Abstract
Chronic active Epstein-Barr virus infection (CAEBV) is a rare disease in which previously healthy persons develop severe, life-threatening illness. Mutations in the perforin gene have been found in familial hemophagocytic lymphohistiocytosis, which shares some features with CAEBV. We studied a patient who died at age 18, 10 years after the onset of CAEBV. The patient had high titers of antibodies to EBV, EBV RNA in lymph nodes, T-cell lymphoproliferative disease, and hemophagocytic lymphohistiocytosis. DNA sequencing showed novel mutations in both alleles of the perforin gene that resulted in amino acid changes in the protein. The quantity of the native form of perforin from the patient's stimulated peripheral blood mononuclear cells (PBMCs) was extremely low and immunoblotting showed accumulation of an uncleaved precursor form of perforin. Stimulated PBMCs from the patient were defective for Fas-independent cytotoxicity. These data imply that mutations in this patient resulted in reduced perforin-mediated cytotoxicity by his lymphocytes. This is the first case in which perforin mutations have been shown to result in accumulation of the uncleaved, immature form of perforin. Mutations in the perforin gene are associated with some cases of CAEBV with hemophagocytic lymphohistiocytosis.
Introduction
Epstein-Barr virus (EBV) is a member of the human herpesvirus family that infects over 95% of the United States population.1 Most infections occur in childhood and are asymptomatic; infection of adolescents and young adults with EBV often results in infectious mononucleosis. EBV is associated with a spectrum of lymphoproliferative diseases in patients with congenital or acquired immunodeficiency.
Chronic active EBV infection (CAEBV) is a rare and often fatal disorder that occurs in previously healthy persons and seemingly immunocompetent persons.2 The disease has been defined by the presence of 3 features.3,4 First, patients have a severe progressive illness that began as a primary EBV infection, or is associated with abnormal EBV-specific antibody titers that include markedly elevated antibodies to viral capsid antigen (VCA) and early antigens (EAs). Second, histology shows evidence of major organ involvement such as lymphadenitis, hemophagocytosis, meningoencephalitis, or persistent hepatitis. Third, elevated EBV DNA, RNA, or proteins are demonstrable by in situ hybridization or immunohistochemical staining of affected tissues. Recent studies showed that patients with CAEBV can also have markedly elevated levels of EBV DNA in the peripheral blood and this criterion has been used diagnostically in some cases.5 Patients with CAEBV often develop a progressive cellular and humoral immunodeficiency with pancytopenia and hypogammaglobulinemia that renders them susceptible to opportunistic infections or B- or T-cell lymphoproliferative disease.3 Therapy for CAEBV is unsatisfactory and, at best, progression of disease is temporarily delayed.
The etiology of CAEBV is unknown. Two studies suggested that persons with CAEBV were infected with unusual lytic strains of virus.6,7 However, the finding of the same lytic strain of EBV in the unaffected father of one of the patients, and in healthy controls,8 suggests that other factors, including inherited abnormalities in the response to EBV, contribute to the pathogenesis of the disease. Four observations favor a genetic cause for CAEBV. First, CAEBV is rare in the United States, but relatively common in Japan, Korea, and China. Most patients reported to have fulminant EBV-positive T-cell lymphoproliferative disease following acute and/or chronic EBV infection have been Asian in origin.9 Second, specific mutations in the signaling lymphocyte activation molecule (SLAM) associated protein (SAP) gene have been identified in boys with a disease that shares many of the features of CAEBV, the X-linked lymphoproliferative disease (XLPD).10-12 Third, 2 studies showed that cytotoxic T lymphocyte (CTL) or natural killer (NK) cell activity was reduced in patients with CAEBV and in their parents.13,14 Fourth, gene mutations and polymorphisms have been associated with severe infections with herpesviruses including EBV.15-17 Taken together, these findings suggest that a genetic abnormality could underlie some cases of CAEBV.
Here we describe a patient with CAEBV in whom we documented a defect in cytotoxic activity in association with markedly reduced levels of the native form of perforin in his stimulated peripheral blood mononuclear cells (PBMCs). Sequence analysis showed him to have compound heterozygous mutations affecting both alleles of the perforin gene. These findings indicate that some cases of CAEBV are due to defects in the perforin gene.
Materials and methods
Cell culture
Blood was drawn after obtaining informed consent from the patient, his parents, and additional patients with CAEBV studied at the National Institutes of Health (Bethesda, MD) under a protocol approved by the institutional review board of the National Institute of Allergy and Infectious Diseases. Blood was also obtained from anonymous healthy blood bank donors and from patients in Japan with CAEBV in whom patient identifiers had been removed from the specimens. Cryopreserved PBMCs were thawed, cultured in RPMI 1640 containing 10% fetal bovine serum supplemented with 10 IU/mL interleukin 2 (IL-2; Roche Molecular Biochemicals, Mannheim, Germany) and 1 × phytohemagglutinin (PHA; Invitrogen, Carlsbad, CA) for 2 days, followed by IL-2 without PHA for 2 weeks. PBMCs and EBV-transformed B cells from patients in Japan with CAEBV5 were also studied.
Cytokine assays
Cells were stimulated for 36 hours with plate-bound anti-CD3 antibody (1 μg/mL, UCHT1; BD Pharmingen, San Diego, CA) plus anti-CD28 antibody (1:5000, CD28.2; BD Pharmingen) and cell culture supernatants were collected as previously reported.18 IL-4, -5, -10, interferon-γ (IFN-γ), and tumor necrosis factor α (TNF-α) levels were all assayed with a Luminex 100 multiplex analyzer, with Luminex IS analysis software (Luminex, Austin, TX) using multianalyte microsphere kits (LINCO Research, St Charles, MO). IL-13 levels were determined by enzyme-linked immunosorbent assay (ELISA; Pierce-Endogen, Pierce Biotechnology, Rockford, IL).
