The possible involvement of germline mutation of the ataxia telangiectasia mutated (ATM) gene in childhood acute leukemia with mixed lineage leukemia (MLL) gene rearrangement (MLL+) was investigated. Of the 7 patients studied, 1 showed a germline missense ATM mutation (8921C>T; Pro2974Leu), located in the phosphatidylinositol-3 (PI-3) kinase domain. In reconstitution assays, the ATM mutant 8921T could only partially rescue the radiosensitive phenotype of AT fibroblasts, and in an in vitro kinase assay, it showed a defective phosphorylation of p53-Ser15. Furthermore, the introduction of 8921T in U2OS cells, characterized by a normal ATM/p53 signal transduction, caused a significant reduction of in vivo p53-Ser15 phosphorylation, suggesting a dominant-negative effect of the mutant ATM over the wild-type protein. Our finding in this patient suggests that altered function of ATM plays some pathogenic roles in the development of MLL+ leukemia.
Introduction
Ataxia telangiectasia (AT) is an autosomal recessive disorder characterized by cerebellar ataxia, oculocutaneous telangiectasia, and immunodeficiency. The responsible gene,ATM (ataxia telangiectasia mutated), is located on chromosome 11q22-23 and encodes a 350-kDa nuclear protein with a carboxy-terminal domain similar to the catalytic subunit of the phosphatidylinositol-3 kinases (PI-3 kinases).1-3 PI-3 kinase–related proteins are known to function in the maintenance of genomic stability, cell cycle control, and cellular responses to DNA damage.4,5 AT cells exhibit hypersensitivity to ionizing radiation and are defective at multiple cell cycle checkpoints.4-7
It is well established that patients with AT are at increased risk of cancer,8 in particular, neoplasms of lymphoid origin.9,10 Somatic mutations in the ATM gene have been identified in patients with T-cell prolymphocytic leukemia (T-PLL)11 and B-cell chronic lymphocytic leukemia (B-CLL) with no family history of AT.12-14 Mutations observed in these reports are missense mutations and most occur in the PI-3 kinase domain. Heterozygous germline missense mutations were also found among B-CLL patients, indicating that such genetic alterations might act as predisposing factors for the development of lymphoid tumors.12,13 The germline missense ATMmutations have been reported in patients with breast carcinoma with early onset disease and positive family history.15
The chromosomal translocation of the mixed lineage leukemia (MLL) gene at 11q23 is involved in a subset of childhood leukemia, most frequently in infantile leukemia.16-18 TheMLL gene rearrangement is also involved in treatment-related leukemias secondary to chemotherapy using topoisomerase II (topo II) inhibitors. This has led to the hypothesis that in utero exposure to chemicals, such as certain antibiotics, laxatives, podophyllin resins, flavonoids, herbal medicines, and benzene metabolites may cause infantile leukemia via an effect on topo II.19,20 Recent epidemiologic data indicate that maternal alcohol exposure and exposure to recreational drugs, pesticides, and anti-inflammatory drugs increase the risk of infantile leukemia.21-23 Thus, environmental factors appear to play an important role in the development of this disease.
Previous studies have shown that AT cells are hypersensitive to topo II inhibitors.24-28 It is plausible that mixed lineage leukemia (MLL) gene rearrangement (MLL+) leukemias such as infantile leukemia could arise in individuals with hypersensitivity to topo II inhibitors, and this hypothesis led us to investigate ATM gene mutation in patients who had developed MLL+ leukemia. In this study, we attempted to determine whether the germline ATM gene mutation could represent one genetic factor for predisposition to MLL+leukemia in children and whether it could play a role in the pathogenesis of this condition.
Patients, materials, and methods
Patients
The study included 7 patients diagnosed as having de novo acute lymphoblastic (4 patients) or myeloid (3 patients) leukemia withMLL gene rearrangement. The patients were aged from 2 months to 1 year 11 months; 2 were aged less than 1 year and classified as infantile leukemia. Normal peripheral blood samples were obtained from these patients during their complete hematologic remission. The clinical features of these patients are summarized in Table1. None of the patients exhibited clinical evidence of AT or a family history of AT or Li-Fraumeni syndrome. Normal control samples (wt1, wt2) were obtained from 2 independent healthy volunteers. Cord blood samples were obtained from a local cord blood bank. Samples were collected after informed consent was obtained, based on the approval of the Showa University of Fujigaoka Hospital ethics committee.
