Specialized cDNA-based microarrays (IronChips) were developed to investigate complex physiological gene-regulatory patterns in iron metabolism. Approximately 115 human cDNAs were strategically selected to represent genes involved either in iron metabolism or in interlinked pathways (eg, oxidative stress, nitric oxide [NO] metabolism, or copper metabolism), and were immobilized on glass slides. HeLa cells were treated with iron donors or iron chelators, or were subjected to oxidative stress (H2O2) or NO (sodium nitroprusside). In addition, we generated a stable transgenic HeLa cell line expressing the HFE gene under an inducible promoter. Gene-response patterns were recorded for all of these interrelated experimental stimuli, and analyzed for common and distinct responses that define signal-specific regulatory patterns. The resulting regulatory patterns reveal and define degrees of relationship between distinct signals. Remarkably, the gene responses elicited by the altered expression of the hemochromatosis protein HFE and by pharmacological iron chelation exhibit the highest degree of relatedness, both for iron-regulatory protein (IRP) and non-IRP target genes. This finding suggests that HFE expression directly affects the intracellular chelatable iron pool in the transgenic cell line. Furthermore, cells treated with the iron donors hemin or ferric ammonium citrate display response patterns that permit the identification of the iron-loaded state in both cases, and the discrimination between the sources of iron loading. These findings also demonstrate the broad utility of gene-expression profiling with the IronChip to study iron metabolism and related human diseases.
Introduction
Iron is a nutrient that plays an essential role in biological functions. It mediates oxygen transport by hemoglobin and constitutes an essential component of the respiratory chain by conferring redox activity on the cytochromes and other enzymes. However, iron can also damage tissues by catalyzing the conversion of hydrogen peroxide to free-radical ions that attack cellular membranes, proteins, and DNA.1,2 It is hence not surprising that both iron deficiency and iron overload cause pathologic changes. Disorders of iron homeostasis are among the most common inherited diseases of humans.3,4 To tightly control iron homeostasis, a complex network of iron transporters, storage molecules, and regulators has evolved. To interface iron metabolism with other metabolic activities of cells, regulators of iron metabolism also respond to noniron signals such as nitric oxide (NO) and oxidative stress.5 6
Iron homeostasis is regulated at the systemic and the cellular level. The expression of central proteins involved in iron uptake and transport, iron storage, and iron usage is controlled by the iron-responsive element (IRE)/iron-regulatory protein (IRP) regulatory system. IREs are RNA elements that function as binding sites for IRP-1 and IRP-2. IRP-1 or IRP-2 bound to a single IRE in the 5′ untranslated region (UTR) of an mRNA controls the translation of, for example, the iron-storage proteins H- and L-ferritin, the erythroid 5-aminolevulinate synthase (eALAS), and mitochondrial aconitase mRNA.7-12 IRPs bound to multiple IREs in the 3′UTR of the transferrin receptor 1 (TfR1) mRNA stabilize the transcript, which encodes a critical receptor for cellular iron uptake (reviewed in Muckenthaler and Hentze,13 Cairo and Pietrangelo,14 and Eisenstein15). The IRE-binding activity of IRP-1 and IRP-2 is itself regulated by the experimentally defined “intracellular chelatable iron pool.”16-21 In addition, H2O2and NO affect IRP activity,22-29 linking the regulation of iron metabolism to the oxidative stress and nitric oxide pathways.
In addition to IRP-mediated posttranscriptional regulation, transcriptional control mechanisms regulate important aspects of cellular iron homeostasis. For example, the transcription of the transferrin-receptor gene is activated by hypoxia-inducible factor 1–α (Hif-1α)30-32 and is down-regulated by tumor necrosis factor–α (TNF-α) and interleukin-1β (IL-1β) in alveolar epithelial cells.33 L-ferritin mRNA transcription is induced by prostaglandin A1,34 and H-ferritin transcription is augmented by c-jun in cultured HeLa cells.35 Both the H-ferritin and IRP-2 genes are targeted by c-myc, and the regulation of these genes is thought to contribute to c-myc–dependent cell proliferation and transformation.36
The positional cloning of the gene affected in hereditary hemochromatosis (HC)37 resulted in the identification of a novel protein with a role in iron homeostasis, termed HFE. HC is characterized by systemic iron overload from increased duodenal iron absorption.38 HFE is a major histocompatibility complex (MHC) class 1–like protein37 that forms a heterodimer with β2-microglobulin (β2M). A missense mutation (Cys282Tyr) in the extracellular domain of HFE alters its conformation and abrogates β2M binding, which results in a loss of HFE-protein presentation on the cell surface.39,40 Other polymorphisms have been found in the HFE gene, but their clinical significance is less clear.37,41,42 A biochemical link between HFE and cellular iron metabolism was established with the finding that HFE can engage in high-affinity interactions with the transferrin receptor.43,44 This interaction interferes with the binding of transferrin to the transferrin receptor, and thus reduces cellular iron uptake.43,45,46 We previously developed a stable HeLa cell line in which HFE is expressed under the control of a tetracycline-responsive promoter.47 We demonstrated that the induction of HFE expression results in decreased iron uptake from diferric transferrin. Moreover, HFE expression activates IRP activity and thus causes reduced synthesis of the iron-storage protein ferritin and an increase in transferrin receptor levels.47 Recently, HFE was overexpressed in Chinese hamster ovary cells.48 Similar to the observations in HeLa cells,47,49 HFE overexpression caused a decrease in transferrin-mediated iron uptake. However, the combined expression of both HFE and β2M increased TfR1-dependent iron uptake and cellular iron levels. It was reported that the HFE-β2M complex enhances the rate of TfR1 recycling and results in an increased steady-state level of TfR1 at the plasma membrane of these stably transfected cells.48 Thus, the availability of β2M may affect HFE function and iron homeostasis.