Real-time PCR for EBV
Real-time quantitative polymerase chain reaction (PCR) assay with a fluorogenic probe was performed using a TaqMan PCR kit and a Model 7700 Sequence Detector (Applied Biosystems, Foster City, CA). A portion of the BamHI W fragment of EBV was amplified using forward primer 5′-GAGGGGGACCACTGCCCCTGG-3′ and reverse primer 5′-CGCTCTGATGCGACCAGA-3′ and detected with fluorogenic probe 5′-(6FAM)-TCCTGCAGCTATTTCTGGTCGCATCA-(TAMRA)-3′. The bcl-2 gene was amplified using forward primer 5′-CCTGCCCTCCTTCCGC-3′ and reverse primer 5′-TGCATTTCAGGAAGACCCTGA-3′ and detected with fluorogenic probe 5′-(6FAM)-CTTTCTCATGGCTGTCC-(TAMRA)-3′.
Reverse transcriptase (RT)–PCR and Southern blotting for EBV
Total RNA was isolated from stimulated PBMCs and cDNA was synthesized. Primers and probes for EBV genes and PCR conditions were used as previously described.19 PCR products were separated by electrophoresis, transferred to nylon membranes, and hybridized with [32P]-labeled oligonucleotide probes.
Sequencing of the perforin gene
DNA was extracted from stimulated PBMCs and the perforin gene was amplified by PCR and sequenced as previously described.20 Nucleotide positions of perforin are numbered according to GenBank accession number X13224. cDNA of perforin exon 3 was amplified by RT-PCR using forward primer (F577) 5′-AACTTTGCAGCCCAGAAG-3′, and reverse primer (ADR)20 5′-TTGCATCTCACCTCATGGGAAC-3′ and the sequence of the cDNA at nucleotides 577 and 1229 of perforin was determined.
Mutagenesis of the perforin gene and expression in cells
RNA was extracted from PBMCs of a healthy donor and cDNA was synthesized with reverse transcriptase. Perforin cDNA was amplified by PCR using forward primer 5′-ATTCTCGAGATGGCAGCCCGTCTGCTCCT-3′ and reverse primer 5′-TATGTCGACTCACCACACGGCCCCACTCC-3′. The PCR product was inserted into the EcoRI site of plasmid pCIneo (Promega, Madison, WI). Site-directed mutagenesis was performed to change nucleotide 577 from T to C (mother's perforin mutation), nucleotide 1229 from G to C (father's perforin mutation), and nucleotide 1122 from G to A (FHL mutation) using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Mutation of each clone was confirmed by DNA sequencing. 293T cells were transfected with plasmid(s) encoding perforin (2 μg/6-cm dish) using the Fugene 6 transfection system (Roche Biochemicals) and cell lysates were prepared 48 hours after transfection for immunoblotting or immunoprecipitation.
Results
Case report
A 7-year-old white boy presented with heterophile-positive infectious mononucleosis followed by persistent splenomegaly and lymphadenopathy. One year later he underwent splenectomy for hypersplenism; at surgery, mesenteric lymphadenopathy and chylous ascites were noted. Histiocytosis with erythrophagocytosis was demonstrated in the spleen. He was treated with adriamycin, vincristine, prednisone, and cyclophosphamide for lymphadenopathy and presumptive lymphoma. Three years later at age 11, at his first evaluation at NIH, he was found to have T-cell lymphoproliferative disease involving the liver, bone marrow, and a supraclavicular lymph node. His EBV serologies were markedly elevated with an anti-EBV VCA immunoglobulin G (IgG) titer of 20 480, anti-EA IgG titer of 20 480, and anti-EBNA IgG titer of 10, indicating the possibility that the lymphoproliferative process was driven by an EBV infection. Hypertriglyceridemia (2.8 mM) and hypofibrinogenemia (1.4 g/L) were noted. He was treated with acyclovir for 4 weeks without clinical benefit. On initial evaluation and over the ensuing several months, progressive pancytopenia and hypogammaglobulinemia were noted and he received intravenous immunoglobulin. The following year he presented with cholestatic hepatitis secondary to T-cell lymphoproliferative disease of the liver. Although he received interferon alpha, the T-cell lymphoproliferative process persisted and cutaneous leukocytoclastic vasculitis, pneumonia, and chronic sinusitis were noted. Computerized tomography showed reduction in the size of his abdominal and peripheral lymphadenopathy on prednisone and cyclophosphamide; these effects were sustained on prednisone and azathioprine. Lymphocyte phenotype of the peripheral blood showed 98.9% T cells and 0.5% B cells. Markedly elevated EBV titers (VCA IgG 20 480, EA-IgG 20 480) persisted, and a monoclonal IgG kappa gammopathy and Coombs positive hemolytic anemia were present. He died at age 18 due to lymphoproliferative disease and disseminated candida infection. Autopsy was limited to the liver, lymph nodes, lungs, and brain and showed polyclonal T-cell infiltration of the liver and mesenteric lymph nodes with loss of cortical, paracortical, and germinal center architecture. Rare T-cell infiltrates were present in the lungs and disseminated candidiasis involving the lungs, liver, and brain was noted.
The patient fit the diagnostic criteria of CAEBV3,4,21 based on (1) symptoms of persistent lymphadenopathy, hepatosplenomegaly, and bone marrow hypoplasia that began after infectious mononucleosis; (2) extremely high EBV-specific antibody titers; and (3) EBV-positive cells in the lymph nodes on in situ hybridization. He also fit the diagnostic guidelines for hemophagocytic lymphohistiocytosis22 based on his fever, splenomegaly, pancytopenia, hypertriglyceridemia, and histologic evidence of hemophagocytosis. However, he did not fit the criteria for familial hemophagocytic lymphohistiocytosis (FHL), since there was no family history of hemophagocytic lymphohistiocytosis. The patient's brother and both parents are EBV seropositive and have no history of EBV-associated or immunologic diseases. None of the other family members whose history could be ascertained, including the patient's aunts, uncles, grandparents, and maternal great-grandparents, have a history of EBV-associated or immunologic diseases. Furthermore, patients with FHL are usually diagnosed in infancy or early childhood and die within a few years of the diagnosis,23,24 whereas our patient lived for 18 years. Therefore, our patient was diagnosed with CAEBV, rather than FHL.