MLL gene rearrangement was determined by the method described previously.29
Epstein-Barr virus (EBV)–immortalized lymphoblastoid cell lines (L-1–L-7, LCL-wt1, LCL-wt2) were established by infecting normal lymphocytes obtained from the patients and 2 healthy volunteers with EBV strain B95-8 as previously described.30 Immortalized lymphoblastoid cell lines (LCLs) were maintained in RPMI 1640 (Gibco BRL Life Technologies, Gaithersburg, MD) with 15% fetal calf serum (FCS; Gibco BRL) at approximately 5 × 105 cells/mL at 37°C in 5% CO2. The SV40-transformed AT cell line GM05849C (7009delTG) was obtained from Coriell Cell Repositories (Camden, NJ). The osteosarcoma cell line U2OS and human embryonic kidney cell line 293 were obtained from Health Science Research Resources Bank (Osaka, Japan).
PCR/DNA sequencing
Mutation analysis of the ATM coding sequence was undertaken by reverse transcription–polymerase chain reaction (Takara LA RT-PCR kit; Takara, Shiga, Japan) and direct sequencing of PCR products. Direct sequencing of PCR products was performed by cycle sequencing using ABI Big Dye Terminator chemistry (Applied Biosystems, Foster City, CA) followed by capillary electrophoresis on an ABI 310 automated sequencer. Mutations identified in reverse transcription–polymerase chain reaction (RT-PCR) products were confirmed by sequencing of genomic DNA-based PCR products from fresh peripheral blood lymphocytes.
Microsatellite analysis
Microsatellite analysis at chromosome 11q neighboring or intragenic to ATM was performed as described.31Briefly, the microsatellite markers D11S2178, D11S1294, D11S1778, D11S2179, D11S2366, and D11S1787 were selected. The forward primer of each pair was 5′-labeled with HEX or 6-FAM dye. Template DNA was amplified by touchdown PCR using AmpliTaq Gold (Applied Biosystems).
PCR/RFLP analysis
Allele frequency of the mutation was analyzed using the PCR/restriction fragment length polymorphism (RFLP) method. For the 8921C>T nucleotide change, digestion with AlwNI (New England Biolabs, Beverly, MA) yielded 2 bands for exon 64 of mutant ATM. For the −787A>G nucleotide change, digestion with SacII (Takara) yielded 2 bands and 3 bands for exon 2 of wild-type ATM and mutant ATM, respectively. This analysis was also used to detect the expression level of 8921TATM mRNA in transfectants and L-4 LCL.
Subcloning of the ATM allele
RNA was obtained from 1 × 106 EB-transformed cells using the Dynabeads RNA extraction kit (Dynabeads, Oslo, Norway).ATM partial sequence (7012-9402) was amplified using LA Taq (Takara) followed by cDNA synthesis using reverse transcriptase XL (Takara). Amplified cDNA was subcloned into the pGEM-T Easy TA cloning system (Promega, Madison, WI). The plasmid, which contained the corresponding nucleotide change of the ATMgene, was digested with PflMI (New England Biolabs) andSalI (Takara), and the insert was subcloned intoPflMI and XhoI (Takara) sites of pcDNA3-YZ5 using Ligation Kit Ver II (Takara). The expression vector thus constructed (pcDNA3/8921T) was subjected to DNA sequencing analysis and was found to have wild-type sequence except for the 8921T change.
Western blot analysis
Cells (1 × 106) were washed with phosphate-buffered saline (PBS) and lysed in 150 mM NaCl, 1.0% NP-40, 0.1% sodium dodecyl sulfate (SDS), 0.1% sodium deoxycholate, 5 mM EDTA (ethylenediaminetetraacetic acid), and 10 mM Tris (tris(hydroxymethyl)aminomethane)–HCl (pH 7.4) containing protease inhibitors. Protein concentrations were measured using the DC protein assay (Bio-Rad, Hercules, CA). After boiling with sample buffer, 30 μg protein was subjected to SDS-polyacrylamide gel electrophoresis (PAGE). After transfer to polyvinylidine difluoride (PVDF) membrane (Millipore, Bedford, MA), the blots were probed with an anti-p53–Ser15 phosphospecific antibody (New England Biolabs), anti-ATM antibody,32 or anti–α-tubulin (Oncogene Science, Cambridge, MA) antibody. Horseradish peroxidase (HRP)–conjugated antirabbit or antimouse antibody (Amersham, Buckinghamshire, England) was used as the secondary antibody.
