Abstract
Human blood platelets are anucleate cells whose response to extracellular stimuli results in actin cytoskeleton rearrangements, thereby providing the critical initial step in the regulation of hemostasis. The serine protease α-thrombin, known to activate platelets by cleavage of a family of protease-activated receptors (PARs), is the most potent physiologic activator of human platelets, though downstream effector proteins uniquely linked to platelet cytoskeletal actin polymerization remain largely uncharacterized. The gene encoding the putative rac1/cdc42 effector protein IQGAP2 was identified within the PAR gene cluster at 5q13, flanked telomeric byPAR1 and encompassing PAR3. Immunofluorescence microscopy demonstrated IQGAP2 expression in filopodial extensions of activated platelets and colocalized with F-actin in lamellipodia and filopodia of IQGAP2-transfected COS1 cells. Platelet activation by α-thrombin, but not saturating concentrations of fibrillar collagen or adenosine 5′-diphosphate, uniquely assemble an IQGAP2/arp2/3–actin cytoplasmic complex, an association regulated by guanosine triphosphate rac1 ([GTP]rac1) but not by [GTP]cdc42. Likewise, only thrombin-activated platelets resulted in rapid translocation of IQGAP2 to the platelet cytoskeleton. These observations identify a physiologic scaffolding function for IQGAP2 and establish the presence of a functional genomic unit in humans uniquely evolved to regulate thrombin-induced platelet cytoskeletal actin reorganization.
Introduction
Remodeling of actin filaments represents the penultimate event in activation-dependent cellular migration and morphogenesis, cytoskeletal changes that are dramatically recapitulated in activated blood platelets. Various extracellular stimuli rapidly transform the normally quiescent discoid platelet to a contractile sphere extruding lamellae and filopodia, physiologic structures that modulate the first key steps in hemostasis by regulating platelet adhesiveness and aggregation. In these cells, specific GTPases appear to be responsible for distinct types of actin assembly that would lead to the formation of either filopodia (presumably involved in fibrin-platelet or platelet-platelet interactions) or lamellae (involved in adhesion for plugging vascular leaks).1 Rho GTPases form a subgroup of the Ras superfamily that is largely involved in the regulation of cytoskeletal organization in response to extracellular stimuli.2 Like all members of the superfamily, the activity of Rho GTPases is determined by the ratio of their guanosine triphosphate–guanosine diphosphate (GTP/GDP)–bound forms, which are regulated by the opposing effects of guanine nucleotide exchange factors (GEFs)—known to enhance the exchange of bound GDP for GTP—and the GTPase-activating proteins (GAPs), which increase the intrinsic rate of hydrolysis of bound GTP. In addition, Rho-like GTPases are regulated further by guanine nucleotide dissociation inhibitors, which can inhibit the exchange of GTP and the hydrolysis of bound GTP.
The serine protease α-thrombin is the most potent physiologic activator of human platelets, specifically leading to GTP-charging of the Rho GTPase rac1.1,2 Although the precise role of cdc42 in platelet activation remains unknown, GTP-bound rac1 can directly stimulate phosphatidylinositol-5-kinase (PI5-K), known to be coactivated in thrombin-stimulated platelets.2 Activated PI5-K results in the generation of polyphosphoinositides, which represent the final stimulus for barbed-end uncapping and actin filament assembly.2,3 Actin filament uncapping can be selectively recapitulated using the constitutively active rac1V12 mutant, with no comparable role for the RhoA GTPase.2Although D3-containing phosphoinositides appear to be necessary for actin assembly mediated through some platelet receptors, their generation does not appear to be required for thrombin-induced actin assembly and platelet shape change,4 suggesting the presence of alternative pathways for thrombin-stimulated actin reorganization. To date, fundamental roles for Gαq5 and Gα12/136in platelet responsiveness and shape change have been identified, though downstream effector proteins linked to platelet cytoskeletal actin polymerization remain largely uncharacterized. We now provide evidence that the rac1/cdc42 effector protein IQGAP2 is expressed in human platelets and functions as a distinctive thrombin-regulated scaffolding protein in platelet activation. Because the gene encoding IQGAP2 was identified within the thrombin-responsive PAR gene cluster at chromosome 5q13,7 8 these observations suggest the emergence of a functional genomic unit uniquely evolved to mediate thrombin-signaling events in humans.
Materials and methods
Molecular studies
Total cellular RNA (HepG2, human erythroleukemia [HEL] cells, or human platelets) was isolated by immediate solubilization in Trizol (Gibco BRL, Carlsbad, CA) and isopropanol precipitation, or poly(A) RNA was isolated by solubilization and oligo(dT)–cellulose chromatography (Invitrogen, Carlsbad, CA). Northern blot analysis, reverse transcription–polymerase chain reaction (RT-PCR), and DNA-PCR were completed as previously described.8 Oligonucleotide primers were IQGAP2001 (sense, bp 4921-4940; [5′-3′, CTGCTGAACAAGTTCTA)]) and IQGAP2002 (antisense, bp 5421-5402; [5′-3′, ATTGATAACTCCCAGAAGAC]). Oligonucleotide primers included G18F (sense, IQGAP2 bp 1581-1604; [5′-3′, AGTTGTAGCTGTAGGGTACATCAA]); G15R (antisense, IQGAP2 bp 1835-1812; [5′-3′, TTGTTTTGCTCTGTCCTCGTCCAC]); P01F (sense, PAR3 bp 1783-1800; [5′-3′, TGCCTTATTGCTACTTCAAGCTCT]); P02R (antisense, PAR3 bp 140-116; [5′-3′, ATGACCTGAGTCCCGTCTCTTAAAC]). The starting point for physical mapping was a size-selected cosmid library generated from the approximately 1.4-megabase YAC 798D11, known to contain all 3 PARgenes.8 Library screening was completed using the [32]P-radiolabeled PAR1, PAR3, or IQGAP2 cDNAs as probes,7,8 and inversion field gel electrophoresis for the separation of large genomic fragments was completed as previously described.8 A 162-bp IQGAP2 exon 12-specific probe was obtained by PCR using oligonucleotide primers G18F and G14R (antisense, IQGAP2 bp 1742-1719, (5′-3′: CCGAGTTTCTGTGATTTAGCATGG]). DNA sequence analysis of PCR or cloned fragments was completed by dideoxy chain termination.