Pathology and immunohistochemistry
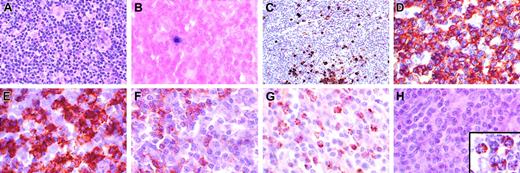
Multiple lymph node biopsies (cervical, mesenteric, splenic hilar) obtained during the course of the patient's disease showed similar histologic findings (Figure 1). The architecture was effaced by an infiltrate composed predominantly of small normal-appearing lymphocytes with admixed histiocytes. A similar infiltrate was seen in the bone marrow and liver. The spleen showed marked histiocytosis with erythrophagocytosis of red pulp.
Characterization of T-cell lymphoproliferative process in lymph node. (A) Architecture is diffusely infiltrated by small lymphocytes with admixed histiocytes. (B) In situ hybridization with an EBV-encoded RNA (EBER) probe shows rare positive small lymphocytes (< 1 per high power field) that correspond to the distribution of B cells in the same area. (C) CD20 stain shows scattered small positive lymphocytes and small aggregates, but absence of follicular structures. The majority of the lymphocytes are CD3+ (D), with a predominance of CD8+ cells (E), over CD4+ cells (F). Numerous lymphocytes are granzyme B positive (G), but negative for perforin (antibody KM585-P1-8; Kamiya Biochemical, Seattle, WA) (H). Inset in panel H shows positive perforin control in a patient with large granular lymphocyte leukemia. Original magnification, × 400, except panel C (× 250) and panel H inset (× 1000).
Characterization of T-cell lymphoproliferative process in lymph node. (A) Architecture is diffusely infiltrated by small lymphocytes with admixed histiocytes. (B) In situ hybridization with an EBV-encoded RNA (EBER) probe shows rare positive small lymphocytes (< 1 per high power field) that correspond to the distribution of B cells in the same area. (C) CD20 stain shows scattered small positive lymphocytes and small aggregates, but absence of follicular structures. The majority of the lymphocytes are CD3+ (D), with a predominance of CD8+ cells (E), over CD4+ cells (F). Numerous lymphocytes are granzyme B positive (G), but negative for perforin (antibody KM585-P1-8; Kamiya Biochemical, Seattle, WA) (H). Inset in panel H shows positive perforin control in a patient with large granular lymphocyte leukemia. Original magnification, × 400, except panel C (× 250) and panel H inset (× 1000).
Immunohistochemical studies in all sites showed predominately CD3+ T cells, with a CD4/CD8 ratio of approximately 1:4. Numerous cells were positive for granzyme B; however, cells were consistently negative for perforin (Figure 1G-H). Germinal centers were absent; however, scattered individual CD20+ cells and small aggregates were seen. In situ hybridization with the EBV encoded RNA (EBER) probe showed only rare positive small lymphocytes (< 1 per high-power field). Southern blot analysis for clonal T-cell receptor beta gene rearrangements showed no evidence of a clonal population (data not shown).
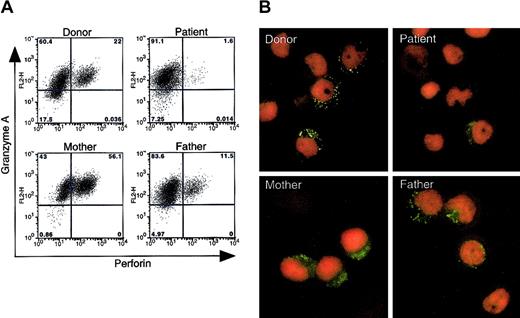
Lymphocyte phenotype, EBV infection, and cytokine profile in PHA and IL-2–stimulated PBMCs from the patient with fatal CAEBV
PBMCs from the patient described above, his parents, and healthy donors were stimulated with PHA and IL-2 for 2 days followed by IL-2 alone for 2 weeks in vitro. More than 50% of the stimulated cells were CD8+ (Figure 2A). While blood from the patient's mother and a healthy donor contained more than 30% CD56+ and CD8– cells (normal range of healthy donors, 7.8%-35%; average, 14.3%; standard error, 2.5%), blood from the patient and his father had less than 5% CD56+ and CD8– cells (Figure 2B). The patient's father also had a small population of CD56+ and CD8+ cells (NK T cells).
Lymphocyte phenotype, EBV DNA levels with latent gene expression, and cytokine levels in stimulated PBMCs from the patient, his parents, and a healthy blood bank donor. (A) CD4/CD8 and (B) CD8/CD56 expression of PBMCs stimulated with PHA and IL-2 for 2 days followed by IL-2 alone for 2 weeks. (C) EBV DNA levels in stimulated PBMCs. DNA from the EBV BamHI W region DNA was quantified using real-time PCR. The EBV BamHI W fragment copy number per cell was calculated by the formula N = 2 × (W/B), where N is the EBV BamHI W copy number/cell, W is the EBV BamHI W copy number, and B is the bcl-2 copy number. Copy numbers of EBV-BamHI W gene per 1×106 cells are indicated. U indicates undetectable. (D) RT-PCR analysis of EBV latency genes in stimulated PBMCs. Southern blots of PCR products were hybridized with [32P]-labeled probes. (E) Cytokine levels were measured in culture supernatants of stimulated PBMCs from a healthy donor (1), an unrelated patient with CAEBV (2), the patient with the perforin mutations (3), the patient's mother (4), and the patient's father (5). Cells were stimulated with anti-CD3 antibody (black bars) or isotype control antibody (white bars). The experiment was performed 3 times with similar results.
Lymphocyte phenotype, EBV DNA levels with latent gene expression, and cytokine levels in stimulated PBMCs from the patient, his parents, and a healthy blood bank donor. (A) CD4/CD8 and (B) CD8/CD56 expression of PBMCs stimulated with PHA and IL-2 for 2 days followed by IL-2 alone for 2 weeks. (C) EBV DNA levels in stimulated PBMCs. DNA from the EBV BamHI W region DNA was quantified using real-time PCR. The EBV BamHI W fragment copy number per cell was calculated by the formula N = 2 × (W/B), where N is the EBV BamHI W copy number/cell, W is the EBV BamHI W copy number, and B is the bcl-2 copy number. Copy numbers of EBV-BamHI W gene per 1×106 cells are indicated. U indicates undetectable. (D) RT-PCR analysis of EBV latency genes in stimulated PBMCs. Southern blots of PCR products were hybridized with [32P]-labeled probes. (E) Cytokine levels were measured in culture supernatants of stimulated PBMCs from a healthy donor (1), an unrelated patient with CAEBV (2), the patient with the perforin mutations (3), the patient's mother (4), and the patient's father (5). Cells were stimulated with anti-CD3 antibody (black bars) or isotype control antibody (white bars). The experiment was performed 3 times with similar results.