Clonogenic assay
Plasmid pcDNA3, pcDNA3-YZ5, or pcDNA3-YZ5/8921T was transfected into GM05849C (Effecten; Qiagen, Hilden, Germany) and stable clones were selected with 500 μg/mL neomycin (Sigma, St Louis, MO). Cells were trypsinized, plated onto 60-mm dishes at a density of 7 to 10 × 103 cells/dish, and incubated for 17 hours. Then cells were exposed to a range of doses of x-irradiation (X-IR) at 0 to 5 Gy followed by incubation for 10 days at 37°C in 5% CO2. Prior to counting colonies, the culture medium was decanted and the cells were fixed in 95% methanol and stained with 0.5% crystal violet, and the numbers of colonies (> 50 cells) from triplicate dishes were counted. Mean colony numbers relative to unirradiated colony numbers were plotted.
Kinase assay
The 293 cells were transiently transfected by FLAG-tagged pcDNA3-YZ5 or pcDNA3-YZ5/8921T. Cells (2 × 107) were washed with PBS and lysed in TGN buffer (150 mM NaCl, 0.3% NP-40, 1% Tween, and 50 mM Tris-HCl [pH 7.5] containing protease inhibitors). Cell lysate (500 μg) was precleared by constant mixing for 1 hour with protein A-Sepharose beads (Pharmacia Biotech, Uppsala, Sweden). The beads were removed by centrifugation, and the supernatant was mixed for 1 hour with anti-FLAG M2 monoclonal antibody (Sigma). Immune complexes were adsorbed onto protein A-Sepharose and then washed twice with TGN buffer, twice with TGN buffer plus 0.5 M LiCl, and twice with kinase buffer (20 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, and 10 mM MnCl2) and incubated with GST-p53 (amino acids 1-100) in 20 mM Tris HCl buffer (pH 7.6; 10 mM MgCl2, 10 mM MnCl2, 50 mM adenosine triphosphate [ATP]), containing 100 μM ATP for 30 minutes at 30°C. Anti-p53 phosphoserine 15 antibody was used to detect phosphorylation of Ser15 of GST-p53 (amino acids 1-100).
Results
Nucleotide change of the ATM gene
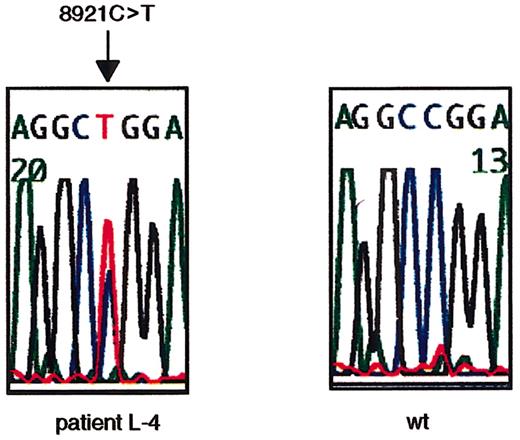
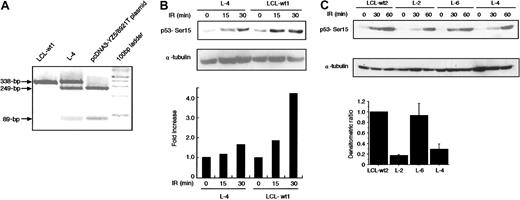
To search for ATM mutations, we sequenced the open reading frame (ORF) of ATM cDNA using RT-PCR–based direct sequencing. L-4 was identified to have a nucleotide change of 8921C>T on one allele (Figure 1). The 8921C>T transversion results in a Pro→Leu amino acid change at codon 2974 within the PI-3 kinase domain. Thus, L-4 was determined to be heterozygous (wt/8921T) at the ATM gene.
Missense nucleotide change of
ATM gene at 8921C>T in L-4. The heterozygous missense nucleotide change at 8921C>T in exon 64 in L-4 is shown. The arrow indicates the position of the nucleotide change. The first codon of the ORF was designated +1 (GenBank no.U33841).
Missense nucleotide change of
ATM gene at 8921C>T in L-4. The heterozygous missense nucleotide change at 8921C>T in exon 64 in L-4 is shown. The arrow indicates the position of the nucleotide change. The first codon of the ORF was designated +1 (GenBank no.U33841).
The same nucleotide change was confirmed in fresh peripheral lymphocyte genomic DNA from patient L-4 (data not shown). Furthermore, the father of patient L-4 was demonstrated to have the same nucleotide change on one of the alleles (data not shown), indicating paternal inheritance of this nucleotide change.