Platelet preparation and immunoprecipitation analyses
Human platelets were obtained from healthy volunteers who were taking no medications for 1 week before phlebotomy. After a 2-mL draw-off, blood obtained by venipuncture was diluted into Aster-Jandl anticoagulant,9 and platelet-rich plasma (PRP) was isolated by centrifugation at 200g for 10 minutes at 22°C. Gel-filtered platelets (GFPs) were isolated by passing PRP supplemented with 0.1 μM prostaglandin E1(PGE1) over Sepharose 2B (Pharmacia LKB, Piscataway, NJ) and eluting with HEPES (N-2-hydroxylethylpiperazine-N'-2-ethanesulfonic acid)–buffered modified Tyrode (HBMT) supplemented with 2 mM MgCl2 to a final concentration of approximately 2.5 × 108platelets/mL HBMT (138 mM NaCl, 2.7 mM KCl, 0.4 mM NaH2PO4, 12 mM NaHCO3, 0.2% bovine serum albumin, 0.1% dextrose, and 10 mM HEPES, pH 7.4). Before activation, platelets were rested for 30 minutes at 37°C to optimize quiescence. For immunoprecipitation studies, platelets (200 μL, 5 × 107 platelets) were activated (without stirring) using 10 nM human α-thrombin (approximately 3500 U/mg; 10 nM approximates 1 U/mL; kindly supplied by Dr J. Jesty, Stony Brook, NY), 20 μM PAR1 activating peptide PAR142-47(SFLLRN),8 10 μg/mL fibrillar collagen (Hormon-Chemie), or 10 μM adenosine 5′-diphosphate (ADP; Sigma), for some experiments preincubated with 10 μM cytochalasin D at 25°C for 10 minutes before activation. For experiments using PRP, platelets were preincubated with 3 mM Gly-Pro-Arg-Pro (GPRP) peptide (Sigma) to inhibit fibrin polymerization. After activation, platelets were immediately solubilized in RIPA buffer (50 mM Tris [tris(hydroxymethyl)aminomethane], 150 mM NaCl, 1 mM EGTA [ethyleneglycotetraacetic acid], 0.25% sodium deoxycholate,1% igepal,1 mM NaF,1 mM orthosodium vanadate, pH 7.4) supplemented with 5 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/mL leupeptin and 10 μg/mL aprotinin, and 200 μL aliquots of the cytoplasmic (soluble) fractions were precleared with a 50 μL slurry of protein G agarose beads, followed by incubation with 4 μg anti-IQGAP2 IgG (Upstate Biotechnology) or preimmune isotypic IgG control at 4°C for 16 hours. Protein samples were affinity-purified using 50 μL protein G agarose beads, washed 3 times with cold RIPA buffer supplemented with 5 mM MgCl2, and eluted into sodium dodecyl sulfate (SDS) loading buffer for analysis by 4% to 15% gradient SDS–polyacrylamide gel electrophoresis (PAGE). Immunoblotting was completed using 1:100 murine anti-rac1 (BD Biosciences Transduction Laboratories), 1:100 murine anti-cdc42 (BD Biosciences Transduction Laboratories), 1:500 rabbit anti-arp3a, 1:500 rabbit anti-arp2 (anti-arp antibodies kindly provided by Dr L. Machesky, United Kingdom), 1:2500 anti-gelsolin (BD Biosciences Transduction Laboratories), 1:500 anti-WASp IgG (Upstate Biotechnology) or 1:1000 anti-actin antibodies (Sigma), and the species-specific horseradish peroxidase-tagged secondary antibody (1:10 000) by enhanced chemiluminescence (ECL; Amersham LifeSciences). For the detection of IQGAP2, samples were loaded in reducing buffer supplemented with 10% β-mercaptoethanol, whereas detection of all other proteins was completed under nonreducing conditions. Platelet cytoskeletons were isolated by solubilization of quiescent or activated platelets in 1% Triton-X, 20 mM HEPES (pH 7.5), 5 mM EDTA (ethylenediaminetetraacetic acid), 5 mM EGTA, and 5 mM MgCl2, followed by centrifugation at 100 000g for 20 minutes at 25°C. Pellets were resuspended in 1.6% SDS/3 mM N-ethylmaleimide for SDS-PAGE and immunoblot analysis. Densitometric analyses were completed using Gel-Pro Analyzer software (Media Cybernetics), and results were expressed as relative changes (fold increase) in integrated optical densities within the linear range of determination. Equivalent loading was confirmed by parallel Coomassie blue staining.
The F-actin content for individual experiments was monitored by parallel determination using flow cytometry.9 In brief, aliquots of GFPs or PRP (stimulated or unstimulated) were fixed in 3.7% formaldehyde followed by incubation with 30 U/mL fluorescein isothiocyanate (FITC)–phalloidin (Molecular Probes) on a rotator for 60 minutes at 25°C. After washing with phosphate-buffered saline (PBS), the fluorescent geometric means of 1 × 104platelets were determined by fluorescence-activated cell sorter (FACS), and results at individual time points were expressed as the ratio to unstimulated labeled platelets.
Complex assembly and activation state of GTPases
GTP-charged complex assembly was completed using a bacterially expressed glutathione-S-transferase (GST) chimer containing a C-terminal PAK1 binding domain (PBD) sequence from the p21-activated kinase (p21PAK, PAK1).10 Commercially available GST-PBD beads with a capacity of 0.5 μg/μL (Upstate Biotechnology) were used interchangeably with a functionally similar product generated from pGEX2TK-PBD or mutant GST-PBD incapable of binding GTP-bound forms of rac1 or cdc42 (PGEX2TK-PBDL83L86) (both kindly provided by Dr L. van Aelst, Cold Spring Harbor Laboratory, NY). Plasmids expressing GST-PBD, mutant GST-PBD, or wild-type GST (pGEX2X) were used for recombinant protein generation in Escherichia coli host strain BL21 with glutathione Sepharose beads essentially as previously described10 11 and were used within 24 hours of preparation. Preparations were standardized using bicinchoninic acid protein determination assays (Pierce) and were confirmed by SDS-PAGE using albumin standards. Platelets (5 × 107) resuspended in 200 μL HBMT/2 mM MgCl2 were activated with individual agonists and immediately solubilized in ice-cold 4 × RIPA/8 mM MgCl2 (supplemented with protease inhibitors outlined above), followed by gentle rocking for 60 minutes at 4°C in the presence of 5 μg GST-PBD, GST, or mutant GST-PBD beads. After 3 washes in ice-cold 1 × RIPA buffer, samples were analyzed by SDS-PAGE and immunoblot analysis. For standardization, equivalent aliquots of unstimulated platelets were lysed in the identical RIPA buffer supplemented with 1 mM dithiothreitol and 1 mM MgCl2 and were loaded with either 100 μM GTPγS or 100 μM GDPβS (Sigma) for 60 minutes at 25°C, before affinity purification onto GST-PBD.
GST-PBD specificity assays for some experiments were completed by COS1 transfections (1 × 107 cells) using dominant-negative rac1N17 and cdc42N17 or constitutively active rac1V12 and cdc42V12 mutants (kindly provided by Dr D. Bar-Sagi, Stony Brook, NY). At 72 hours, cells were directly solubilized and incubated with 5 μg GST-PBD in RIPA buffer as outlined above, followed by SDS-PAGE and immunoblot analysis using rac1 or cdc42 antibodies.
Platelet immunofluorescence
GFP resuspended in TSE (10 mM Tris [pH 7.4], 150 mM NaCl, 10 mM EDTA) were immediately fixed in 2% paraformaldehyde (PFA)–PBS followed by centrifugation at 1500g onto glass slides (unactivated) or were directly centrifuged onto glass slides at 1500g before fixation using 2% PFA/PBS (activated). Agonist-activated platelets were stimulated in suspension using 10 nM thrombin or 20 μM PAR142-47 for 60 seconds, followed by immediate fixation and centrifugation onto glass slides. Cells were permeabilized using 0.1% Triton/HS (20 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol), blocked by washing 3 times with 0.1% bovine serum albumin (BSA)–PBS at 25°C, and stained using anti-IQGAP2 (1:50) for 60 minutes at 25°C, followed by FITC-conjugated antimouse IgG (1:500) (Jackson Immunolabs) for 60 minutes at 25°C. Fluorescent images were obtained using a Nikon Diaphot inverted confocal microscope equipped with fluorescence optics.
Transfection studies and immunofluorescence analysis
COS1 cells were grown in Dulbecco minimal essential medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin sulfate. Transient transfections with full-length IQGAP2 cDNA7 were completed using diethylaminoethyl (DEAE)–dextran. Immunofluorescence staining was completed 48 to 72 hours after transfection in 8-well chamber slides after fixation and permeabilization using ice-cold 100% acetone for 60 seconds. After a 16-hour blocking step at 4°C using PBS/0.1% BSA, cells were stained for 60 minutes at 25°C with murine anti-IQGAP2 (1:100), followed by immunofluorescence detection for 60 minutes at 25°C using 1:750 dilution of the FITC-conjugated antimouse antibody (Jackson Immunolabs). Parallel immunostaining demonstrated no cross-reactivity or fluorescence bleed-through of the species-specific secondary antibodies. F-actin staining was completed using 66 nM Alexa Fluor 568 phalloidin (Molecular Probes) for 60 minutes at 25°C.