Gene-expression profiling using DNA microarrays has allowed gene-expression analyses to be broadened from the study of single genes to the investigation of complex regulatory networks.50-52 Here, we report the development of the IronChip, a cDNA-based microarray that represents human genes directly involved in iron metabolism or in interlinked pathways such as oxidative stress, NO metabolism, or copper metabolism. We analyzed the genetic response patterns of HeLa cells to iron perturbation as well as exposure to oxidative stress and the NO+ donor sodium nitroprusside (SNP). We demonstrate that the resulting regulatory patterns reflect degrees of relationship between the different signals. Remarkably, the gene responses elicited by HFE induction and by pharmacological iron chelation exhibit the highest degree of relatedness, for both IRP and non-IRP target genes. This finding suggests that HFE expression directly targets the regulatory iron pool(s) of the transfected cells.
Materials and methods
Selection of cDNA clones
The genes that are immobilized on the IronChip were selected on the basis of (1) literature searches, (2) microarray experiments performed on filters that contain approximately 20 000 human nonredundant expressed sequence tags (ESTs) comparing hemin- and desferrioxamine-treated Caco-2 cells, and (3) gene lists from published microarray studies that address metabolic pathways of interest.
For the IronChip (version 2.0) (http://www.embl-heidelberg.de/ExternalInfo/hentze/suppinfo.html; accessed January 16, 2003), 113 human EST clones that were sequence-verified from both ends were chosen. The ESTs were selected to contain the 3′ end of a cDNA (ie, the polyadenylation signal) and to extend for at least 300 bp toward the 5′ end. The clone-finder software, developed by the HUSAR Biocomputing Service Group at the German Cancer Research Center (Heidelberg, Germany) (http://genome.dkfz-heidelberg.de; accessed January 16, 2003) facilitated the selection. The clones were purchased from the German Resource Center and Primary Database (RZPD; Berlin and Heidelberg).
Preparation of the IronChip microarray platform
The preparation of the IronChip microarray platform, which includes amplification, spotting, and attachment of the cDNAs, is described elsewhere.53 The same reference outlines the use of positive and negative hybridization controls integrated into the analysis to determine the cutoff signals for noise as well as the cutoff ratio for differential expression on the IronChip.
Synthesis of fluorescent cDNA probes
Fluorescent cDNA probes were synthesized from 5 μg total RNA by means of a linear mRNA amplification protocol, exactly as described in http://cmgm.stanford.edu/pbrown/protocols/ampprotocol_3.html(accessed January 16, 2003). Subsequently, 3 μg T7 RNA polymerase–amplified antisense RNA was subjected to a direct labeling reaction by incorporation of cyanin 3 (Cy3) and Cy5 fluorescent dyes (Cy3 or Cy5) with the use of random primers (http://cmgm.stanford.edu/pbrown/protocols/4_human_RNA.html; accessed January 16, 2003).53
At least 2 independent cell culture experiments were performed for each experimental condition tested. Cy3 fluorescent dyes were incorporated into the cDNA synthesized from the control sample, and Cy5 fluorescent dyes into cDNA synthesized from the experimental sample and vice versa. This “dye switch” helps to eliminate technical artifacts that derive from the biophysical properties of the 2 different dyes. Genes were scored as differentially expressed only if they displayed a consistent regulatory pattern in such dye-switch experiments.
Microarray analysis
The microarrays were immersed at 42°C in 6 × standard saline citrate (SSC)/0.5% sodium dodecyl sulfate (SDS)/1% bovine serum albumin (BSA) for 40 minutes and subsequently washed extensively with ddH2O at room temperature. Prior to hybridization, the spotted polymerase chain reaction (PCR) products were denatured by immersing the slides at 95°C in double-distilled (dd) H2O for 2 minutes. Excess of liquid was removed from the slides by centrifuging them briefly at 715g in a microtiter plate centrifuge (Z320; Hermle, Wehingen, Germany). Prior to hybridization, the purified Cy3- and Cy5-labeled cDNAs were mixed; 5 μg polydeoxyadenosine (poly(dA)) and 1 μg human Cot1 DNA (both Gibco Invitrogen, Carlsbad, CA) were added and subsequently evaporated in a vacuum Concentrator 5301 (Eppendorf, Hamburg, Germany) at 60°C. The resulting pellet was dissolved in 12 μL hybridization buffer (50% formamide/6 × SSC/0.5% SDS/5 × Denhardt) and denatured by incubating at 95°C for 2 minutes. The probe was then transferred onto the array under a 24 × 24 mm coverslip and incubated in a humid chamber (GeneMachines, San Carlos, CA) containing 2 × SSC drops for providing humidity. Hybridization was performed for 12 to 16 hours in a 42°C water bath (GFL, Burgwedel, Germany).