Patients with CAEBV frequently have elevated levels of EBV DNA in their PBMCs, T cells, NK cells, or tissues.5,25,26 Real-time PCR showed that stimulated PBMCs from the patient contained high levels of EBV DNA (Figure 2C). EBV DNA was not detected in PHA and IL-2–stimulated PBMCs from the patient's parents and healthy donors; however, stimulated PBMCs from an unrelated patient with CAEBV had elevated levels of EBV DNA. EBV-infected cells from patients with CAEBV consistently express the latency-associated genes EBV nuclear antigen 1 (EBNA-1) and EBERs; however, other latency-associated genes such as EBNA-2 and latent membrane protein 1 (LMP-1) are expressed in cells from some, but not all, patients.26 Stimulated PBMCs from our patient expressed EBNA-1, but not EBNA-2, LMP-1, or LMP-2A (Figure 2D, data not shown). These results indicate that the stimulated PBMCs from the patient had a type I EBV latency program.
Some patients with CAEBV have been reported to have dysregulation of cytokines with increased expression of both T-helper (Th) 1 and Th2 cytokine transcripts, termed an “unbalanced cytokine profile.”25 Stimulation of cells from the patient with antibodies to CD3 and CD28 resulted in high levels of both Th1 (IFN-γ) and Th2 (IL-4, IL-10, and IL-13) cytokines (Figure 2E). These cytokines are involved in regulation of cytokine production, activation, and proliferation of lymphocytes and monocytes. Cells from the patient's father showed similar elevations in some (IL-4, IL-13), but not all (IFN-γ, IL-10) of the cytokines found to be elevated in his son. In contrast, cells from an unrelated patient with CAEBV did not show cytokine elevations. These data suggest that stimulated PBMCs from the patient had an unbalanced cytokine profile.
Mutations in the perforin gene of the patient and his parents
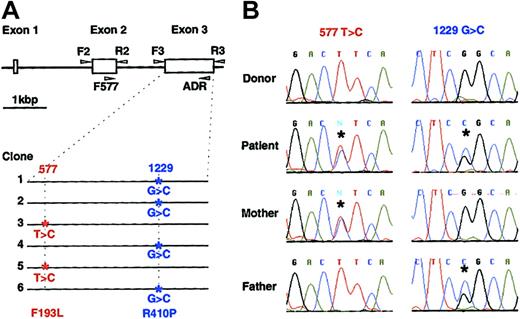
Since many patients with CAEBV have hemophagocytic syndrome, and since perforin mutations have been found in patients with FHL,20 we determined the sequence of the entire coding region of the perforin gene (exons 2 and 3) in our patient as well as 14 other patients with CAEBV (12 patients from the United States, 2 patients from Japan). Whereas none of the 14 other patients with CAEBV showed any mutations in their perforin genes, sequence analysis showed that our patient had mutations in both alleles of his perforin gene. Of 6 cloned PCR products from the patient's genomic DNA, 2 clones had a T to C mutation at nucleotide 577 of the perforin gene, and 4 had a G to C mutation at nucleotide 1229 (Figure 3A). The change in nucleotides 577 and 1229 are predicted to change amino acid 193 from phenylalanine to leucine, and amino acid 410 from arginine to proline, respectively. To verify that these changes were due to mutations and not to polymorphisms, DNAs were obtained from 54 blood bank donors (predominantly white, like our patient) and PCR analyses (corresponding to 108 chromosomes) showed that none had the nucleotide changes noted in the patient. Thus, these nucleotide changes were likely due to mutations and not to polymorphisms in the perforin gene. These 2 nucleotide changes in perforin have not been reported previously.27
Perforin mutations in the patient with CAEBV and in his parents. (A) The perforin gene contains 3 exons and the open reading frame is encoded by the second and third exons. Exons 2 and 3 were amplified by PCR from genomic DNA using the indicated primers (arrowheads),20 cloned in plasmid pCR2.1, and sequenced. Mutations were found in both alleles of exon 3 in the patient. Four of 6 clones had a G to C mutation at nucleotide 1229 and 2 clones had a T to C mutation at nucleotide 577, indicating that each allele had a separate mutation. (B) Chromatograms of RT-PCR products. RT-PCR was performed using mRNA isolated from stimulated PBMCs of the patient, his parents, and a healthy donor. RT-PCR products were directly sequenced (not having been cloned) and asterisks indicate mutations.
Perforin mutations in the patient with CAEBV and in his parents. (A) The perforin gene contains 3 exons and the open reading frame is encoded by the second and third exons. Exons 2 and 3 were amplified by PCR from genomic DNA using the indicated primers (arrowheads),20 cloned in plasmid pCR2.1, and sequenced. Mutations were found in both alleles of exon 3 in the patient. Four of 6 clones had a G to C mutation at nucleotide 1229 and 2 clones had a T to C mutation at nucleotide 577, indicating that each allele had a separate mutation. (B) Chromatograms of RT-PCR products. RT-PCR was performed using mRNA isolated from stimulated PBMCs of the patient, his parents, and a healthy donor. RT-PCR products were directly sequenced (not having been cloned) and asterisks indicate mutations.
The nucleotide sequence of the perforin genes from the patient's mother and father were also determined. Sequence analysis of DNA from the patient's mother showed a T to C change in nucleotide 577 in one allele of the perforin gene, whereas analysis of DNA from the patient's father showed a G to C change in nucleotide 1229 in one allele of the gene. These data indicate that mutations in nucleotides 577 and 1229 in the patient's perforin gene were inherited from his mother and father, respectively.
To determine if transcripts of mutant and wild-type perforin were expressed at similar levels in the patient and his parents, RT-PCR products of exon 3 in the perforin gene were sequenced. Direct sequencing and analysis of the chromatogram showed that T and C at nucleotide 577, and the G and C at nucleotide 1229 produced peaks of similar heights, indicating that the perforin mRNAs were expressed at similar levels in the patient (Figure 3B). In addition, the similar heights of peaks for these nucleotides in the chromatograms of the cDNAs from the patient's mother and father indicates that both the wild-type and mutant transcripts were expressed at similar levels in the patient's parents.