We also analyzed for the presence or absence of wild-type allele in leukemic cells of patient L-4 by microsatellite analysis, and no loss of heterozygosity (LOH) was identified (supplemental data on theBlood website; see the Supplemental Figure link at the top of the online article).
We screened 166 samples from an archive of DNA stock of unselected neonatal cord blood for detection of this nucleotide change using RT-PCR/RFLP analysis and direct sequencing. One sample was heterozygous for 8921C>T, resulting in an allele frequency of 0.3%.
Biologic activity of 8921T ATM
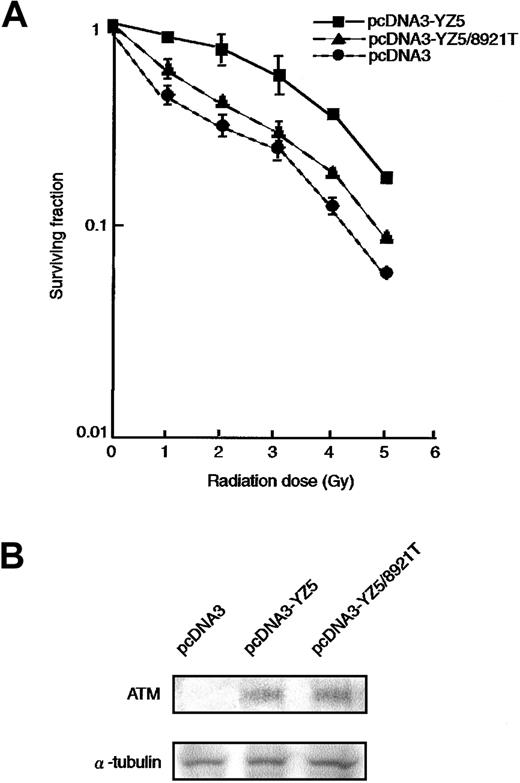
It is well established that AT cells are hypersensitive to ionizing radiation.6,7 33 To determine whether the 8921T nucleotide change, which is located within the PI-3 kinase domain, can rescue the radiosensitive phenotype of an AT cell line (ATM−/−), we transfected pcDNA3 (vector alone), pcDNA3-YZ5 (wild-type ATM), or pcDNA3-YZ5/8921T (mutant ATM) into the AT fibroblast cell line GM05849C. Stable clones expressing comparable amounts of wild-type or 8921T ATM protein were isolated (Figure2B) and analyzed for clonogenic cell survival after exposure to variable doses of irradiation. pcDNA3-YZ5/8921T showed only partial rescue of the radiosensitive phenotype of GM05849C, when compared with pcDNA3-YZ5 (Figure2A).
Radiosensitivity of AT fibroblast cell transfectants.
(A) Radiosensitivity of GM05849C cells transfected with pcDNA3-YZ5 and pcDNA3-YZ5/8921T was determined by clonogenic assay. Cells were exposed to 0, 1, 2, 3, 4, or 5 Gy X-IR and then cultured for 10 days prior to fixation, staining, and assessment of colony formation. The assay was performed in triplicate; mean colony numbers relative to unirradiated controls were plotted and the SE was shown as an error bar.(B) Expression of ATM protein was equivalent between GM05849C cells transfected with pcDNA3-YZ5 and pcDNA3-YZ5/8921T. Western blot analysis was performed with anti-ATM antibody. α-Tubulin was used as a control for protein loading.
Radiosensitivity of AT fibroblast cell transfectants.
(A) Radiosensitivity of GM05849C cells transfected with pcDNA3-YZ5 and pcDNA3-YZ5/8921T was determined by clonogenic assay. Cells were exposed to 0, 1, 2, 3, 4, or 5 Gy X-IR and then cultured for 10 days prior to fixation, staining, and assessment of colony formation. The assay was performed in triplicate; mean colony numbers relative to unirradiated controls were plotted and the SE was shown as an error bar.(B) Expression of ATM protein was equivalent between GM05849C cells transfected with pcDNA3-YZ5 and pcDNA3-YZ5/8921T. Western blot analysis was performed with anti-ATM antibody. α-Tubulin was used as a control for protein loading.