After hybridization, the microarrays were washed in 0.1 × SSC/0.1% SDS for 10 minutes and twice with 0.1 × SSC for 5 minutes (on an orbital shaker), followed by a brief immersion of the slides in ddH2O. Finally, the washed slides were dried by centrifuging them briefly at 715g in a microtiter plate centrifuge (Z320, Hermle). All washing steps were performed at room temperature.
Scanning and data analysis
All microarrays were scanned on a GenePix 4000B Microarray Scanner (Axon Instruments, Union City, CA). For each microarray, individual laser power and photomultiplier settings were used, allowing all signals to remain in the linear range of the scanner. Separate scan images for Cy3 and Cy5 were produced and analyzed by means of the ChipSkipper microarray data evaluation software (http://pc-ansorge11.embl-heidelberg.de/ chipskipper; accessed January 16, 2003). Intensity values for each spot were calculated by subtraction of the local background surrounding the spot. All spots were used for the calculation of a linear regression line. The regression line's parameters (offset, slope) were used for normalization. The resulting data were analyzed in Excel (Microsoft, Redmond, WA). At least 2 independent cell culture experiments were performed for each experimental condition tested and analyzed on the IronChip (version 2.0). For the bioinformatic analysis of the data, ratios of all the triplicate spots representing one cDNA were averaged. For those genes that are represented by multiple cDNA clones on the IronChip, the average of the ratios of those different clones was calculated. The standard deviation for each resulting ratio was determined. Genes listed in Table 6 represent those that have been scored as differentially expressed in all the experiments performed for a specific treatment. Genes are scored as differentially expressed if the calculated ratios exceed the ratio cutoff value, defined by the use of positive spike-in controls,53 For most experiments, this value lies between 1.4- and 1.7-fold.
Cell culture, RNA extraction, and Northern analysis
The maintenance of cultured HeLa cells and the treatments with 100 μM hemin, 100 μM ferric ammonium citrate, 100 μM desferrioxamine, 100 μM H2O2, and 100 μM SNP were performed as described previously.27 All treatments were performed for 8 hours. The establishment and maintenance of the HFE-overexpressing cell line as well as the experimental conditions of HFE overexpression are described in Riedel et al.47 Total RNA from HeLa cells was extracted by means of RNAclean (Hybaid-AGS, Heidelberg, Germany) according to the manufacturer's instructions.
For Northern analysis, 10 μg total RNA was separated on a 1% formaldehyde agarose gel and blotted onto a Nylon membrane (Nytran N; Schleicher and Schuell, Dassel, Germany). The membrane was subsequently hybridized to radioactively labeled probes in Church buffer.54 The signals obtained were quantified on a Fluoroimager (Molecular Dynamics, now Amersham Biosciences, Piscataway, NJ).
Sucrose gradient analysis
Results
Validation of the human IronChip
We first established a cDNA microarray platform that represents a selection of human genes that are directly involved in iron metabolism or that play a role in interlinked pathways such as copper metabolism, NO metabolism, the redox pathway, stress responses, selenium metabolism, or cell growth (IronChip). In addition, we included control genes that are not expected to be affected by the experimental conditions, as well as genes that are not represented in the human genome and hence serve as negative (background) controls and that can be used as so-called spike-in controls for standardization purposes.53 For version 2.0 of the IronChip, 113 different human genes represented by up to 3 independent cDNA clones were selected. The names of these genes and their GenBank accession numbers are shown athttp://www.embl-heidelberg.de/ExternalInfo/hentze/suppinfo.html. In addition, GenBank accession numbers are included in the text for all IronChip genes mentioned.
To assess whether the IronChip reflects changes in mRNA levels in response to iron perturbations, HeLa cells were either iron loaded by treatment with 100 μM hemin (H) for 8 hours, or made iron deficient by incubation with 100 μM desferrioxamine (D) for 8 hours. Total RNA was purified from the treated cells, labeled with Cy3 (D) and Cy5 (H), or vice versa (“Materials and methods”), and analyzed on the IronChip. The results of these experiments are shown in Figure 1 and Table1. As expected, TfR1 (NM_003234) mRNA levels are increased in iron-deficient cells, consistent with a stabilization of the TfR1 mRNA by IRP binding to its 3′UTR.57,58 We further observe an approximately 2-fold increase in the IRE-containing splice variant of DMT1/DCT1/Nramp2 (AB004857) mRNA, consistent with the notion that IRP binding to the IRE in the 3′UTR stabilizes this mRNA.59 In hemin-treated cells, we observe a strong increase of heme oxygenase-1 (HO-1) (X06985) mRNA, which encodes a critical enzyme in heme breakdown.60This result confirms earlier findings in cultured pig alveolar macrophages and in a human leukemia cell line.61,62L-ferritin (M11147) mRNA expression is also increased in hemin-treated cells, while H-ferritin (M11146) mRNA levels remain unchanged. A comparable result was obtained in rat liver after iron administration.56 Housekeeping genes, like glyceraldehyde phosphate dehydrogenase (GAPDH) (M33197) or β-actin (X00351), are not affected (Figure 1; Table 1). These results show that the microarray analysis on the IronChip accurately reflects the cellular responses to iron perturbations that have been observed earlier. To further validate this approach, Northern blots were performed for 8 selected genes and quantitated by phosphoimaging (Figure 1C). The close correlation between the results obtained by microarray analysis and Northern blotting confirms that the IronChip provides a reliable tool for the qualitative and quantitative analysis of gene expression in human iron metabolism.