Expression of the native form of perforin is impaired in the patient with CAEBV
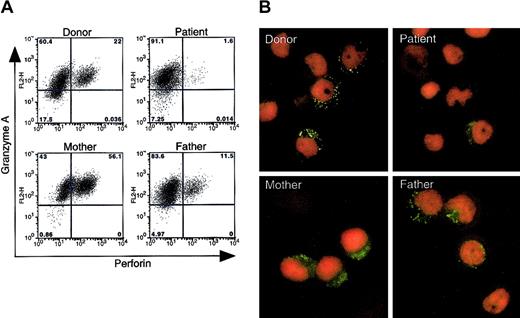
To determine whether the mutations in the perforin gene alter the level of expression of perforin protein in the patient's cells, we performed flow cytometry on permeabilized cells from the patient, his parents, and from blood bank donors. PHA- and IL-2–stimulated PBMCs were tested, since perforin is expressed in CD8+ T cells and NK cells. Incubation of the cells with fluorescein isothiocyanate (FITC)–conjugated antiperforin antibody dG9,28 which recognizes only the native form of perforin,28,29 showed markedly reduced levels of perforin in cells from the patient, whereas perforin was readily detected in stimulated cells from his mother and father, 2 blood bank donors, and another (unrelated) patient with CAEBV (Figure 4A, data not shown). In contrast, granzyme A expression was observed in more than 80% of cells from all of the individuals tested. Immunofluorescence assay of stimulated PBMCs using FITC-conjugated antiperforin antibody dG9 showed punctate staining in the cytoplasm in 40% to 60% of stimulated cells from the patient's mother and father and from healthy blood bank donors; however, very rare cells from the patient stained with the perforin antibody (Figure 4B). Perforin usually shows a bright, punctate staining pattern in the cytoplasm, since the protein is present in granules in the cytoplasm.20 In contrast to the punctate distribution of perforin seen in his parent's cells, the patient's cells showed a weaker and more diffuse pattern of staining with the antibody. Expression of perforin in the mother's cells demonstrated not only the punctate staining pattern, but also weak diffuse staining in the cytoplasm. Since antibody dG9 recognizes the native form of perforin, these data suggest that expression of the native form of perforin is markedly reduced in the patient with the perforin mutations.
Expression of the native form of perforin in stimulated PBMCs from the patient, his parents, and a healthy blood bank donor. (A) PHA- and IL-2–stimulated PBMCs were fixed and permeabilized, stained with FITC-conjugated antiperforin antibody (dG9; Ancell, Bayport, MN) or phycoerythrin (PE)–conjugated anti–granzyme A antibody (CB9; BD Pharmingen), and analyzed by flow cytometry. The experiment was performed 3 times with similar results. The normal range of perforin-positive cells in 3 healthy donors was 3.4% to 56% (average 14%) in 4 separate experiments. (B) Immunofluorescence assay shows expression of perforin in PBMCs stimulated with PHA and IL-2, fixed, permeabilized, incubated with FITC-conjugated antiperforin antibody dG9 (green), and counterstained with propidium iodide (red). Original magnification, × 1000.
Expression of the native form of perforin in stimulated PBMCs from the patient, his parents, and a healthy blood bank donor. (A) PHA- and IL-2–stimulated PBMCs were fixed and permeabilized, stained with FITC-conjugated antiperforin antibody (dG9; Ancell, Bayport, MN) or phycoerythrin (PE)–conjugated anti–granzyme A antibody (CB9; BD Pharmingen), and analyzed by flow cytometry. The experiment was performed 3 times with similar results. The normal range of perforin-positive cells in 3 healthy donors was 3.4% to 56% (average 14%) in 4 separate experiments. (B) Immunofluorescence assay shows expression of perforin in PBMCs stimulated with PHA and IL-2, fixed, permeabilized, incubated with FITC-conjugated antiperforin antibody dG9 (green), and counterstained with propidium iodide (red). Original magnification, × 1000.
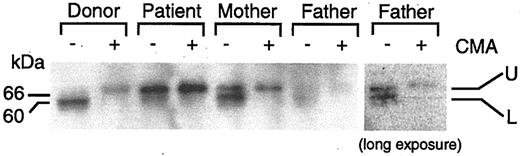
Proteolytic cleavage of perforin is inhibited in the patient with fatal CAEBV
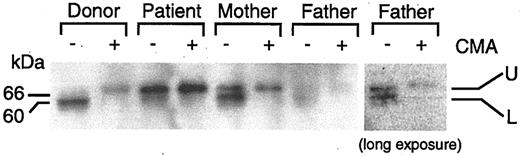
Perforin is synthesized as a 70-kDa inactive glycosylated precursor which is subsequently cleaved at the C-terminus to yield a 60-kDa active, mature form.29 To determine the level of expression of the precursor and mature forms of perforin, we performed immunoblot analyses using the 2d4-perf monoclonal antibody to perforin, which recognizes both mature and precursor forms of perforin.29,30 Stimulated PBMCs from the patient expressed predominantly the 70-kDa precursor protein that reacted with the antibody that was larger in size than the 60-kDa mature protein from the healthy donor that bound to the antibody (Figure 5). To verify that the perforin molecule from the patient corresponds to the 70-kDa inactive precursor form, concanamycin A, which inhibits the cleavage of the precursor form of perforin, was added to the cells to block cleavage of perforin. Incubation of cells from the healthy blood bank donor or from the patient with CAEBV with concanamycin A followed by immunoblotting with antibody 2d4-perf resulted in detection of only the 70-kDa precursor form of perforin. Thus, cells from the patient with fatal CAEBV expressed predominantly the precursor form of perforin, indicating that proteolytic cleavage of perforin is inhibited. Since the cleavage site in perforin is located at amino acids 520-521,29 and the perforin mutations are located at amino acids 193 and 410, these findings suggest that his mutations in perforin result in a conformational change in the protein that prevents cleavage to the mature protein.
