to be mutually exclusive with BCL6 rearrangements.
Discussion
Mutations affecting the 5′ noncoding region of the BCL6gene are found in 30% to 40% of GC B cells and in all GC or post–GC-derived B-cell non-Hodgkin lymphomas.22-27Despite this high frequency, the functional consequences of the mutations in normal and transformed B cells are unknown. The present study elucidates one mechanism by which specific BCL6 mutations associated with DLBCL can deregulate its own expression, and it identifies a negative autoregulatory circuit that controls BCL6 transcriptional activity in vivo. These results have direct implications for the regulation of BCL6 expression in normal and neoplastic B cells and for the role of BCL6 in lymphomagenesis.
The first observation emerging from this study is that the pattern of BCL6 mutations can differ among normal GC cells and transformed cells from various GC-derived lymphoma subtypes. In fact, BCL6 mutations affecting the 2 BCL6 binding sites in exon 1 (BSE1A and BSE1B) were found at sizable frequency among DLBCL-derived alleles, but not in GC cells or in indolent lymphomas. Because BCL6 mutations occur physiologically in GC B cells, most likely by the same mechanism that hypermutates IgV sequences,22-24 it is conceivable that their distribution may be stochastically determined in different cells. Thus, the finding of BSE1 mutations specifically restricted to DLBCL suggests a selection for these mutations in association with lymphoma development.
Our results also indicate that the basis for this selection is the ability of BSE1 mutations to disrupt the negative autoregulatory mechanism that controls BCL6 expression. The possibility that BCL6 regulates its own expression was previously proposed based on the identification of a BCL6 binding motif in exon 1 that mediates BCL6-dependent transcriptional repression of a reporter plasmid containing a heterologous promoter.31 The present study provides conclusive evidence for the presence of 2 BCL6 binding sites, one of which was not previously reported, and demonstrates that both are required for BCL6-mediated transcriptional repression in the context of the physiological BCL6 promoter region. Most notably, our results show that these 2 sites bind BCL6 in vivo and that this binding is lost in DLBCL-associated mutant alleles. Taken together, these findings are consistent with the existence of a negative autoregulatory feedback loop in which BCL6 regulates its own expression by direct binding and repression of its promoter region. The existence of this mechanism is further supported by a number of independent observations: (1) endogenous BCL6 expression is down-regulated in cell lines (Figure 5A-B) and in transgenic mice (our unpublished results) expressing exogenous BCL6; (2) the expression of truncated BCL6 alleles is increased in mice expressing a nonfunctional BCL6 protein due to inactivation of the BCL6 gene by homologous recombination (Ye et al, 1997, and our unpublished results); (3) the normal BCL6 allele is not expressed in DLBCL cases in which the second allele is deregulated by chromosomal translocation.20
Autoregulatory circuits represent a common, evolutionarily conserved mechanism for the modulation of gene expression and are typically used by genes encoding transcription factors.32 These circuits have homeostatic functions by limiting the range over which expression levels of a gene can fluctuate.32 In mammalian cells, autoregulatory circuits have been described for well-known proto-oncogenes encoding transcription factors that need to be tightly regulated (eg, c-MYC).33,34 In the case of BCL6, loss of proper autoregulation could be especially relevant considering that this protein can repress the transcription of genes that induce cell cycle arrest, apoptosis, and differentiation.13,14 For at least some of these genes, the extent of repression depends on the relative levels of BCL6 versus other transcriptional activators, such as STAT6, which regulate the same genes.12 Thus, aberrantly high levels of BCL6 may lead to constitutive repression of genes necessary for GC exit and further differentiation, thereby contributing to lymphomagenesis.
The results herein identify the 2 BSE1 motifs within BCL6 exon 1 as the critical targets for BCL6 deregulation in DLBCL. This notion is supported by several observations: (1) exon 1 sequences spanning the BCL6 binding sites are highly conserved between human and mouse (100% identity over 120 bp; see also Figure 3A); (2) this region is separated from the BCL6 coding exons in most DLBCL cases that carry a 3q27 translocation (approximately 40%); and (3) this region is lost by internal deletions in a small fraction of cases.35-37Thus, one common consequence of most translocations and DLBCL-associated BSE1 mutations or deletions is the removal of the sequences involved in BCL6 autoregulation.
The identification of a fraction of DLBCL (approximately 13% overall, corresponding to 16% of the nontranslocated cases) in which BCL6 expression is deregulated by exon 1 mutations suggests a broader involvement of the BCL6 proto-oncogene in the pathogenesis of DLBCL than previously suspected based on the frequency of chromosomal translocations (40%).1,18 19 In fact, the frequency of DLBCL-associated mutations that alter BCL6 expression may be even higher if we consider that the assay used here can identify deregulated alleles only if the mutations affect constitutive BCL6 expression levels. Thus, tumor-associated mutations that alter the response of theBCL6 gene to regulatory signals may also exist but would not be identified by our assay. This possibility is supported by the observation that, analogous to cases carrying BCL6 translocations, a number of DLBCLs that display BCL6 mutations, but not translocations, are resistant to the physiologic BCL6 down-regulation induced by CD40 signaling (G. Cattoretti et al, manuscript in preparation). The experimental strategies used in this study can be adapted to investigate the effect of mutations on the ability of various signals to down-regulate BCL6 expression.