Gene-expression profiles from iron-manipulated HeLa cells.
HeLa cells were treated with 100 μM hemin (H) or with 100 μM desferrioxamine (D) for 8 hours, and total RNA was purified from the cells. Fluorescent probes synthesized from total RNA derived from hemin-treated cells were labeled with Cy5-modified deoxyuridine 5-triphosphates (dUTPs), and those synthesized from total RNA derived from desferrioxamine-treated cells were labeled with Cy3-modified dUTPs; all were analyzed on the IronChip. (A) Virtual IronChip. Colors correspond to the calculated compensated ratios. Red spots represent genes with increased mRNA levels in hemin-treated cells. Green spots represent genes with increased mRNA levels in desferrioxamine-treated cells. Yellow spots represent genes that are equally expressed in both conditions tested. Selected genes are annotated. (B) Scatter plot analysis. Signals corresponding to the desferrioxamine-treated sample are represented on the y-axis. Signals corresponding to the hemin-treated sample are represented on the x-axis. In the experiment shown here, a gene is considered to be differentially expressed if the H/D ratio is calculated above 1.4 or below 0.7. Genes with a calculated H/D ratio above 1.4 (1.4-fold) are shown in red. Genes with a calculated H/D ratio below 0.7 (−1.4-fold) are represented in green. Housekeeping genes, like GAPDH and actin, are represented in yellow, and negative controls are shown in blue. Positive spike-in controls53 that have been added in equal amounts to the total RNA of hemin- and desferrioxamine-treated cells and thus by definition should not appear regulated are shown in pink. (C) Northern blot analysis of selected mRNAs. The ratios of signals obtained in H- and D-treated cells (as quantified on a Fluoroimager) are indicated.
Gene-expression profiles from iron-manipulated HeLa cells.
HeLa cells were treated with 100 μM hemin (H) or with 100 μM desferrioxamine (D) for 8 hours, and total RNA was purified from the cells. Fluorescent probes synthesized from total RNA derived from hemin-treated cells were labeled with Cy5-modified deoxyuridine 5-triphosphates (dUTPs), and those synthesized from total RNA derived from desferrioxamine-treated cells were labeled with Cy3-modified dUTPs; all were analyzed on the IronChip. (A) Virtual IronChip. Colors correspond to the calculated compensated ratios. Red spots represent genes with increased mRNA levels in hemin-treated cells. Green spots represent genes with increased mRNA levels in desferrioxamine-treated cells. Yellow spots represent genes that are equally expressed in both conditions tested. Selected genes are annotated. (B) Scatter plot analysis. Signals corresponding to the desferrioxamine-treated sample are represented on the y-axis. Signals corresponding to the hemin-treated sample are represented on the x-axis. In the experiment shown here, a gene is considered to be differentially expressed if the H/D ratio is calculated above 1.4 or below 0.7. Genes with a calculated H/D ratio above 1.4 (1.4-fold) are shown in red. Genes with a calculated H/D ratio below 0.7 (−1.4-fold) are represented in green. Housekeeping genes, like GAPDH and actin, are represented in yellow, and negative controls are shown in blue. Positive spike-in controls53 that have been added in equal amounts to the total RNA of hemin- and desferrioxamine-treated cells and thus by definition should not appear regulated are shown in pink. (C) Northern blot analysis of selected mRNAs. The ratios of signals obtained in H- and D-treated cells (as quantified on a Fluoroimager) are indicated.
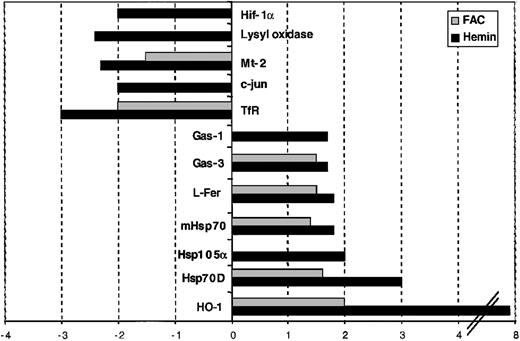
In addition to those genes that are directly involved in iron metabolism, we found some additional genes to be regulated. In iron-replete cells, 3 members of the heat-shock protein (hsp) family (hsp70D [M11717], hsp105α [ΑΒ003334], and the mitochondrial [m] hsp70 [L11066]) and 3 genes mediating growth effects (c-myc [V00568] and growth arrest–specific [Gas]–1 [L13698] and Gas-3 [L03203]) show increased mRNA levels. In iron-deficient cells, we observe a robust increase in the amount of mRNA encoding metallothionein (mt)–2 (X97260) and in lysyl oxidase (lox) (M94054), and a small but consistent increase in the mRNA level of hypoxia-inducible factor (Hif)–1α (NM_001530). Furthermore, the expression of c-jun (J04111), a gene involved in cell proliferation, is increased in iron-deficient cells (Table 1). Note that the quantitative changes in gene expression in this experiment are monitored as the sum of increased and decreased expression for a given gene in both conditions tested.
Gene-expression profiles derived from hemin- and ferric ammonium citrate (FAC)–treated HeLa cells
We next assessed HeLa cells that were treated with 2 different sources of iron, hemin (ferric protoporphyrin IX) or ferric ammonium citrate (FAC), to address 2 questions: first, whether the iron-loaded state resulting from both treatments elicited a common pattern in the expression profiles; second, whether these 2 similar treatments could be discriminated by diagnostic features of the gene-response patterns.