Inhibition of proteolytic cleavage of perforin in the patient with CAEBV. Stimulated PBMCs were cultured in the presence (+) or absence (–) of 200 nM concanamycin A (CMA) for 5 hours. Lysates were prepared from the cells and perforin was detected by immunoblotting with 2d4-perf antibody under nonreducing conditions.29,30 Two bands corresponding to perforin are seen. The upper (U) and lower (L) bands indicate precursor and mature form of perforin, respectively. Protein concentrations were equal in each sample. The right panel indicates a darker exposure from a repeat experiment using the father's cells.
Inhibition of proteolytic cleavage of perforin in the patient with CAEBV. Stimulated PBMCs were cultured in the presence (+) or absence (–) of 200 nM concanamycin A (CMA) for 5 hours. Lysates were prepared from the cells and perforin was detected by immunoblotting with 2d4-perf antibody under nonreducing conditions.29,30 Two bands corresponding to perforin are seen. The upper (U) and lower (L) bands indicate precursor and mature form of perforin, respectively. Protein concentrations were equal in each sample. The right panel indicates a darker exposure from a repeat experiment using the father's cells.
Immunoblotting of perforin in stimulated PBMCs from the patient's mother showed 2 forms of perforin, the 70-kDa inactive precursor form and the 60-kDa mature form. This is consistent with heterozygosity in the mother. Immunoblotting of perforin in PBMCs from the patient's father showed 2 faint bands of 70 kDa and 60 kDa, even though equal amounts of cell lysates were used.
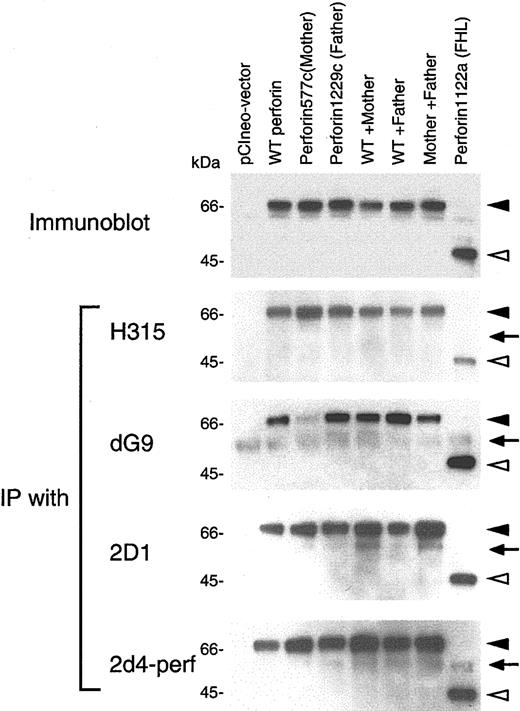
Mutation in the patient's perforin changes the reactivity with an antiperforin antibody
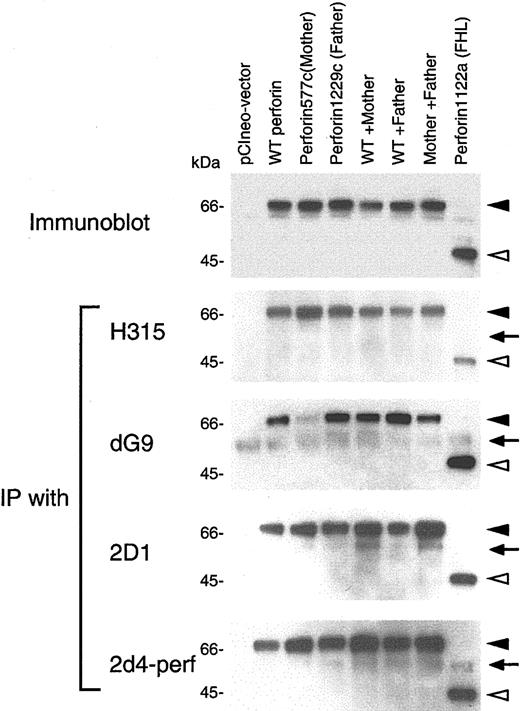
The inhibition of proteolytic cleavage of mutant perforin in cells from the patient suggested that a dynamic conformational change in perforin occurred in his stimulated PBMCs due to the mutations in perforin. To provide further evidence for this hypothesis, we cloned cDNA for perforin derived from a healthy donor and inserted the gene into expression vector pCIneo. Site-directed mutagenesis of the plasmid was performed to construct perforin genes with each of the patient's mutants as well as a perforin mutant with stop codons previously identified in a patient with FHL.20 These plasmids were transfected either individually or in combinations into 293T cells. Immunoblotting using antiperforin antibody 2d4-perf, which recognizes both mature and precursor forms of perforin, showed that cells transfected with plasmids encoding perforin with the mother's or father's mutation, or cotransfected with 2 plasmids together (wild-type and mother's mutant perforin, wild-type and father's mutant perforin, or mother's and father's mutant perforin), expressed perforin in 293T human embryonic kidney cells (Figure 6, top panel). Since perforin is not processed to its mature form in non-T cells,29 the 70-kDa bands represent the precursor form of perforin. Transfection of cells with the plasmid encoding mutant perforin from a patient with FHL produced a truncated form of perforin. To see if mutations in perforin might induce conformational changes, transfected cell lysates were immunoprecipitated with various antibodies recognizing different portions of perforin. Rabbit polyclonal antibody H315 recognizes the carboxy terminal region (amino acids 241-555) of perforin; murine monoclonal antibodies 2D1 and 2d4-perf recognize a domain within amino acids 189-320 of mouse perforin and amino acids 264-279 of human perforin, respectively.30,31 Murine monoclonal antibody dG9 recognizes only the native form of perforin, but the epitope detected by the antibody is unknown.28,29 Mutant perforin from the patient's mother was weakly immunoprecipitated from 293T cells with antibody dG9, whereas the wild-type and other mutant perforins were immunoprecipitated to a similar extent (Figure 6). Immunoprecipitation of wild-type perforin and each of the mutant forms of perforin were similar with antibodies H315, 2D1, and 2d4-perf. These data suggest the mutant perforin from the patient's mother may have a conformational change, resulting in its reduced ability to bind antibody dG9. Alternatively, the perforin epitope recognized by antibody dG9 might overlap the mutation in the mother's perforin, resulting in reduced immunoprecipitation of the protein.