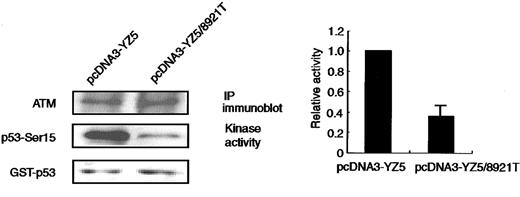
ATM directly phosphorylates p53 at Ser15 in response to X-IR–induced DNA damage.34,35 Phosphorylation of Ser15 is a necessary step for p53 accumulation and p53-dependent transactivation.36,37 Rapid phosphorylation of p53 at Ser15 in response to X-IR is reduced in AT cells and cells from some obligate AT heterozygous carriers.32 To examine the ability of the 8921T ATM to phosphorylate p53-Ser15, we transiently transfected 293 cells with pcDNA3-YZ5 or pcDNA3-YZ5/8921T and in vitro kinase activity was assessed using anti-FLAG–tagged immunoprecipitates. The 8921T exhibited about 60% less p53-Ser15 phosphorylation activity (Figure 3), suggesting that this is a genuine mutation rather than a polymorphic variant.
In vitro phosphorylation of p53-Ser15 by immunoprecipitated ATM.
The catalytic activity of ATM protein immunoprecipitated with anti-FLAG antibody from 293 cells transiently transfected with pcDNA3-YZ5 or pcDNA3-YZ5/8921T was assayed in vitro against the GST-p53 (amino acids 1-100) substrate. Kinase reactions were Western blotted and tested with an anti-p53–Ser15 phosphospecific antibody, or with an anti-ATM antibody, to verify the amount of immunoprecipitated ATM protein per lane. Equal amounts of GST-p53 were loaded. Representative data from 3 independent experiments are shown (left panel). Phosphorylation of recombinant GST-p53 (amino acids 1-100) by pcDNA3-YZ5/8921T was standardized for kinase activity of pcDNA3-YZ5 (= 1), and was shown as fold increase. Data were expressed as the mean (± SE) of 3 independent experiments (right panel).
In vitro phosphorylation of p53-Ser15 by immunoprecipitated ATM.
The catalytic activity of ATM protein immunoprecipitated with anti-FLAG antibody from 293 cells transiently transfected with pcDNA3-YZ5 or pcDNA3-YZ5/8921T was assayed in vitro against the GST-p53 (amino acids 1-100) substrate. Kinase reactions were Western blotted and tested with an anti-p53–Ser15 phosphospecific antibody, or with an anti-ATM antibody, to verify the amount of immunoprecipitated ATM protein per lane. Equal amounts of GST-p53 were loaded. Representative data from 3 independent experiments are shown (left panel). Phosphorylation of recombinant GST-p53 (amino acids 1-100) by pcDNA3-YZ5/8921T was standardized for kinase activity of pcDNA3-YZ5 (= 1), and was shown as fold increase. Data were expressed as the mean (± SE) of 3 independent experiments (right panel).
Dominant-negative effect of the 8921T mutation
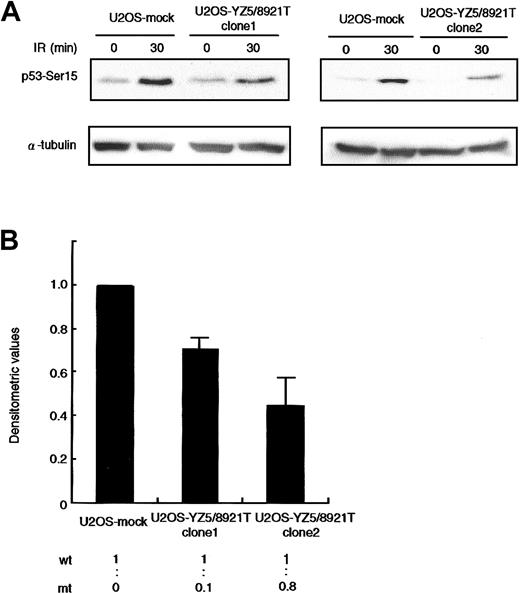
An issue of great interest was to determine whether the 8921T mutant protein interferes with the normal ATM function. We transfected U2OS cells, which exhibit normal ATM-dependent p53-Ser15 phosphorylation activity, with a mock or pcDNA3-YZ5/8921T mutant ATM expression vector, and in vivo p53-Ser15 phosphorylation was examined by Western blotting. We isolated 2 independent U2OS clones expressing wild-type and 8921T ATM at a ratio of 1:0.1 (clone 1) and 1:0.8 (clone 2), and assayed for p53-Ser15 phosphorylation 30 minutes after X-IR. The phosphorylation level was reduced by approximately 30% and 50% in clone 1 and clone 2, respectively, when compared with mock transfectant (Figure 4).