Subconfluent HeLa cells were treated with either 100 μM hemin or 100 μM FAC for 8 hours. Untreated HeLa cells were used as a control for both. As can be seen in Figure2 and Table2, the expression profiles derived from hemin- and FAC-treated cells closely resemble each other. Genes that are differentially expressed after the treatment with both iron sources include HO-1, Hsp70D, mhsp70, L-ferritin, Gas-3, TfR-1, and Mt-2. The increased expression of the first 5 and the decreased expression of the last 2 genes appear to define a common denominator that is the hallmark of the iron-loaded state. In general, the magnitude of the expression change is lower in the FAC-treated cells. Note that the induction of HO-1 mRNA is highly pronounced in hemin-treated cells, consistent with the function of HO-1 in heme breakdown.63 Furthermore, a more than 1.4-fold change in mRNA levels (which we used as the minimum defining cutoff between regulated and unregulated genes; “Materials and methods”) of hsp105α, c-jun, lysyl oxidase, Gas-1, and Hif-1α mRNAs was unique to the hemin-treated cells and was not observed following FAC administration. These data show that microarray analysis with the IronChip can identify both the common features that identify cellular iron load as well as the distinct features that allow the discrimination between the sources of iron.
Gene-expression profiles derived from hemin- and ferric ammonium citrate (FAC)–treated HeLa cells.
Comparison of the gene-expression profiles of hemin- and FAC-treated HeLa cells. Genes that show increased expression in hemin- and/or FAC-treated cells are shown in positive numbers and those with decreased expression in negative numbers.
Gene-expression profiles derived from hemin- and ferric ammonium citrate (FAC)–treated HeLa cells.
Comparison of the gene-expression profiles of hemin- and FAC-treated HeLa cells. Genes that show increased expression in hemin- and/or FAC-treated cells are shown in positive numbers and those with decreased expression in negative numbers.
Gene-expression profiles of H2O2- and sodium nitroprusside (SNP)–treated HeLa cells
H2O2 treatment and iron deficiency both activate IRP-122-28 and trigger posttranscriptional changes in the expression of IRE-regulated mRNAs. As a consequence, H- and L-ferritin mRNA translation is repressed, and transferrin receptor mRNA levels increase in both conditions.64 We next recorded the broader gene-expression profile from H2O2-treated HeLa cells and assessed whether it can be distinguished from the gene-expression profile derived from iron-deficient cells. HeLa cells were exposed to 100 μM H2O2 for 8 hours, and total RNA was subsequently analyzed on the IronChip in comparison with total RNA from untreated control cells. H2O2 treatment induced increased HO-1 and TfR-1 mRNA levels (Table3). The induction of HO-1 by H2O2 has been reported previously.65 By contrast, we observe neither the regulation of the IRE-containing DMT1/DCT1/Nramp2 mRNA nor any regulation of those genes that are seen regulated in iron-deficient HeLa cells (Table 4). Thus, the expression profile derived from H2O2-treated HeLa cells is clearly distinct from the gene-expression profiles obtained from iron-deficient HeLa cells.
In addition to iron perturbation and oxidative stress, nitric oxide also affects IRP activity and the regulation of IRE-containing mRNAs.22,24,25,27,28 Thus, we also tested the effect of sodium nitroprusside (SNP), which releases nitrosonium ions (NO+), on the regulation of the genes immobilized on the IronChip. NO+ has been suggested to cause theS-nitrosylation of critical thiol groups, to prevent the binding of IRP-2 to IREs, and to result in TfR-1 mRNA degradation.66
HeLa cells were treated with 100 μM SNP for 8 hours; total RNA was extracted and analyzed on the IronChip in comparison with an untreated control sample. As expected, SNP treatment reduces TfR mRNA levels (Table 5). In addition, the mRNA levels of the IRE-containing splice variant DMT1/DCT1/Nramp2 are also reduced. The approximately 2-fold increase of DMT1/DCT1/Nramp2 mRNA in iron deficiency (when compared with hemin-treated cells; Figure 1 and Table 1) and its reduced expression in response to SNP are consistent with an IRP-mediated regulatory mechanism of DCT1/DMT1/Nramp2 mRNA stability. In contrast to iron-manipulated HeLa cells, hsp70D is coregulated with TfR-1 and DCT-1 in SNP-treated cells. SNP treatment strongly induces HO-1 and affects the expression of the metallothioneins 1 and 2 (Table 5). The regulation of heterogeneous nuclear ribonucleoprotein D–like protein JKTBP (D89092) and of the prion protein (M13899) detected after SNP treatment of HeLa cells was not observed in iron-perturbed or H2O2-treated HeLa cells. With the exception of IRP target genes, the expression profile obtained in SNP-treated HeLa cells is clearly distinct from those of iron-manipulated or H2O2-treated HeLa cells.
HFE expression and iron deficiency yield highly similar gene-expression profiles
Previous work showed that induced HFE expression in mammalian cells resulted in decreased iron uptake from diferric transferrin, IRP activation, and the regulation of IRP target mRNAs.43,46,47 67-70 These findings suggested that HFE expression in transfected cells affects the regulatory “labile iron pool” in a way that is similar to desferrioxamine-induced iron starvation. To more globally record the responses of transfected HeLa cells to induced HFE expression and to compare these with the response of desferrioxamine-treated cells, additional microarray analyses were performed.