Binding activity of perforin mutants in 293T cells with antibodies. 293T cells were transfected with plasmid(s) expressing wild-type (WT) and/or mutant perforin genes, cell lysates were prepared, and immunoblots were performed using 2d4-perf antibody (top panel). Lysates were immunoprecipitated with perforin antibodies H315 (Santa Cruz Biotechnology, Santa Cruz, CA; second panel), dG9 (Endogen, Woburn, MA; third panel), 2D1 (US Biological, Swampscott, MA; fourth panel) and 2d4-perf (the bottom panel). Immunoprecipitates were immunoblotted under reducing conditions and stained with 2d4-perf antibody. Solid and open arrowheads indicate full-length and truncated forms of perforin, respectively. Arrows correspond to the heavy chain of immunoglobulin (IgH).
Binding activity of perforin mutants in 293T cells with antibodies. 293T cells were transfected with plasmid(s) expressing wild-type (WT) and/or mutant perforin genes, cell lysates were prepared, and immunoblots were performed using 2d4-perf antibody (top panel). Lysates were immunoprecipitated with perforin antibodies H315 (Santa Cruz Biotechnology, Santa Cruz, CA; second panel), dG9 (Endogen, Woburn, MA; third panel), 2D1 (US Biological, Swampscott, MA; fourth panel) and 2d4-perf (the bottom panel). Immunoprecipitates were immunoblotted under reducing conditions and stained with 2d4-perf antibody. Solid and open arrowheads indicate full-length and truncated forms of perforin, respectively. Arrows correspond to the heavy chain of immunoglobulin (IgH).
Cytotoxicity is impaired in cells from the patient with fatal CAEBV
Perforin is important for the cytotoxic activity of CD8+ T and NK cells. Since the patient with CAEBV had mutations in both perforin genes that resulted in reduced expression of the native form of the protein and that inhibited maturation of the protein, we postulated that cytotoxicity by his CD8+ T and NK cells might be reduced. Incubation of PHA- and IL-2–stimulated PBMCs from the patient with Fas-deficient target cells showed a marked diminution in cytotoxicity compared with cells from blood bank donors (Figure 7A). Cells from the patient's father demonstrated an intermediate level of cytotoxicity, whereas cells from the patient's mother or from an unrelated patient with CAEBV did not show a reduction in cytotoxicity. The reduced level of cytotoxicity with the father's cells may be related to the lower number of NK cells in his stimulated PBMCs (Figure 2B). Although stimulated PBMCs from the patient contained a larger number of NK cells than PBMCs from his father, the cytotoxicity of the patient's PBMCs was markedly lower than PBMCs from his father. Similar cytotoxicity results for cells from the patient, his parents, and the blood bank donors were observed in 3 separate experiments performed on different days. Taken together, these results suggest that the mutations in the patient's perforin genes result in defective Fas-independent cytotoxic killing of target cells.
Cytotoxicity and granule exocytosis assays using stimulated PBMCs from the patient, his parents, an unrelated patient with CAEBV, and healthy donors. (A) PBMCs were stimulated with PHA and IL-2, cultured in IL-2 (effector cells), and flow cytometry showed lymphocyte phenotypes described in Figure 2B. The cells were incubated with [51Cr]-labeled L1210 target cells (Fas-deficient target cells) in the presence of anti-CD3 (UCHT-1; BD Pharmingen) and anti-Fas blocking antibody (ZB4; Beckman Coulter, Fullerton, CA) in a Fas-independent CTL killing assay. Cytotoxicity in this assay system is CD3-dependent, since T-cell receptor activation is required to trigger release of granules containing perforin and granzymes.20 The effector-to-target (E/T) ratio is indicated on the x axis and the percent specific lysis is indicated on the y axis. The percent lysis was calculated as follows: % lysis = (E–S)/(M–S) × 100, where E is the release from experimental samples, S is the spontaneous release, and M is the maximum release upon lysis with 2% NP-40. The gray band indicates the normal range of cytotoxicity seen in 4 healthy donors (data not shown). Cells from a healthy blood bank donor incubated in the absence of anti-CD3 antibody were assayed as a negative control. Flow cytometry showed the following lymphocyte phenotypes: donor 1: CD8 = 49%, CD56 = 42%; donor 2: CD8 = 55%, CD56 = 42%; patient: CD8 = 54%, CD56 = 25%; mother: CD8 = 50%, CD56 = 68%; father: CD8 = 91%, CD56 = 4%; unrelated patient with CAEBV: CD8 = 55%, CD56 = 40%. (B) PBMCs were stimulated with PHA and IL-2 followed by incubation in the presence (black bars) and absence (white bars) of plate-bound anti-CD3/anti-CD28 antibody. Secretion of β-hexosaminidase in the cell culture supernatant was measured to quantify the level of granule exocytosis.33 β-hexosaminidase release was expressed as a percentage of the enzyme in the supernatant divided by the total enzyme from cells lysed in 0.1% Triton X-100. Experiments in panels A and B were performed 3 times and a representative result is shown.
Cytotoxicity and granule exocytosis assays using stimulated PBMCs from the patient, his parents, an unrelated patient with CAEBV, and healthy donors. (A) PBMCs were stimulated with PHA and IL-2, cultured in IL-2 (effector cells), and flow cytometry showed lymphocyte phenotypes described in Figure 2B. The cells were incubated with [51Cr]-labeled L1210 target cells (Fas-deficient target cells) in the presence of anti-CD3 (UCHT-1; BD Pharmingen) and anti-Fas blocking antibody (ZB4; Beckman Coulter, Fullerton, CA) in a Fas-independent CTL killing assay. Cytotoxicity in this assay system is CD3-dependent, since T-cell receptor activation is required to trigger release of granules containing perforin and granzymes.20 The effector-to-target (E/T) ratio is indicated on the x axis and the percent specific lysis is indicated on the y axis. The percent lysis was calculated as follows: % lysis = (E–S)/(M–S) × 100, where E is the release from experimental samples, S is the spontaneous release, and M is the maximum release upon lysis with 2% NP-40. The gray band indicates the normal range of cytotoxicity seen in 4 healthy donors (data not shown). Cells from a healthy blood bank donor incubated in the absence of anti-CD3 antibody were assayed as a negative control. Flow cytometry showed the following lymphocyte phenotypes: donor 1: CD8 = 49%, CD56 = 42%; donor 2: CD8 = 55%, CD56 = 42%; patient: CD8 = 54%, CD56 = 25%; mother: CD8 = 50%, CD56 = 68%; father: CD8 = 91%, CD56 = 4%; unrelated patient with CAEBV: CD8 = 55%, CD56 = 40%. (B) PBMCs were stimulated with PHA and IL-2 followed by incubation in the presence (black bars) and absence (white bars) of plate-bound anti-CD3/anti-CD28 antibody. Secretion of β-hexosaminidase in the cell culture supernatant was measured to quantify the level of granule exocytosis.33 β-hexosaminidase release was expressed as a percentage of the enzyme in the supernatant divided by the total enzyme from cells lysed in 0.1% Triton X-100. Experiments in panels A and B were performed 3 times and a representative result is shown.