Dominant-negative effect on p53-Ser15 phosphorylation in U2OS transfectants.
(A) Western blot analysis of p53-Ser15 phosphorylation after X-IR in clone 1 (left panel) and clone 2 (right panel). Cells were harvested at 0 and 30 minutes after 5-Gy X-IR and analyzed by Western blotting with anti-p53–Ser15 phosphospecific antibody. α-Tubulin was used as a control for protein loading. (B) Fold decrease of p53-Ser15 phosphorylation in clone 1 and clone 2, standardized for the level in irradiated/nonirradiated U2OS-mock (= 1.00). Data were expressed as the mean (± SE) of 2 independent experiments. U2OS cells were transfected with vector alone or pcDNA3-YZ5/8921T. Two independent clones (1 and 2) were isolated and analyzed for expression of wild-type and mutant ATM mRNA by RT-PCR/RFLP analysis using theAlwNI restriction site specific for the 8921T mutation. Clones 1 and 2 expressed wild-type and mutant ATM at a ratio of 1:0.1 and1:0.8, respectively.
Dominant-negative effect on p53-Ser15 phosphorylation in U2OS transfectants.
(A) Western blot analysis of p53-Ser15 phosphorylation after X-IR in clone 1 (left panel) and clone 2 (right panel). Cells were harvested at 0 and 30 minutes after 5-Gy X-IR and analyzed by Western blotting with anti-p53–Ser15 phosphospecific antibody. α-Tubulin was used as a control for protein loading. (B) Fold decrease of p53-Ser15 phosphorylation in clone 1 and clone 2, standardized for the level in irradiated/nonirradiated U2OS-mock (= 1.00). Data were expressed as the mean (± SE) of 2 independent experiments. U2OS cells were transfected with vector alone or pcDNA3-YZ5/8921T. Two independent clones (1 and 2) were isolated and analyzed for expression of wild-type and mutant ATM mRNA by RT-PCR/RFLP analysis using theAlwNI restriction site specific for the 8921T mutation. Clones 1 and 2 expressed wild-type and mutant ATM at a ratio of 1:0.1 and1:0.8, respectively.
In vivo phosphorylation of p53 at Ser15 in L-4
The finding that 8921T has a dominant-negative activity in transfection assay lead us to study in vivo p53-Ser15 phosphorylation in L-4 immediately after X-IR. RT-PCR/RFLP analysis allowed us to determine that the expression level of 8921T allele was comparable to that of wild-type allele in L-4 (Figure5A). At 15 and 30 minutes after 5-Gy X-IR, L-4 exhibited a 40% to 80% reduction in phosphorylation compared with 2 normal control LCLs (LCL-wt1, LCL-wt2) and L-6 with noATM mutation (Figure 5B-C). We noted an unexpected finding that L-2, with no detectable mutation within the ORF of ATM(data not shown), also demonstrated a significant decrease in p53-Ser15 phosphorylation (Figure 5C). ATM and Rad-3–related (ATR) kinase also phosphorylates p53-Ser15 1 hour after X-IR, whereas ATM acts immediately.38 Therefore, the level of p53-Ser15 phosphorylation at 60 minutes in this assay was less informative with regard to ATM activity.
Reduction of X-IR–induced phosphorylation of p53 in L-4.
(A) PCR products of exon 64 (338 bp) from L-4, LCL-wt1, and pcDNA3-YZ5/8921T plasmid were digested with AlwNI. The 249-bp and 89-bp fragments are expected in the 8921C>T mutation, but not in the wild-type sequence. (B-C) Cells were harvested at various minute intervals after 5-Gy X-IR and analyzed by Western blotting with anti-p53–Ser15 phosphospecific antibody. α-Tubulin was used as a control for protein loading. Levels of phosphorylated p53–Ser15 as detected by Western blotting were quantified by scanning densitometry and were corrected for α-tubulin content, and standardized to those of LCL-wt1 (= 1) (B) and LCL-wt2 (C). Data were expressed as the mean (± SE) of 2 independent experiments (bottom graph in C). In panel C, the histogram is shown only for the data at 30 minutes because the data at 60 minutes were less informative due to the activity of ATR.
Reduction of X-IR–induced phosphorylation of p53 in L-4.