Finally, by identifying an expanded fraction of DLBCL cases carrying deregulated BCL6 expression, these findings have diagnostic and therapeutic implications. The prognostic significance of BCL6 deregulation has been the object of intense controversy, with some studies claiming it as a favorable indicator and others disclaiming the finding.38-42 This controversy can be explained by the observation herein that the fraction of DLBCLs carrying chromosomal translocations, the one considered by the above prognostic studies, does not account for the entire fraction of DLBCLs carrying deregulated BCL6 and that those with deregulating mutations should also be considered. On the therapeutic side, these results have implications for the clinical trials in which BCL6 is under evaluation as a therapeutic target for histone deacetylase inhibitors.17
We thank G. Cattoretti for the isolation of GC B cells, V. Miljkovic and AnaMaria Babiac for excellent assistance in DNA sequencing, and R. Baer for critically reading the manuscript.
Prepublished online as Blood First Edition Paper, December 19, 2002; DOI 10.1182/ blood-2002-11-3387.
Supported by National Institutes of Health grants CA-072699 (R.D.-F.) and CA-34775 and CA-66999 (R.S.K.C.). L.P. is a Special Fellow of the Leukemia and Lymphoma Society of America.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Riccardo Dalla-Favera, Institute for Cancer Genetics, Columbia University, 1150 St Nicholas Ave, New York, NY 10032; e-mail: rd10@columbia.edu.


![Fig. 7. Deregulated BCL6 expression in DLBCL cases carrying BSE1 mutations. / (A) Northern blot analysis of BCL6 expression in various DLBCL-derived cell lines. Equal amounts (12 μg) of total RNA were loaded on a formaldehyde-agarose gel, blotted, and sequentially hybridized with a full-length BCL6 cDNA probe and with GAPDH as control for the amount and integrity of the RNA. Quantitative analysis of the data was performed using PhosphorImager (bottom panel). For each sample, the presence of 3q27 chromosomal translocations [t(3q27)], mutations affecting the BSE1 sites [M (BSE1)], and mutations located outside the BSE1 sites [M (others)] are indicated. The lymphoblastoid cell line CB33, which does not express BCL6, was included as a negative control. (B) Western blot analysis of BCL6 protein expression in the same cell lines, using the anti-BCL6 (N3) antibody and an anti–β-tubulin antibody as control for protein loading. (C) Nucleotide sequencing of RT-PCR products generated from the mutated lymphoma cell lines Ly1 and BJAB. Only the allele A, identified by the 257T>C mutation in the BSE1 site, but not the allele B (containing a normal sequence in exon 1) is expressed in Ly1, whereas BCL6 is biallelically expressed in the control line BJAB, heterozygous for nonderegulating mutations. Ramos cells are shown as a control for unmutated exon 1 sequences.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/8/10.1182_blood-2002-11-3387/4/m_h80834161007.jpeg?Expires=1766002121&Signature=5IvdFwKpkDrv0MPT6TBJnXOdJBA4IaEYvbeyK46OluhJV-kLMdoZcgsXziw2Q7WESK~zgjifhYqtPbuSeSFeeX3NvCCyCZf1U4dZ5VLa20MCbIMbAmAv38QkH67CeBseWYvpopPzDaXG~1p1gZK5uyA26gwbZn62GmqNXzP5w5MJrolvj~5k7vlawU8sU44nQ6V33sUdA6ERF5FITq-u-QsK8hVXfaUwkDQja0wh9v~OSVYA550lEJ9n1BaKCCj5toGoo-w0Aaq9QmoCWVb06BITp4QzlMtHwjbHPooCipsx0xNR7tqOHiDxodRoqUDiII3V0OXF77V4WzvbJ3vKrA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 7. Deregulated BCL6 expression in DLBCL cases carrying BSE1 mutations. / (A) Northern blot analysis of BCL6 expression in various DLBCL-derived cell lines. Equal amounts (12 μg) of total RNA were loaded on a formaldehyde-agarose gel, blotted, and sequentially hybridized with a full-length BCL6 cDNA probe and with GAPDH as control for the amount and integrity of the RNA. Quantitative analysis of the data was performed using PhosphorImager (bottom panel). For each sample, the presence of 3q27 chromosomal translocations [t(3q27)], mutations affecting the BSE1 sites [M (BSE1)], and mutations located outside the BSE1 sites [M (others)] are indicated. The lymphoblastoid cell line CB33, which does not express BCL6, was included as a negative control. (B) Western blot analysis of BCL6 protein expression in the same cell lines, using the anti-BCL6 (N3) antibody and an anti–β-tubulin antibody as control for protein loading. (C) Nucleotide sequencing of RT-PCR products generated from the mutated lymphoma cell lines Ly1 and BJAB. Only the allele A, identified by the 257T>C mutation in the BSE1 site, but not the allele B (containing a normal sequence in exon 1) is expressed in Ly1, whereas BCL6 is biallelically expressed in the control line BJAB, heterozygous for nonderegulating mutations. Ramos cells are shown as a control for unmutated exon 1 sequences.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/8/10.1182_blood-2002-11-3387/4/m_h80834161007.jpeg?Expires=1766321701&Signature=gZJV2RLTfHrJTeuPu23545wdb5CABhw~psVvNlN8g8YWCHyo8j2ZDD4TwMRTSk-EFjTkbLRb5x2Vi8xKN1011fm04kbHJxI3ezmIDiX3y8BsNae858dAaO~aedeoN-HRXzq5-hqUYBa9pLsWG1g6aFMp54EeevUcmZ6HFFRQj5bL44ufJAB8PB4PjsJpCAX3FYMtk-1-vv5LkryCJHHQDYHKgPBu6evdQ1uGj6Ty-oLRdYtD94BVv6zOhpTn3Wn~opxmTc50jqe~G1ee5Ts85LI2V9eWEV6J5u5kLBm0E021Geyy8GmXuQo5Dlz01MQDCrnrS0ZT4HQb38u2UECWew__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)