Results
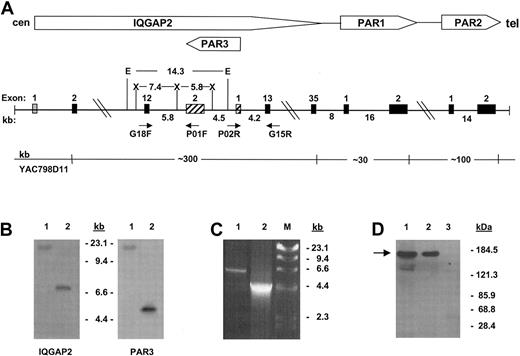
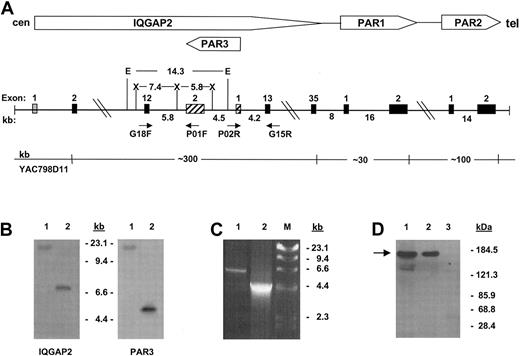
The gene encoding IQGAP2 is identified within the PAR gene cluster. α-Thrombin is known to cleave 3 G-protein–coupled protease-activated receptors (PAR1, PAR3, and PAR4),8,12-14 though current evidence suggests that human platelets signal through a dual-receptor thrombin activation system involving PAR1 and PAR4,15 with ancillary or modulatory roles for platelet glycoproteins Ibα16 and V.17 Previous work from this laboratory had identified a discrete PAR gene cluster at 5q13 encompassing PARs 1 to 3.8 Initial genomic scanning of this region identified the putative rac1/cdc42 effector protein IQGAP2 within the PAR gene cluster, uniquely situated proximate to the PAR1 and PAR3 genes. Nucleotide sequence analysis using Celera Genomics and the publicly funded international Human Genome Project (HGP) databases placed humanPAR3 within the twelfth intron of the large approximately 300-kb IQGAP2 gene, transcribed 5′→ 3′ off the negative strand.18,19 Because this region of the genome continues to contain gaps, and to definitively resolve previously suggested gene ordering discrepancies between large-scale assembly efforts and those elucidated by radiation hybrid mapping,20 more extensive characterization was completed using cosmid clones isolated from YAC798D11, known to encompass all 3 PARs.8 Physical mapping confirmed that both PAR3 and IQGAP2 exon 12 were contained not only within identical approximately 100-kb genomic fragments,8 but also within a smaller approximately 14-kb EcoRI fragment (Figure 1A-B). Long-range PCR and partial sequence analysis of cosmid clone 3-2 (identified as encompassing the entire PAR3 gene) confirmed these genomic contextual relationships (Figure 1C). Thus, these data support those reported by both genomic assembly groups and firmly establish a definitiveIQGAP2/PAR3 gene structure and organization as outlined in Figure 1.
Characterization of platelet IQGAP2.
(A) Schema of the IQGAP2/PAR gene cluster integrating consensus data from Celera18 and public databases19 with those generated by PCR, restriction analysis, and sequence analysis. Approximate distances in kilobases are outlined, as are selected restriction enzymes used for genomic mapping (E indicatesEcoRI; X, XbaI). Exons are depicted as solid boxes, except for IQGAP2 exon 1 (fine stipples), which has not been identified within either database, and PAR3 exons (cross-hatches) (cen, centromere; tel, telomere). (B) Southern blot analysis using 100 ng YAC798D11 DNA and an IQGAP2 exon 12-specific probe or the PAR3 cDNA as probe. Lane 1 isEcoRI-digested, and lane 2 is XbaI-digested. Note that PAR3 exon 2 and IQGAP2 exon 12 are contained within the identical EcoRI fragment. (C) PCR using 100 ng cosmid 3-2 DNA and primer pair G18F/P01F (lane 1) or P02R/G15R (lane 2). DNA sequence analysis confirmed the identity of the PCR fragments. Fragment sizes corresponding to HindIII-digested bacteriophage λ DNA are shown in panels B and C. (D) Platelet immunoprecipitations were completed using 1 × 108RIPA-solubilized gel-filtered platelets (GFP), and anti-IQGAP2 monoclonal antibody (mAb) (lane 2) or preimmune mouse IgG (lane 3), followed by immunoblot analysis using the IQGAP2-specific mAb (arrow). The corresponding SDS-solubilized GFP lysate is shown in lane 1.
Characterization of platelet IQGAP2.
(A) Schema of the IQGAP2/PAR gene cluster integrating consensus data from Celera18 and public databases19 with those generated by PCR, restriction analysis, and sequence analysis. Approximate distances in kilobases are outlined, as are selected restriction enzymes used for genomic mapping (E indicatesEcoRI; X, XbaI). Exons are depicted as solid boxes, except for IQGAP2 exon 1 (fine stipples), which has not been identified within either database, and PAR3 exons (cross-hatches) (cen, centromere; tel, telomere). (B) Southern blot analysis using 100 ng YAC798D11 DNA and an IQGAP2 exon 12-specific probe or the PAR3 cDNA as probe. Lane 1 isEcoRI-digested, and lane 2 is XbaI-digested. Note that PAR3 exon 2 and IQGAP2 exon 12 are contained within the identical EcoRI fragment. (C) PCR using 100 ng cosmid 3-2 DNA and primer pair G18F/P01F (lane 1) or P02R/G15R (lane 2). DNA sequence analysis confirmed the identity of the PCR fragments. Fragment sizes corresponding to HindIII-digested bacteriophage λ DNA are shown in panels B and C. (D) Platelet immunoprecipitations were completed using 1 × 108RIPA-solubilized gel-filtered platelets (GFP), and anti-IQGAP2 monoclonal antibody (mAb) (lane 2) or preimmune mouse IgG (lane 3), followed by immunoblot analysis using the IQGAP2-specific mAb (arrow). The corresponding SDS-solubilized GFP lysate is shown in lane 1.
Characterization of platelet IQGAP2
IQGAP2 was initially classified as a liver-specific GTPase activating protein (GAP) because its primary translation product predicted the presence of a RasGAP-GTPase binding domain (GBD) found in the orthologous Schizosaccharomyces pombe sar1 gene.7 Nonetheless, IQGAP2 lacks inherent GAP activity toward Ras or other GTPases in vitro, presumably related to lack of an arginine finger residue within its GBD known to be essential for GAP activity.21 Rather, IQGAP2 interacts with GDP- or GTP-bound rac1 and cdc42 (but not RhoA) with comparable affinities in vitro,7 though its function remained largely enigmatic. The colocalization of IQGAP2 with the thrombin receptors PAR1 and PAR3—coupled with thrombin's ability to activate an overlapping subset of Rho GTPases—prompted subsequent investigations into a potential role for IQGAP2 in platelet-activation events. Northern analyses using megakaryocyte-like HEL cells demonstrated the identical approximately 6.5-kb IQGAP2 transcript as that found in HepG2 cells, and RT-PCR demonstrated IQGAP2 mRNA in HEL cells and human platelets (not shown). These transcript analyses were consistent with results of protein expression studies that demonstrated a single approximately 180-kDa protein in HEL and platelet lysates analyzed under reducing and nonreducing conditions and identical to the immunoreactive species seen in HepG2 cell lysates7; furthermore, an anti-IQGAP2 IgG specifically immunoprecipitated the protein from platelet lysates (Figure 1D).