(A) PCR products of exon 64 (338 bp) from L-4, LCL-wt1, and pcDNA3-YZ5/8921T plasmid were digested with AlwNI. The 249-bp and 89-bp fragments are expected in the 8921C>T mutation, but not in the wild-type sequence. (B-C) Cells were harvested at various minute intervals after 5-Gy X-IR and analyzed by Western blotting with anti-p53–Ser15 phosphospecific antibody. α-Tubulin was used as a control for protein loading. Levels of phosphorylated p53–Ser15 as detected by Western blotting were quantified by scanning densitometry and were corrected for α-tubulin content, and standardized to those of LCL-wt1 (= 1) (B) and LCL-wt2 (C). Data were expressed as the mean (± SE) of 2 independent experiments (bottom graph in C). In panel C, the histogram is shown only for the data at 30 minutes because the data at 60 minutes were less informative due to the activity of ATR.
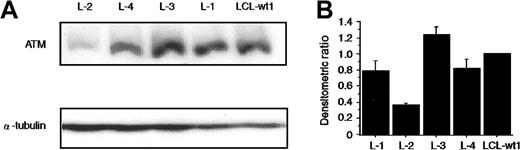
Normal expression of ATM protein in L-4
In most heterozygous AT carriers, the ATM protein level is reduced due to truncation mutations.32 However, in the case of missense mutation (7271T>G), the ATM protein is expressed at normal level.39 Thus, ATM protein expression was studied by Western blotting in L-4 and L-2, the latter showing no mutation ofATM gene but poor p53 phosphorylation activity. The level of ATM protein expression in L-4 was comparable with those seen both in normal LCL-wt1 and in the remaining LCLs, except for L-2 (data not shown for L-5, L-6, and L-7). In L-2, the ATM protein was approximately 37% of normal LCL-wt1 (Figure 6A-B).ATM mRNA expression in L-2 was reduced by 30% as analyzed by semiquantitative RT-PCR assay (data not shown). L-2, however, showed no mutation in ORF of ATM gene. Thus, we sequenced the 5′ and partial 3′ untranslated region (UTR) of the ATM gene and identified the −787A>G nucleotide change (data not shown). Subsequent analysis in 100 Japanese cord blood samples revealed a 61% allele frequency of −787A>G and this was considered as a normal polymorphism (data not shown). Thus, the reason for the decreased ATM expression in L-2 requires further investigation.
Normal expression of ATM protein in L-4.
(A) The levels of ATM protein present in the lysates from the indicated cell lines were determined by Western blotting using anti-ATM antibody32 and normalization with an α-tubulin antibody. Lanes 1 to 4 are LCLs from patients L-2, L-4, L-3, and L-1, respectively. Lane 5 is a normal control LCL (LCL-wt1). (B) The histograms were obtained by the densitometric analysis of the Western blot autoradiographs, after correction of each lane for α-tubulin content. Values are normalized to those of the normal control LCL-wt (= 1). Although not shown, ATM protein levels in L-5, L-6, and L-7 were comparable to those of LCL-wt1 and of another control (LCL-wt2). Data were expressed as the mean (± SE) of 3 independent experiments.
Normal expression of ATM protein in L-4.
(A) The levels of ATM protein present in the lysates from the indicated cell lines were determined by Western blotting using anti-ATM antibody32 and normalization with an α-tubulin antibody. Lanes 1 to 4 are LCLs from patients L-2, L-4, L-3, and L-1, respectively. Lane 5 is a normal control LCL (LCL-wt1). (B) The histograms were obtained by the densitometric analysis of the Western blot autoradiographs, after correction of each lane for α-tubulin content. Values are normalized to those of the normal control LCL-wt (= 1). Although not shown, ATM protein levels in L-5, L-6, and L-7 were comparable to those of LCL-wt1 and of another control (LCL-wt2). Data were expressed as the mean (± SE) of 3 independent experiments.
Discussion
AT homozygotes and heterozygotes are reportedly hypersensitive to topo II inhibitors, such as etoposide (VP16).40 The finding that the plant isoflavenoid genistein (that has topo II inhibitor activity), activates p53 and Chk2 in an ATM-dependent manner41 further suggests an important link between topo II inhibitors and ATM. Epidemiologic data indicate a significant association between infantile leukemia and maternal exposures to topo II inhibitors or topo II inhibitor-like chemicals,42 which are known to cause intracellular rearrangement of the MLLgene.19 20 These findings led us to hypothesize that at least some leukemias with MLL gene rearrangement may develop in individuals with a combination of hypersensitivity to topo II inhibitors as characterized by genetic ATM dysfunction and environmental effects, such as exposure to topo II inhibitors.