HeLa cells were stably transfected with the human HFE cDNA under the control of a doxycyclin-responsive promoter.47 The absence of doxycyclin induces HFE expression,47 whereas the transgene is not transcribed following the addition of doxycyclin to the culture medium. Total RNA extracted from doxycyclin-treated and untreated HeLa cells bearing the HFE transgene was used for fluorescent cDNA synthesis and subsequent analysis on the IronChip. As expected, the HFE mRNA is strongly induced in the absence of doxycyclin (Table4). When HFE expression is induced, TfR-1, c-jun, lysyl oxidase, and Mt-2 mRNAs are increased, whereas the mRNA levels of HO-1, Hsp70D, Hsp105a, mHsp70, L-ferritin (L-fer), and Gas-3 decrease. These data were confirmed by Northern analysis (Figure3A). Doxycyclin treatment of nontransfected HeLa cells did not affect the regulation of IronChip genes (data not shown). When HeLa cells were treated with 100 μM desferrioxamine, TfR-1, c-jun, and lysyl oxidase mRNA levels increased. Decreased mRNA levels were found for HO-1, Hsp70D, Hsp105a, mHsp70, L-fer, Gas-1, Gas-3, and c-myc (Table 4).
Gene-expression profiles obtained from desferrioxamine-treated HeLa cells and HFE-expressing HeLa cells.
(A) Total RNA derived from HeLa cells treated with 100 μM desferrioxamine (D) for 8 hours or total RNA derived from HeLa cells that express HFE from an inducible transgene was analyzed on the IronChip in comparison with control samples. The average ratios of differentially expressed genes and their standard deviations are shown. (B) Comparison of the gene-expression profiles from desferrioxamine-treated and HFE-expressing HeLa cells. Genes with increased expression in desferrioxamine-treated and/or HFE-expressing cells are shown in positive numbers and those with decreased expression in negative numbers.
Gene-expression profiles obtained from desferrioxamine-treated HeLa cells and HFE-expressing HeLa cells.
(A) Total RNA derived from HeLa cells treated with 100 μM desferrioxamine (D) for 8 hours or total RNA derived from HeLa cells that express HFE from an inducible transgene was analyzed on the IronChip in comparison with control samples. The average ratios of differentially expressed genes and their standard deviations are shown. (B) Comparison of the gene-expression profiles from desferrioxamine-treated and HFE-expressing HeLa cells. Genes with increased expression in desferrioxamine-treated and/or HFE-expressing cells are shown in positive numbers and those with decreased expression in negative numbers.
These data sets reveal striking similarities between the gene responses elicited by HFE expression and desferrioxamine treatment. This conclusion applies both to IRP target genes and non-IRP target genes (Figure 3B; Table 4). Both gene-response patterns differ significantly from those elicited by, for example, SNP and H2O2 treatment (Figure 3; Table 6). Thus, we conclude that HFE expression in transfected HeLa cells triggers cellular iron deficiency.
Monitoring translational responses to iron perturbations by microarray analyses
Traditionally, microarray analyses are employed to monitor changes in steady-state mRNA levels. Because gene regulation in response to iron perturbations also prominently involves translational control in the absence of concomitant changes in mRNA levels, we wanted to adapt our experimental approach to reveal regulation at the translational level.
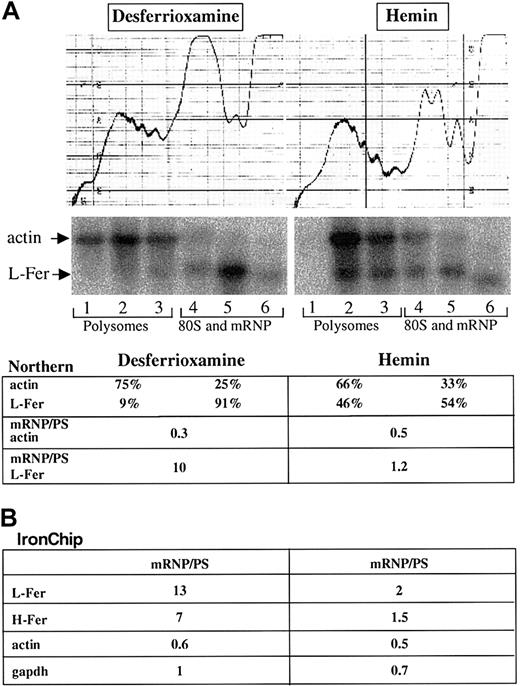
Cytoplasmic extracts were prepared from HeLa cells that were treated with either hemin or desferrioxamine, and subjected to linear sucrose gradient centrifugation (“Materials and methods”). Six fractions were prepared from each sample; total RNA was extracted and initially analyzed by Northern blotting. As shown in Figure4, L-ferritin mRNA is enriched in the polysomal fractions at the bottom of the sucrose gradient in hemin-treated cells, as previously observed.56 In iron-deficient cells, L-ferritin mRNA is enriched in the fractions that contain monosomes (80S) and messenger ribonucleic proteins (mRNPs), consistent with previous findings that IRP inhibits the translation of ferritin mRNAs by interfering with the first steps of the translation-initiation process.71 As a control, actin mRNA remains localized in the polysomal fractions in both hemin- and desferrioxamine-treated cells.