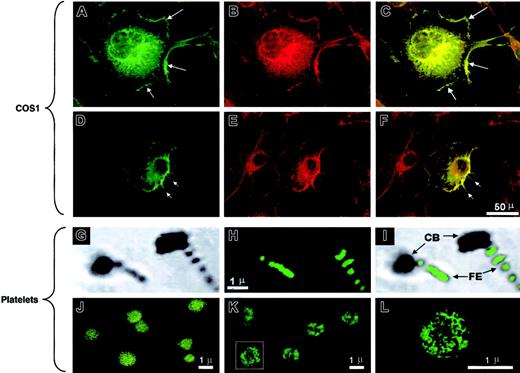
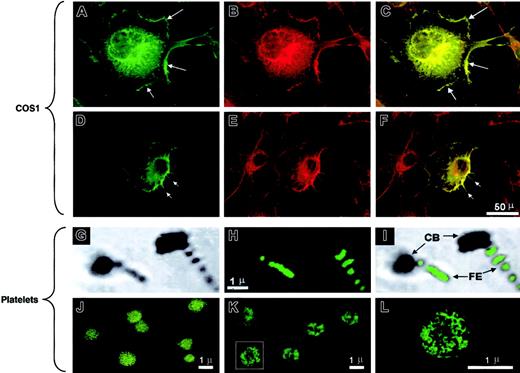
Quiescent gel-filtered platelets demonstrated diffuse cytoplasmic IQGAP2 staining without a clear cell-surface expression pattern, results that were dramatically different from those seen using glass- or thrombin-activated platelets. Gentle cytocentrifugation of platelets onto glass slides results in platelet activation with the generation of lamellae and filopodia. In these cells, IQGAP2 staining was most readily identifiable in platelet filopodia, with less prominent staining evident in the contractile platelet body (Figure 2G-I). Furthermore, selective activation using thrombin or PAR142-47 demonstrated a pattern most consistent with cytoskeletal IQGAP2 translocation (Figure 2J-L).
IQGAP2 interacts with actin and arp2/3
The pattern of IQGAP2 redistribution in activated platelets suggested that IQGAP2 might have a physiologic role in cytoskeletal actin assembly, consistent with its predicted domain structure that encompasses an N-terminal actin-binding calponin homology (CH) domain.22 Transient transfection of a full-length IQGAP2 cDNA in COS1 cells demonstrated diffuse cytoplasmic staining and intense cell-surface membrane staining 48 hours after transfection. Simultaneous phalloidin staining demonstrated that IQGAP2 colocalized with filamentous (F)–actin in both lamellipodia and filopodia (Figure 2A-F). Thus, the patterns of IQGAP2 expression mirrored those expected for a rac1/cdc42 effector protein—that is, expression appeared localized to areas of filopodial/lamellipodial extensions (demonstrable in platelets and transfected COS1 cells). These patterns are consistent with the previously identified GTPase specificity as determined by in vitro nitrocellulose pull-down assays,7 and they suggested a function for IQGAP2 in cytoskeleton actin assembly similar to that previously described for its closely related homologue, IQGAP1.23
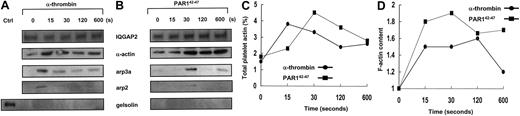
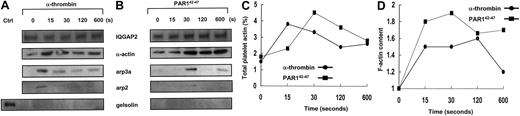
Actin assembly is initiated on the exposure of actin filament barbed ends, occurring by uncapping of barbed ends of preexisting filaments, by severing filaments, or by de novo nucleation.24,25 The arp2/3 complex is a 7-member protein complex found in human platelets in nearly equivalent stoichiometric ratios, and it is the only known cellular factor that nucleates the formation of actin filaments.26 Arp2/3 promotes the addition of actin monomers to the barbed ends of actin filaments,26 a reaction that requires a scaffolding protein such as the Wiskott-Aldrich syndrome protein (WASp)27,28 or theListeria monocytogenes ActA protein.29 To determine whether IQGAP2 scaffolding functions involved biochemical interactions with F-actin and arp2/3 in a physiologic platelet model system, confirmatory association studies were completed using either gel-filtered platelets or platelet-rich plasma, with essentially identical results (Figure3). Platelet activation with α-thrombin was followed by IQGAP2 immunoprecipitation and subsequent immunodetection using α-actin and arp2/3-specific antibodies. In most experiments α-actin was identified as part of an IQGAP2/actin complex, presumably related to baseline actin binding through IQGAP2 CH domains. On thrombin stimulation, however, F-actin and arp2/3 were rapidly and reversibly recruited to the IQGAP2 complex in a time frame typical of that seen in platelet aggregation—recruitment was evident at 15 seconds, followed by a slow decline to baseline. In quiescent platelets, there was no evidence of IQGAP2—arp2/3 protein interactions—despite the presence of an IQGAP2/actin complex—suggesting that the association of arp2/3 with IQGAP2 was not solely dependent on its binding to actin. Furthermore, neither WASp (not shown) nor another actin-binding protein (gelsolin) was detected in the immunoprecipitated complex, suggesting that the complex was not related to nonspecific protein trapping within a fibrin/actin meshwork. These initial observations were confirmed by specifically activating PAR1 using PAR142-47 (bypassing thrombin-induced fibrin polymerization), with essentially similar results except for a delay in actin and arp2/3 recruitment (30 seconds). Finally, there was no change in the cytosolic concentration of IQGAP2, suggesting that the mechanism of assembly was most likely related to an IQGAP2 conformational change.
IQGAP2 cellular localization.
(A-F) COS1 cells transfected with the IQGAP2 cDNA were fixed and permeabilized, followed by confocal microscopic detection of F-actin (B,E) or IQGAP2 (A,D). The overlay images (C,F) demonstrate colocalization (yellow) readily observed in the lamellipodia (A,C arrows) or filopodia (D,F arrows). Note that the IQGAP2/actin colocalization pattern reflects that expected for rac1/cdc42-mediated filopodial/lamellipodial structures. (G-I) Platelets activated by cytocentrifugation onto glass slides were stained with the anti-IQGAP2 mAb (H), with the corresponding phase-contrast image displayed in panel G and the phase/overlay image in panel I. Note that IQGAP2 expression is essentially restricted to platelet filopodial extensions (FE), to the exclusion of the platelet contractile bodies (CB). (J-L) Unactivated platelets (J) or thrombin-activated platelets (K,L) were stained with anti-IQGAP2 mAb after fixation and permeabilization. Note the redistribution of IQGAP2 within the platelet cytoskeleton in thrombin-stimulated platelets (panel L is a higher magnification of the boxed platelet in panel K); identical results were seen using PAR142-47-stimulated platelets (not shown). Scale bars for corresponding COS1 or platelet immunofluorescence staining are shown.
IQGAP2 cellular localization.
(A-F) COS1 cells transfected with the IQGAP2 cDNA were fixed and permeabilized, followed by confocal microscopic detection of F-actin (B,E) or IQGAP2 (A,D). The overlay images (C,F) demonstrate colocalization (yellow) readily observed in the lamellipodia (A,C arrows) or filopodia (D,F arrows). Note that the IQGAP2/actin colocalization pattern reflects that expected for rac1/cdc42-mediated filopodial/lamellipodial structures. (G-I) Platelets activated by cytocentrifugation onto glass slides were stained with the anti-IQGAP2 mAb (H), with the corresponding phase-contrast image displayed in panel G and the phase/overlay image in panel I. Note that IQGAP2 expression is essentially restricted to platelet filopodial extensions (FE), to the exclusion of the platelet contractile bodies (CB). (J-L) Unactivated platelets (J) or thrombin-activated platelets (K,L) were stained with anti-IQGAP2 mAb after fixation and permeabilization. Note the redistribution of IQGAP2 within the platelet cytoskeleton in thrombin-stimulated platelets (panel L is a higher magnification of the boxed platelet in panel K); identical results were seen using PAR142-47-stimulated platelets (not shown). Scale bars for corresponding COS1 or platelet immunofluorescence staining are shown.
Thrombin receptor-dependent assembly of the IQGAP2/arp2/3-actin scaffold.