In the present study, a heterozygous germline mutation (8921C>T) in the PI-3 kinase domain of the ATM gene was identified in a case (L-4) of MLL+ leukemia. L-4 expressed an apparently normal level of ATM protein. 8921T is associated with amino acid substitution of Pro2974Leu, which leads to a change in the secondary structure of the ATM protein, namely, loss of random coil between α-helices.43 The in vitro kinase assay showed that 8921T ATM was defective in mediating p53-Ser15 phosphorylation following X-IR. Thus, this change in the secondary structure of the PI-3 kinase domain may disrupt its enzymatic function. Expression of 8921T ATM in AT fibroblasts rescued the radiosensitive phenotype only partially. Furthermore, its expression in U2OS cells revealed an interfering effect with the normal function of ATM. DNA damage–induced in vivo p53-Ser15 phosphorylation activity was reduced in L-4, which was compatible with dominant-negative activity against p53-Ser15 phosphorylation in U2OS cells transfected with 8921T. Thus, it is strongly suggested that 8921T is not just a polymorphism but a pathogenic mutation. LOH was not identified in leukemia cells of L-4. This finding is not incompatible with dominant-negative activity of this mutation and suggests an activity distinct from the tumor suppressor function, which has been suggested in some B-CLL patients with ATM mutation-associated p53 dysfunction.44
The ATM protein is known to multimerize and to associate with other proteins, forming a functional complex.45 Gatti et al have hypothesized that there are 2 types of ATM heterozygotes, those with truncating mutations making no protein, and those with missense mutations making the mutant protein.46 Presence of the mutant protein in such a complex may disrupt its function, leading to a dominant-negative effect against wild-type ATM protein.46,47 In addition, it has been reported that overexpression of a kinase dead ATM cDNA has a dominant interfering effect on ATM kinase activity.48 During the preparation of this manuscript, Scott et al have shown that a missense mutation (7775C>G) of ATM gene identified in a breast cancer patient has a dominant-negative effect against wild-type ATM.45Furthermore, Spring et al reported that mice heterozygous forAtm mutation (7636del9) but not for truncation mutation, showed an increased susceptibility to developing tumors.49
In a segregation analysis, the father of patient L-4 was heterozygous for the 8921T mutation, excluding the possibility that the alteration arose de novo in the patient. The father, however, remains free from malignant disease to date. These results suggest that the 8921T mutation has a relatively low penetrance, and the environmental stresses, such as exposure to topo II inhibitors at certain developmental stages, played a critical role in the development of MLL+ leukemia in this case. This is important in light of the finding that allele frequency of 8921T was 0.3% in normal cord blood samples.
Thus, although further extensive studies are required to justify our hypothesis, our findings suggest that dysregulation of ATM/p53 signal transduction cascade in response to environmental stresses may be one of the possible genetic factors underlying the pathogenesis of MLL+ leukemia.
Further study is required to better understand the role of ATM dysfunction with regard to the disease susceptibility and exposures to DNA-damaging agents.
We are grateful to Dr M. Kastan for the kind gift of pcDNA3-YZ5.
Prepublished online as Blood First Edition Paper, January 2, 2003; DOI 10.1182/blood-2002-02-0570.
Supported by a Grant-in-Aid for Pediatric Research from the Ministry of Health and Welfare, Japan, by a Grant-in-Aid for Cancer Research from the Ministry of Health and Welfare, Japan, and by a Grant-in-Aid from the Ministry of Health and Welfare, Japan, as part of a comprehensive 10-year strategy for Cancer Control; by a Grant-in-Aid from the Ministry of Education, Science and Culture, Japan; by a Grant from the Human Science Foundation, Japan; by a Grant from the Japan Leukemia Research Fund; and by the Italian Association for Cancer Research (AIRC) and Telethon grants E764 and GPO205/01. L.Z. is supported by a fellowship from the Italian Foundation for Cancer Research (FIRC).
K.O. and M.T. contributed equally to this work.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Shuki Mizutani, Department of Pediatrics and Developmental Biology, Postgraduate Medical School, Tokyo Medical and Dental University, 1-5-45, Yushima, Bunkyo-ku, Tokyo 113-8519, Japan; e-mail: smizutani.ped@tmd.ac.jp.