Microarray assessment of iron-mediated translational control.
Cytoplasmic extracts from hemin- and desferrioxamine-treated HeLa cells were sedimented through a 10%-40% sucrose gradient (see “Materials and methods”). The profile on the top denotes the A254 absorption profile. The positions of polysomes, monosomes (80S), and mRNPs are indicated. (A) Northern blot analysis was performed with total RNA extracted from the 6 individual fractions obtained from the sucrose gradient. The Northern blot was sequentially probed with radiolabeled probes for actin and L-ferritin. The signals obtained from the individual fractions were quantified on Fluoroimager (Molecular Dynamics) and the signals in the polysomes and the 80S and mRNP fractions were calculated as a percentage of the sum of signals in all lanes. The ratio of the 80S and mRNP fractions to the polysomal fractions (PS) is indicated. (B) The 3 fractions containing polysomal (PS) and the 3 fractions containing the monosomal and mRNP-derived RNA were pooled for each condition and analyzed on the IronChip. The regulatory ratio between mRNP and polysomal fractions is indicated for L-ferritin (L-fer), H-ferritin (H-fer), actin, and GAPDH.
Microarray assessment of iron-mediated translational control.
Cytoplasmic extracts from hemin- and desferrioxamine-treated HeLa cells were sedimented through a 10%-40% sucrose gradient (see “Materials and methods”). The profile on the top denotes the A254 absorption profile. The positions of polysomes, monosomes (80S), and mRNPs are indicated. (A) Northern blot analysis was performed with total RNA extracted from the 6 individual fractions obtained from the sucrose gradient. The Northern blot was sequentially probed with radiolabeled probes for actin and L-ferritin. The signals obtained from the individual fractions were quantified on Fluoroimager (Molecular Dynamics) and the signals in the polysomes and the 80S and mRNP fractions were calculated as a percentage of the sum of signals in all lanes. The ratio of the 80S and mRNP fractions to the polysomal fractions (PS) is indicated. (B) The 3 fractions containing polysomal (PS) and the 3 fractions containing the monosomal and mRNP-derived RNA were pooled for each condition and analyzed on the IronChip. The regulatory ratio between mRNP and polysomal fractions is indicated for L-ferritin (L-fer), H-ferritin (H-fer), actin, and GAPDH.
For microarray analyses, we pooled the total RNA derived from the polysomal fractions 1 through 3 and the total RNA derived from the monosomal/mRNP fractions 4 and 5. For each condition (hemin and desferrioxamine treatment), the polysomal and monosomal/mRNP fractions were labeled with different fluorescent dyes and hybridized to the IronChip. Similarly to the Northern blots, the data (Figure 4B) clearly reveal the translational regulation of L-ferritin mRNA and the lack of actin mRNA regulation. Similarly to L-ferritin, the IronChip also reflects the translational control of the H-ferritin mRNA in response to iron perturbation (Figure 4). Mitochondrial aconitase mRNA and eALAS mRNA, which have also been shown to be translationally regulated by an IRE in their mRNAs,9,11 are not expressed at sufficiently high levels in HeLa cells to allow a reliable assessment of their ribosome association. Likewise, the IRE-containing ironregulated transporter (IREG)–1/ferroportin/metal transporter protein–1 (MTP-1) is expressed mainly in duodenal enterocytes, macrophages, and the placenta,72-74 and its mRNA is undetectable in HeLa cells. No additional genes represented on the IronChip (version 2.0) show an altered translation of their mRNAs. We conclude that microarray analyses with the IronChip can also be used to monitor iron-induced changes in mRNA translation.
Discussion
We have studied the genetic responses of a human cell line to changes in iron metabolism employing a newly developed cDNA-based microarray platform (IronChip). With this approach, novel insights into human iron metabolism were obtained. In addition, the results show the utility of the IronChip as a versatile tool to investigate a broad range of questions regarding the physiology of human iron metabolism and diseases that result from its aberrations.
New insights into human iron metabolism
Human HeLa cells have served as an intensively characterized model system for the investigation of iron metabolism. We therefore chose HeLa cells to explore the utility of a specialized DNA microarray that represents 113 different human genes and that was expected to reveal insights into regulatory responses of human cells to iron deficiency, iron overload, HFE expression, and small signaling molecules. Table 6 provides a synopsis of these responses. Importantly, for the genes known to be regulated by iron, the microarray data are consistent with the existing literature. Moreover, many of the emerging results were confirmed by Northern blotting (Figures 1C and 3A), which in addition ascertained a surprisingly good performance of the microarray platform in yielding quantitatively accurate data.