(A-B) PRP (5 × 107 platelets/lane corresponding to 100 μg) supplemented with 3 mM GPRP (thrombin activation only) were stimulated with 10 nM α-thrombin (1 U/mL) (A) or 20 μM PAR142-47 (B) for indicated time points, and total soluble lysates were immunoprecipitated with the anti-IQGAP2 mAb, followed by immunoblot detection for IQGAP2, arp3a, arp2, actin, and gelsolin. Note the immediate and specific assembly of arp2/3 and actin without any evidence for gelsolin involvement (for PAR1,42-47 arp2 was maximally detected at 30 seconds with low-level detection at 120 and 600 seconds). Ctrl indicates 5 μg SDS-solubilized platelet lysate control. Comparable results were seen using GFP solubilized in HBMT/1% Triton-X (not shown). (C) Percentage of platelet actin immunoprecipitated at individual time points was determined by densitometric comparison to 5 μg SDS-solubilized platelets. (D) F-actin content was monitored as described in “Materials and methods.” Results represent one complete set of experiments duplicated on 2 occasions.
Thrombin receptor-dependent assembly of the IQGAP2/arp2/3-actin scaffold.
(A-B) PRP (5 × 107 platelets/lane corresponding to 100 μg) supplemented with 3 mM GPRP (thrombin activation only) were stimulated with 10 nM α-thrombin (1 U/mL) (A) or 20 μM PAR142-47 (B) for indicated time points, and total soluble lysates were immunoprecipitated with the anti-IQGAP2 mAb, followed by immunoblot detection for IQGAP2, arp3a, arp2, actin, and gelsolin. Note the immediate and specific assembly of arp2/3 and actin without any evidence for gelsolin involvement (for PAR1,42-47 arp2 was maximally detected at 30 seconds with low-level detection at 120 and 600 seconds). Ctrl indicates 5 μg SDS-solubilized platelet lysate control. Comparable results were seen using GFP solubilized in HBMT/1% Triton-X (not shown). (C) Percentage of platelet actin immunoprecipitated at individual time points was determined by densitometric comparison to 5 μg SDS-solubilized platelets. (D) F-actin content was monitored as described in “Materials and methods.” Results represent one complete set of experiments duplicated on 2 occasions.
IQGAP2 scaffolding functions require [GTP]rac1/cdc42
Although IQGAP2 has been shown to bind rac1 and cdc42 using in vitro nitrocellulose binding assays, the functional role(s) of such associations in a primary cell (such as platelets) endogenously expressing all these proteins remains unknown. Human platelets contain approximately 2 to 3 μM rac1 and 0.2 to 0.3 μM cdc42,9both of which are primarily (more than 90%) found in the cytosolic fractions of quiescent platelets.9,30 In unstimulated platelets, IQGAP2-specific immunoprecipitation studies demonstrated that cytosolic IQGAP2 was complexed to rac1 but not to cdc42, presumably related to the relative concentrations of these 2 GTPases in human platelets.9 Reciprocal immunoprecipitation analyses using either anti-rac1 or anti-cdc42 antibodies confirmed these initial observations (data not shown). Despite evidence for this IQGAP2/rac1 complex in quiescent platelets, most (more than 90%) of rac1 appeared to be free of IQGAP2, as demonstrated by immunoblot analysis of the post-IQGAP2 immunoprecipitate sample. In parallel experiments using thrombin-stimulated platelets, immunoprecipitation studies failed to demonstrate increased rac1 or cdc42 recruitment to the cytoplasmic IQGAP2–arp2/3-actin complex (not shown). Because IQGAP2 associates with GDP- and GTP-bound GTPases,7 this lack of quantitative difference could be explained by the exchange of GDP- for GTP-bound rac1/cdc42, with no total change in overall stoichiometric associations.
To more specifically dissect the molecular mechanisms whereby rac1 and cdc42 could effect IQGAP2-associated arp2/3-actin complex assembly, the activation state of bound GTPases was studied using a recombinantly expressed PAK1 binding domain (PBD). This PBD, containing a highly conserved 14–amino acid cdc42/rac1 interactive binding region (CRIB) sequence,31 specifically binds and distinguishes active (GTP-bound) from inactive (GDP-bound) forms of rac1 and cdc42,10 and it has been used as a sensitive means of demonstrating GTPase charging in thrombin-stimulated platelets9 or formyl-methionyleucylphenylalanine (fMLP)–activated neutrophils.10 To determine whether PBD could be used to study IQGAP2/GTPase interactions, we cotransfected IQGAP2 with dominant-negative rac1N17 and cdc42N17 or constitutively active rac1V12 and cdc42V12 mutants into COS1 cells, followed by affinity purification using GST-PBD agarose beads (or GST beads alone). As shown in Figure 4A, IQGAP2 was specifically identified only in the presence of [GTP]rac1 or [GTP]cdc42, suggesting that the PBD could specifically bridge a [GTP]GTPase/IQGAP2 complex. Although the efficiency for this interaction was no more than 5% of cellular IQGAP2, it suggested a means of discriminating between the activation-dependent states of IQGAP2/GTPase interactions.
GTP-dependent assembly of an IQGAP2/arp2/3-actin scaffold.
(A) The ability of GST-PBD to specifically bind IQGAP2 was determined by cDNA cotransfection studies in COS1 cells using IQGAP2 and constitutively active rac1V12 (lane 1) or cdc42V12 (lane 3) or the dominant-negative rac1N17 (lane 2) or cdc42N17 (lane 4) mutants, followed by bead-affinity assays and immunoblot analysis with anti-IQGAP2 or anti-GTPase antibodies. GST-conjugated agarose beads lacking the PBD domain failed to precipitate [GTP]rac1/IQGAP2 or [GTP]cdc42/IQGAP2 complex (not shown). (B) GFPs were resuspended to 2.5 × 108/mL, and 200-μL aliquots were activated with individual agonists for various time points as outlined. Samples were then solubilized in RIPA (final concentration, 100 μg/sample), incubated with GST-PBD, and evaluated by SDS-PAGE and immunoblot analysis for the presence of IQGAP2, arp3a, arp2, or actin. (C) F-actin content determinations for individual agonists; F-actin content is inhibited in thrombin-stimulated platelets preincubated with 10 μM cytochalasin D (Cyt D). (D) Percentage (of total) of individual proteins identified within the thrombin-induced IQGAP2 scaffold was determined by densitometric comparison to 5 μg total platelet lysates, adjusted for nonspecific binding using parallel samples incubated with equivalent concentrations of GST control beads. Note that neither gelsolin nor WASp is identified within these complexes and that preincubation of platelets with 10 μM cytochalasin D abrogates thrombin-induced IQGAP2/actin association. Results are representative of one complete set of experiments completed on 2 separate occasions from 2 different healthy volunteers.
GTP-dependent assembly of an IQGAP2/arp2/3-actin scaffold.
(A) The ability of GST-PBD to specifically bind IQGAP2 was determined by cDNA cotransfection studies in COS1 cells using IQGAP2 and constitutively active rac1V12 (lane 1) or cdc42V12 (lane 3) or the dominant-negative rac1N17 (lane 2) or cdc42N17 (lane 4) mutants, followed by bead-affinity assays and immunoblot analysis with anti-IQGAP2 or anti-GTPase antibodies. GST-conjugated agarose beads lacking the PBD domain failed to precipitate [GTP]rac1/IQGAP2 or [GTP]cdc42/IQGAP2 complex (not shown). (B) GFPs were resuspended to 2.5 × 108/mL, and 200-μL aliquots were activated with individual agonists for various time points as outlined. Samples were then solubilized in RIPA (final concentration, 100 μg/sample), incubated with GST-PBD, and evaluated by SDS-PAGE and immunoblot analysis for the presence of IQGAP2, arp3a, arp2, or actin. (C) F-actin content determinations for individual agonists; F-actin content is inhibited in thrombin-stimulated platelets preincubated with 10 μM cytochalasin D (Cyt D). (D) Percentage (of total) of individual proteins identified within the thrombin-induced IQGAP2 scaffold was determined by densitometric comparison to 5 μg total platelet lysates, adjusted for nonspecific binding using parallel samples incubated with equivalent concentrations of GST control beads. Note that neither gelsolin nor WASp is identified within these complexes and that preincubation of platelets with 10 μM cytochalasin D abrogates thrombin-induced IQGAP2/actin association. Results are representative of one complete set of experiments completed on 2 separate occasions from 2 different healthy volunteers.
Pull-down assays of thrombin-stimulated platelets using GST-PBD agarose beads were then pursued, with end points focusing on the ability of IQGAP2 to interact with actin and arp2/3. Consistent with the evidence for biochemical interactions outlined above, thrombin stimulation again demonstrated assembly of the entire IQGAP2/arp2/3-actin complex in a time-course essentially identical to that seen using IQGAP2-specific immunoprecipitations. The complex was assembled within 15 seconds, with fall-off 60 seconds after agonist stimulation (Figure 4B). Minimal to no complex assembly was evident using mutant GST-PBD or GST beads alone (not shown), and the assay failed to pull-down the [GTP]cdc42 effector protein WASp or gelsolin, again supporting the IQGAP2-dependent specificity of the complex assembly scaffold. Furthermore, preincubation of platelets with cytochalasin D abrogated thrombin-induced IQGAP2/actin interaction and F-actin content (Figure 4C-D). Maximal IQGAP2 association was evident at the most immediate time point, with subsequent fall-off by 60 seconds, and entirely consistent with the rapid movement of IQGAP2 into the cytoskeletal fraction (see below). Although low-level IQGAP2 remained evident at 60 seconds and beyond, as demonstrated by long exposure, arp2/3 and actin were found at disproportionately higher concentrations. At this point, we could not exclude the possibility that another, as yet uncharacterized, rac1/cdc42 effector protein was a component of the scaffolding complex (though it did not appear to be platelet WASp).
IQGAP2 scaffolding functions are α-thrombin restricted
Physiologic platelet activation is triggered by a variety of endogenous stimulants, including epinephrine, fibrillar collagen, thromboxane A2, and ADP. To further dissect the agonist specificity of the IQGAP2-mediated arp2/3-actin assembly complex, assays were repeated using collagen and ADP. Platelet adhesion to subendothelial collagen is mediated by binding to the glycoprotein Ia/IIa (α2β1) receptor complex,32 though activation is mediated by intracellular signaling through a platelet glycoprotein VI/FcRγ-chain receptor tyrosine kinase complex.33 ADP-induced platelet aggregation is mediated through the activation of PY21 and PY212 purinergic receptors.34 Unexpectedly, the ability to assemble the complex was uniquely agonist dependent, with no evidence that fully aggregative doses of either fibrillar collagen (10 μg/mL) or ADP (10 μM) could effect IQGAP2/arp2/3-actin complex formation (Figure 4B).
Divergent IQGAP2 effector pathways mediated by [GTP]rac1 or [GTP]cdc42 binding
Given that these assays do not distinguish [GTP]rac1-from [GTP]cdc42-induced complex assembly, dissection of the GTPases regulating agonist-dependent IQGAP2 scaffold formation was pursued in greater detail. Consistent with the results outlined in Figure 4, complex assembly correlated with GTP-charging capabilities of individual agonists, demonstrating changes restricted to thrombin-stimulated platelets (Figure 5). Interestingly, rac1 and cdc42 demonstrated distinctly divergent responses on thrombin stimulation. The cytosolic concentration of [GTP]rac1 increased substantially (maximally accounting for approximately 45% of cellular [GTP]rac1), first evident by 5 seconds and reaching peak concentrations at 15 seconds, with rapid subsequent decay. In contrast, cytosolic concentrations of [GTP]cdc42 rapidly decreased and became asymptotically undetectable beyond 30 seconds. These initial responses were clearly evident at 2 seconds (preceding [GTP]rac1 charging) and were consistent with the previously described rapid translocation of nearly 30% of [GTP]cdc42 to the platelet actin cytoskeleton.9 30 We hypothesized that if the activation of IQGAP2 required the binding of [GTP]rac1 or [GTP]cdc42, its translocation to the actin cytoskeleton would only be evident in thrombin-stimulated platelets. This was confirmed by isolating the Triton-X insoluble actin cytoskeletons of agonist-treated platelets and probing for the presence of IQGAP2 (Figure 5C). The rapid translocation of IQGAP2 was readily seen only on thrombin stimulation and as evaluated by parallel studies using total platelet lysates, estimated to account for approximately 20% of total cellular IQGAP2. Neither collagen- nor ADP-stimulated platelets demonstrated IQGAP2 cytoskeletal translocation, even with extension of the experiments to 5 minutes to ensure maximal platelet activation.
Agonist-dependent changes in [GTP]rac1, [GTP]cdc42, and cytoskeletal IQGAP2.
(A-B) GFP (5 × 107 platelets/lane; 100 μg starting protein) were preincubated with GST-PBD and stimulated with 10 nM α-thrombin, 10 μg/mL collagen, or 10 μM ADP for various time points, followed by lysis and immunoblot analysis for the presence of GTP[rac1] (upper panel) or GTP[cdc42] (middle panel). Data are presented as percentage of [GTP]-bound GTPase determined by densitometric comparison with 100 μg platelet lysates loaded with saturating concentrations (100 μM) of GTPγS or GDPβS (lanes marked accordingly). Nonspecific binding by GST-PBD was adjusted for by subtracting the “background” GDPβS component. Note the rapid increase of cytosolic GTP[rac1] with a concomitant decrease in [GTP]cdc42, demonstrable only in α-thrombin-stimulated platelets. (C) Triton-X insoluble cytoskeletons from agonist-stimulated platelet (8.5 × 106 platelets/lane corresponding to 17 μg total protein) were immunoblotted for the presence of IQGAP2, and data were reported as the percentage of total platelet IQGAP2 determined by densitometric comparison to platelet control lysates. F-actin contents were essentially as outlined in Figure 4. All results are representative of one complete set of experiments, repeated on 2 occasions with 2 healthy volunteers.
Agonist-dependent changes in [GTP]rac1, [GTP]cdc42, and cytoskeletal IQGAP2.
(A-B) GFP (5 × 107 platelets/lane; 100 μg starting protein) were preincubated with GST-PBD and stimulated with 10 nM α-thrombin, 10 μg/mL collagen, or 10 μM ADP for various time points, followed by lysis and immunoblot analysis for the presence of GTP[rac1] (upper panel) or GTP[cdc42] (middle panel). Data are presented as percentage of [GTP]-bound GTPase determined by densitometric comparison with 100 μg platelet lysates loaded with saturating concentrations (100 μM) of GTPγS or GDPβS (lanes marked accordingly). Nonspecific binding by GST-PBD was adjusted for by subtracting the “background” GDPβS component. Note the rapid increase of cytosolic GTP[rac1] with a concomitant decrease in [GTP]cdc42, demonstrable only in α-thrombin-stimulated platelets. (C) Triton-X insoluble cytoskeletons from agonist-stimulated platelet (8.5 × 106 platelets/lane corresponding to 17 μg total protein) were immunoblotted for the presence of IQGAP2, and data were reported as the percentage of total platelet IQGAP2 determined by densitometric comparison to platelet control lysates. F-actin contents were essentially as outlined in Figure 4. All results are representative of one complete set of experiments, repeated on 2 occasions with 2 healthy volunteers.