Heme oxygenase 1 (HO-1) emerges as the most strongly responsive gene in our data set. HO-1 mRNA levels decrease in iron-deficient and in HFE-expressing cells, and increase in response to iron loading as well as to SNP and H2O2exposure (Table 6). HO-1 may thus represent a central stress-response gene in the iron-regulatory network. Hsp70D appeared as another strongly responsive gene. Although it did not show regulation in H2O2-treated cells, it responded to all other experimental perturbations. It is conceivable that additional genes (perhaps including Hsp70D) might have responded to higher concentrations of H2O2 or might have responded if a different cell line had been tested. Only 2 genes displayed H2O2 regulation under our experimental conditions (Table 6). However, Hsp70D mRNA levels responded to iron deficiency (and to HFE expression) and iron overload more strongly than TfR1 mRNA levels, which are often considered to be the classic regulatory response to these challenges. It is also notable that Gas-3 and mHsp70 are consistently regulated by altered cellular iron supply. The latter result establishes mitochondrial Hsp70 (mHsp70) as a human iron-responsive gene and reveals that this regulation appears to be conserved between human and yeast: yeast mutants (ssc2-1) of mHsp70 show increased cellular iron uptake, and the excess iron accumulates in the mitochondria.75 It will be important to explore whether the induction of mHsp70 in iron-loaded cells fulfills a protective or a regulatory function in human cells.
Previous studies indicated that the heterologous or induced expression of the HFE protein negatively affects cellular iron uptake via the transferrin receptor43,46,47,67-70 and hence triggers an iron-deficiency response by the IRE/IRP regulatory network.47,69 We find that desferrioxamine treatment and the induction of HFE expression yield nearly identical responses of the 113 genes represented on the IronChip. This allows the conclusion that not only does HFE expression in this experimental system trigger an iron-deficiency response by the IRE/IRP network, but that the iron-deficiency state induced by HFE affects every regulatory system that is also reached by desferrioxamine.76-78 Since the list of regulated genes includes non-IRP target genes such as the growth-effect genes (eg, c-jun, Gas-3) and stress-response genes (eg, HO-1, Hsp70D, Hsp105α, mHSP70), we suspect that at least a part of these responses may be mediated transcriptionally. Regarding the putative function of HFE as an inducer of cellular iron deficiency, one needs to consider that we expressed HFE without induced coexpression of its heterodimerization partner β2-microglobulin. It has been reported that the coexpression of both yields effects opposite to those arising from the induction of HFE alone.48 From a more methodological perspective, the close resemblance of the desferrioxamine- and HFE-induced gene regulatory responses highlights an important application of microarray analyses in studying iron metabolism: the identification of similar profiles elicited by 2 different stimuli makes it possible to reveal common effector functions.
The similarity between the genetic responses to iron overload by hemin and ferric ammonium citrate (FAC) was predicted and has been confirmed with the IronChip (Figure 2; Table 2). Most genes that respond to FAC also respond to hemin, and the latter response is usually slightly stronger. As an exception to this, the HO-1 mRNA response to hemin is far stronger than to FAC. Considering the biological role of HO-1 in heme breakdown, a more profound induction of HO-1 mRNA by hemin is not surprising. We suggest that the magnitude of the HO-1 response in relation to the responses by mHsp70, L-fer, Gas-3, TfR1, and Mt-2 is diagnostic for hemin-induced versus FAC-induced iron overload. More generally, this analysis provides an example for the possibility of discriminating between 2 related stimuli by IronChip analysis.
The IronChip provides a versatile tool for the analysis of iron metabolism
As illustrated, the human IronChip was validated as a reliable assay system to identify the responses of more than 100 genes involved in iron metabolism and interlinked biological pathways. We also show that in combination with sucrose gradient analysis, the IronChip successfully identifies genetic responses at the translational level (Figure 4). This is particularly pertinent for the study of mammalian iron metabolism.13
Compared with far more comprehensive cDNA or oligonucleotide-based microarrays, the IronChip offers only limited chances to identify “new genes” that are regulated by particular stimuli. For this reason, we believe that more comprehensive microarrays can offer helpful entry points for microarray studies and help to identify genes to be included on the IronChip, as was done during the original design and as is being used for updated versions. On the other hand, we believe that both the gene verification and technical performance parameters of the IronChip compare favorably with those of larger arrays. All the genes represented on our arrays have been sequence verified from both ends. Owing to the limited number of genes, each gene can be spotted multiple times and in different locations of the chip, and many genes are represented by up to 3 independent cDNA clones. This redundancy offers additional controls for gene specificity.
A major application of the IronChip lies in the identification of gene regulatory patterns that provide a characteristic “fingerprint” of a particular treatment or genetic alteration. For this application, the technical quality of the data is critical, particularly the ability to score limited quantitative differences reliably and reproducibly. The recognition of similarities or differences in the genetic responses to different stimuli can be highly informative, and we suggest that the IronChip could also prove useful in the analysis of human patient samples. Recently, we increased the number of different relevant genes that are represented on the IronChip to nearly 300 (version 3.0) (data not shown). This should further enhance its utility in defining precise gene-response patterns and hence the ability to ultimately understand the networks that operate within human iron metabolism. Moreover, we also established an analogous microarray platform with murine cDNAs (data not shown). The murine IronChip will not only facilitate cross-species comparisons, but in particular facilitate access to the growing pool of genetic murine model systems for human diseases of iron metabolism and to integrate findings from animal models into our understanding of human iron physiology and pathophysiology.
We thank the Resource Center and Primary Database (RZPD) for the supply of IMAGE clones.
Prepublished online as Blood First Edition Paper, September 26, 2002; DOI 10.1182/blood-2002-07-2140.
Supported by funds from the Gottfried Wilhelm Leibniz Prize to M.W.H., which were used to establish the IronChip.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Matthias W. Hentze, European Molecular Biology Laboratory, Meyerhofstrasse 1, D-69117 Heidelberg, Germany; e-mail: hentze@embl.de.