Discussion
A functional transcriptosome composed of its receptor(s) and intracellular effector protein has been identified that uniquely regulates an agonist-specific human platelet activation pathway. Recent evidence35 from human genomic analyses has identified discrete chromosomal regions that demonstrate clustered expression patterns in healthy and malignant tissues. Such patterns of expression were suggested as a means of identifying novel genes that may be differentially associated with a particular (malignant) phenotype, but developmental functional relationships within these regions were not anticipated. Computational genomic approaches have suggested that so-called gene adjacencies may be used to predict functional coupling, though the model systems for such contextual-based approaches are best exemplified in prokaryotes.36,37 To our knowledge, this is the first example of such a functional relationship in humans, raising the possibility that comparably developed computational methods may be adapted for human genomic analysis. Although IQGAP2 is contextually adjacent to 2 thrombin receptors, it remains unestablished if its scaffolding functions are achievable through the activation of all 3 PARs individually, only the contiguous PAR1 and PAR3, or the dual PAR1/PAR4 receptor system evident in human platelets.8,12 14
Previous observations have outlined distinct roles for WASp25 and Scar38 as endogenous effectors of actin polymerization mediated through arp2/3 assembly and activation. IQGAP2 thus represents an additional protein implicated in such a function, though direct evidence for an actin nucleating function remains to be demonstrated. Alternatively, IQGAP2 activation may generate specific cross-linked F-actin structures unrelated to a function in the initiation of actin polymerization.23,39In the case of WASp, the activating signal appears to be cdc42-dependent.25 It is clear, however, that WASp-deficient platelets demonstrate essentially normal patterns of collagen and thrombin-responsiveness, consistent with the role of another scaffolding protein, such as IQGAP2 in cytoskeletal actin reorganization.40 In preliminary data, we have demonstrated by RT-PCR and immunoblot analysis that IQGAP1 is also expressed in human platelets. IQGAP1 and IQGAP2 are approximately 60% homologous, but they display distinct abilities to bind GTPases—that is, whereas IQGAP2 demonstrates comparable interactions with [GDP]cdc42/rac1 and [GTP]cdc42/rac1, IQGAP1 interactions appear to be restricted to [GTP]rac1 and [GTP]cdc42.41 It remains unknown at this time whether these 2 homologous proteins demonstrate overlapping, complementary, or interactive functions in platelet cytoskeletal actin reorganization.
Components of cell-signaling pathways are likely to assemble into multimolecular complexes held together by scaffolding proteins, as has been specifically demonstrated for β-arrestin42 or more generally for multifunctional protein complexes in yeast.43 Our data suggest that IQGAP2 could have comparable scaffolding functions, based not only on its predicted domain structure (the presence of calmodulin-binding IQ motifs and a single WW domain) but also on the evidence for an IQGAP2/GTPase complex able to interact with the PAK1 binding domain. Indeed, the latter observation raises the intriguing hypothesis that IQGAP2 may directly interact with PAK1 (or its 3 related homologues44) to generate a ternary complex bridged by [GTP]rac1 or [GTP]cdc42. It is well established from structural studies that PAK and other CRIB-containing proteins (such as WASp and ACK) bind to the G-protein switch 1 and 2 domains and to the C-terminus in a GTP-dependent manner.45 Because the switch domains change conformation between the GTP- and GDP-bound forms, a priori considerations argue against GTPase binding to more than one effector. Nonetheless, in the case of ras, for example, the binding sites and functions of the switch 1 and 2 domains are clearly distinct in their capabilities for guanine nucleotide exchange.46 Furthermore, IQGAP2 is distinct among GAPs in its comparable affinity for binding GDP- and GTP-bound forms of the rac1/cdc42 GTPases.7 Thus, these collective observations suggest that IQGAP2 recognizes GTPase segments that may not undergo a major conformational change on GTP hydrolysis (ie, switch domains) and resolve the apparent paradox of such a ternary complex. Although IQGAP2 fails to retain any inherent GAP activity, preliminary observations suggest that it stabilizes GTP-bound rac1 or cdc42.7 This stabilization of GTP-bound GTPases by IQGAP2 may enhance recruitment and cellular compartmentalization of other members of a larger scaffolding complex. Whether such interaction(s) involve PAKs during physiologic platelet activation is suggested by our data, though identification of the individual PAK remains an ongoing area of investigation.
What is the significance of IQGAP2 interacting with rac1 and cdc42 GTPases, and could differential binding mediate distinct effector functions? The integration of data presented here and elsewhere demonstrates that rac1 and cdc42 display divergent fates during platelet activation, with most rac1 remaining in the cytoplasm and a larger fraction of cdc42 translocating to the actin cytoskeleton.9,47 In contrast to the data of Azim et al,9 our data did not demonstrate significant cytosolic increases of [GTP]cdc42 but are more consistent with rapid translocation to the cytoskeleton. In this respect, the results from both laboratories are in general agreement and entirely consistent with prior evidence for an αIIbβ3-dependent translocation of cdc42 to the actin cytoskeleton.30 Furthermore, [GTP]cdc42 has been specifically identified in the actin cytoskeleton as one component of a ternary complex with F-actin and IQGAP1, specifically implicated in the assembly and regulation of higher order actin complexes.23 Taken collectively, these results are most consistent with a model system in which IQGAP2 effector functions would display divergent pathways based on interaction with distinct GTPases—that is, IQGAP2 interaction with [GTP]cdc42 results in actin cytoskeletal localization (with actin assembly)—whereas a cytoplasmic IQGAP2-[GTP]rac1 complex functions as a scaffold for arp2/3-actin complex formation. Interestingly, IQGAP2/GTPase binding appears to be insufficient for IQGAP2 functional effects; preliminary experiments in COS1 cells using IQGAP2 and the 4 GTPase mutants (rac1N17, cdc42N17, rac1V12, and cdc42V12) failed to recapitulate IQGAP2 functional endpoints involving complex assembly and F-actin generation. Although preliminary, these data suggest that a second thrombin-induced signal may be necessary for coordinated IQGAP2 functions. Finally, our observations suggest that selective IQGAP2 inhibition represents a novel target for abrogating thrombin-induced platelet aggregation.
We thank Drs L. Van Aelst, D. Bar-Sagi, N. Nasser, and I. Spector for reagents and helpful discussions, Dr Machesky for anti-arp antibodies, Dr D. Gnatenko for assistance with densitometric calculations, and David Colflesh (University Microscopy Imaging Center) for assistance with the confocal microscopy.
Prepublished online as Blood First Edition Paper, December 19, 2002; DOI 10.1182/blood-2002-09-2807.
Supported by grants from the National Institutes of Health (NHL49141 and HL53665), a Veteran's Administration Medical Center Research Enhancement Award Program (REAP), and an American Heart Association Postdoctoral Fellowship and NIH DK62040 grants (V.S.). W.F.B. is an Established Investigator of the American Heart Association.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Wadie F. Bahou, Division of Hematology, HSCT15-040, State University of New York, Stony Brook, NY 11794-8151; e-mail: wbahou@notes.cc.sunysb.edu.

![Fig. 4. GTP-dependent assembly of an IQGAP2/arp2/3-actin scaffold. / (A) The ability of GST-PBD to specifically bind IQGAP2 was determined by cDNA cotransfection studies in COS1 cells using IQGAP2 and constitutively active rac1V12 (lane 1) or cdc42V12 (lane 3) or the dominant-negative rac1N17 (lane 2) or cdc42N17 (lane 4) mutants, followed by bead-affinity assays and immunoblot analysis with anti-IQGAP2 or anti-GTPase antibodies. GST-conjugated agarose beads lacking the PBD domain failed to precipitate [GTP]rac1/IQGAP2 or [GTP]cdc42/IQGAP2 complex (not shown). (B) GFPs were resuspended to 2.5 × 108/mL, and 200-μL aliquots were activated with individual agonists for various time points as outlined. Samples were then solubilized in RIPA (final concentration, 100 μg/sample), incubated with GST-PBD, and evaluated by SDS-PAGE and immunoblot analysis for the presence of IQGAP2, arp3a, arp2, or actin. (C) F-actin content determinations for individual agonists; F-actin content is inhibited in thrombin-stimulated platelets preincubated with 10 μM cytochalasin D (Cyt D). (D) Percentage (of total) of individual proteins identified within the thrombin-induced IQGAP2 scaffold was determined by densitometric comparison to 5 μg total platelet lysates, adjusted for nonspecific binding using parallel samples incubated with equivalent concentrations of GST control beads. Note that neither gelsolin nor WASp is identified within these complexes and that preincubation of platelets with 10 μM cytochalasin D abrogates thrombin-induced IQGAP2/actin association. Results are representative of one complete set of experiments completed on 2 separate occasions from 2 different healthy volunteers.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/8/10.1182_blood-2002-09-2807/4/m_h80834153004.jpeg?Expires=1765934951&Signature=EeYiMbXMnWKRIDWhKJBI3-dvN1nRpvFZbvp1Mr4WlXeLSCB9dLQTK3spjVkjnSajZO9y1wB-dSHmll10ap~gQgxLtSGyX-8HSgFYdgZVULtUSLWmDdb2ZPgwr~pd0a11XqoRdun3mlXG8gKh3Ap9kOiyLD5h7ExJkCu695XSWRI-EjZDsy66sEaE9k2hnpVv5~WthSP50QbbyiwVG-IkCy38oCGLXLiwQ~-XxpkYwCVgWWBTPTdZCUqFdJNZd4O~cVFsfhD8yNz82Uh7GgQteOrEP72NKe15G7R7qfnRRL4VTCgLzoFbRWNS6xTykAGGfbw91qy2w0Zh6PnyBEEFiA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Agonist-dependent changes in [GTP]rac1, [GTP]cdc42, and cytoskeletal IQGAP2. / (A-B) GFP (5 × 107 platelets/lane; 100 μg starting protein) were preincubated with GST-PBD and stimulated with 10 nM α-thrombin, 10 μg/mL collagen, or 10 μM ADP for various time points, followed by lysis and immunoblot analysis for the presence of GTP[rac1] (upper panel) or GTP[cdc42] (middle panel). Data are presented as percentage of [GTP]-bound GTPase determined by densitometric comparison with 100 μg platelet lysates loaded with saturating concentrations (100 μM) of GTPγS or GDPβS (lanes marked accordingly). Nonspecific binding by GST-PBD was adjusted for by subtracting the “background” GDPβS component. Note the rapid increase of cytosolic GTP[rac1] with a concomitant decrease in [GTP]cdc42, demonstrable only in α-thrombin-stimulated platelets. (C) Triton-X insoluble cytoskeletons from agonist-stimulated platelet (8.5 × 106 platelets/lane corresponding to 17 μg total protein) were immunoblotted for the presence of IQGAP2, and data were reported as the percentage of total platelet IQGAP2 determined by densitometric comparison to platelet control lysates. F-actin contents were essentially as outlined in Figure 4. All results are representative of one complete set of experiments, repeated on 2 occasions with 2 healthy volunteers.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/8/10.1182_blood-2002-09-2807/4/m_h80834153005.jpeg?Expires=1765934951&Signature=FyWrkthGLfB3JqIgrmFM45MMHPnm-7uje3gqxH5Nc85e7zNAyiA3YVgVbcvyRpAH8sGcxcUFhVy4xFov9VOV~GolDoflYzKFK5Jx4wT~Gm~lqIR2UagxEsXR28-EBRoBZoqm2FC1LxDoCj0g8eeRLN~EYjqg0rR-SG4GwZXGMjPYW4R3lUvBmdzv4qMYmgpn7DLMcoxObYihkLtAAuTgv3ytIvyzK-NddirY6sBmNNqGGW0hA~U1KwTwK2LCyRJNPfHPFi4rPnuTFopLsokQX76wWW1tg4C2YLbzDPbXPv7SGNX-swn33T7XiznuTfLVXXFIsDroK4ZHi6In0Qj7PQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 4. GTP-dependent assembly of an IQGAP2/arp2/3-actin scaffold. / (A) The ability of GST-PBD to specifically bind IQGAP2 was determined by cDNA cotransfection studies in COS1 cells using IQGAP2 and constitutively active rac1V12 (lane 1) or cdc42V12 (lane 3) or the dominant-negative rac1N17 (lane 2) or cdc42N17 (lane 4) mutants, followed by bead-affinity assays and immunoblot analysis with anti-IQGAP2 or anti-GTPase antibodies. GST-conjugated agarose beads lacking the PBD domain failed to precipitate [GTP]rac1/IQGAP2 or [GTP]cdc42/IQGAP2 complex (not shown). (B) GFPs were resuspended to 2.5 × 108/mL, and 200-μL aliquots were activated with individual agonists for various time points as outlined. Samples were then solubilized in RIPA (final concentration, 100 μg/sample), incubated with GST-PBD, and evaluated by SDS-PAGE and immunoblot analysis for the presence of IQGAP2, arp3a, arp2, or actin. (C) F-actin content determinations for individual agonists; F-actin content is inhibited in thrombin-stimulated platelets preincubated with 10 μM cytochalasin D (Cyt D). (D) Percentage (of total) of individual proteins identified within the thrombin-induced IQGAP2 scaffold was determined by densitometric comparison to 5 μg total platelet lysates, adjusted for nonspecific binding using parallel samples incubated with equivalent concentrations of GST control beads. Note that neither gelsolin nor WASp is identified within these complexes and that preincubation of platelets with 10 μM cytochalasin D abrogates thrombin-induced IQGAP2/actin association. Results are representative of one complete set of experiments completed on 2 separate occasions from 2 different healthy volunteers.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/8/10.1182_blood-2002-09-2807/4/m_h80834153004.jpeg?Expires=1766003993&Signature=EnmO2e0lF7QsDLL6ZEhl1EyOCgaq12n3GVq5fEsvhwsaLVQtPvNZ0PA9xt4Fpthx4YENGjy8yZ4QH2s0Nju9pWnRsKsQDfR96ubVL4V0xnfv1uU4mxX2VZClbckwLVRlAhEabpdkM9dLbjqLoNa92XasdapCBjondvZ5oJ2D93ziSWjrg1ksM6T0jY3yVoFAwrfys8CUnkp0go-cKw5Iqoarg5jPWuouYe5BrKKClUzDbA-R2QvosDhgNLu1C4lCtVkPwZDR0Cem~UTdrx9ps4e3~0e6p0hYmU88Mzj0mFmqKkFsgLZTMDJEjNMobGybUJuuFaeUh~9c-Lf5JakGig__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Agonist-dependent changes in [GTP]rac1, [GTP]cdc42, and cytoskeletal IQGAP2. / (A-B) GFP (5 × 107 platelets/lane; 100 μg starting protein) were preincubated with GST-PBD and stimulated with 10 nM α-thrombin, 10 μg/mL collagen, or 10 μM ADP for various time points, followed by lysis and immunoblot analysis for the presence of GTP[rac1] (upper panel) or GTP[cdc42] (middle panel). Data are presented as percentage of [GTP]-bound GTPase determined by densitometric comparison with 100 μg platelet lysates loaded with saturating concentrations (100 μM) of GTPγS or GDPβS (lanes marked accordingly). Nonspecific binding by GST-PBD was adjusted for by subtracting the “background” GDPβS component. Note the rapid increase of cytosolic GTP[rac1] with a concomitant decrease in [GTP]cdc42, demonstrable only in α-thrombin-stimulated platelets. (C) Triton-X insoluble cytoskeletons from agonist-stimulated platelet (8.5 × 106 platelets/lane corresponding to 17 μg total protein) were immunoblotted for the presence of IQGAP2, and data were reported as the percentage of total platelet IQGAP2 determined by densitometric comparison to platelet control lysates. F-actin contents were essentially as outlined in Figure 4. All results are representative of one complete set of experiments, repeated on 2 occasions with 2 healthy volunteers.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/8/10.1182_blood-2002-09-2807/4/m_h80834153005.jpeg?Expires=1766003993&Signature=0D-C4LFy0LSVN8M0o8VWI3kfROhs6Mz-IBGl41WT8Vxz2FQrGIBASa6hjw1pw9V-cPnkNbIFLc0gCUcByZCWjXrs52rSzoQLgVQk24sAAQLFFAQtbM97i11WyvVSJvLA104IIcUdgqpHTCn2gcQuUy95PNSBBUZ3mBRTSq~IrvQQbeP2U7bZtY61FgSfer8R4tHh2~469sTkQVxa6mJOiHLYMEolmxnQdtdxF6V7v3cGNzGLxzZ0lfxq6MxmMFCD9hd-rrM0cewdmtb99Wo8MQxTLVMER1If67-y6o5t0JSTbkvgSICiUevS-syk5BPHDEzUeBpVaIH14NRh3NqwGA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)