Granule exocytosis is another important process required for CTL activity. Reduced CTL activity has been demonstrated in humans and mice that have mutations in the Rab27A gene that controls granule exocytosis.32,33 In addition, hemophagocytic syndrome has been noted in patients with Rab27A mutations.34 To determine if granule exocytosis is impaired in cells from the patient with fatal CAEBV, PHA- and IL-2–stimulated PBMCs were stimulated with antibodies to CD3 and CD28 and the level of β-hexosaminidase secreted into the culture media, a marker of degranulation, was measured (Figure 7B). Elevated levels of β-hexosaminidase were detected in culture supernatants of cells from the patient with perforin mutations, compared with cells from the patient's parents, 2 blood bank donors, and an unrelated patient with CAEBV. These results suggest that granule exocytosis was up-regulated in stimulated cells from the patient.
Discussion
We have identified mutations in both alleles of the perforin gene in a patient with fatal CAEBV. The patient's PBMCs showed reduced expression of the native form of perforin and the unprocessed precursor was the predominant form detected. Expression of perforin mutants in 293T cells suggested that the mother's mutant perforin might have a conformational change. Stimulated PBMCs from the patient were markedly impaired for cytotoxic activity in vitro. Taken together, these data imply that the reduced cytotoxicity of CTL and NK cells in the patient is due to the mutations in his perforin genes. This is the first case of CAEBV in which genetic mutations have been linked to the disease.
Perforin is present in cytolytic granules of CTL and NK cells and has a critical role in their cytotoxic activity. Perforin comprises one of 2 major pathways that CTLs use to kill virus-infected cells.35 In the Fas pathway, Fas ligand on CTLs binds to Fas on virus-infected cells, which initiates caspase-mediated killing of the cells. In the perforin pathway, engagement of the T-cell receptor on the surface of CTLs with viral peptides associated with major histocompatibility complex (MHC) class I molecules triggers release of granules containing perforin and granzymes. Perforin is known to be important for control of certain virus infections by the immune system.36 Perforin knock-out mice have a normal phenotype, but when they are infected with lymphocytic choriomeningitis virus they show immune defects.35,37 Similarly, perforin-deficient mice show increased mortality after infection with herpes simplex virus and ectromelia virus compared with control mice.38,39
Perforin has been implicated in the killing of EBV-infected cells. EBV-specific T-cell cytotoxicity is mediated through the perforin pathway in patients with lymphoproliferative disorders after allogeneic bone marrow transplantation.40 The perforin/granzyme pathway, rather than the Fas or TRAIL pathways, is the major pathway for MHC class II–restricted lysis of EBV-transformed lymphoblastoid cell lines by virus-specific CD4+ T cells.41,42 Recent studies indicate that expression of perforin is impaired in CD8+ cells from patients with EBV-positive nasopharyngeal carcinoma and HIV infection, suggesting that low expression of perforin in CD8+ T cells may constitute an important mechanism for immune escape by tumors or virus-infected cells.43-45 Perforin-deficient mice also have a higher incidence of T- or B-cell lymphomas, confirming that perforin is critical in immune surveillance against cancer.46
Perforin mutations have been identified in patients with FHL.20 Patients with FHL may share certain clinical findings seen in some patients with CAEBV such as fever, splenomegaly, lymphadenopathy, hepatomegaly, neurologic abnormalities, and hemophagocytosis.47,48 Patients with FHL usually present in infancy or very early in childhood and in the absence of bone marrow transplantation most patients die within 1 year after diagnosis.23,27,49 Patients with FHL and nonsense mutations tend to present within the first few months of life, whereas those with missense mutations usually present by 2 years of age.23 Although our patient showed many of the features of FHL early in the course of his disease,22 he presented at an older age (7 years) than most patients with FHL, survived for 10 years after diagnosis, and did not have a family history of hemophagocytic lymphohistiocytosis. While a variety of virus infections have been reported in patients with FHL at the time of diagnosis, including cytomegalovirus, parvovirus, EBV, hepatitis B virus, and adenovirus, no single agent has been identified that triggers the disorder in most cases. FHL and EBV-associated hemophagocytic lymphohistiocytosis (EBV-HLH) have overlapping clinical manifestations, and CAEBV is often associated with EBV-HLH at some point during the course of disease. HLH is considered the prototype of hemophagocytic syndrome.50 In a study of patients with EBV-associated hemophagocytic syndrome from Japan, no mutations were detected in the perforin gene of 14 patients.51 Thus, whereas perforin defects have been associated with FHL, mutations in the gene do not appear to be responsible for most cases of EBV-associated hemophagocytic syndrome.
Perforin is synthesized as an inactive precursor that must be cleaved at its carboxy terminus to yield the active, mature form of the protein.29 The precursor form of perforin consists of 555 amino acids and contains 2 N-linked glycosylation sites and an approximately 300 amino acid C2 domain that is homologous to the C2 domain of protein kinase C. The C2 domain of perforin is important for binding of the protein to the phospholipid bilayer of the cytoplasmic membrane.29 The C2 domain in the uncleaved form of perforin cannot bind to the membrane, due to the presence of a bulky N-linked glycan on the carboxy terminal domain of the protein. Proteolytic cleavage of the carboxy terminal domain allows the C2 domain to bind to the membrane. We demonstrated that the inactive precursor form of perforin accumulated in the patient's PBMCs, implying that the proteolytic cleavage was inhibited. Reduced expression of the active, mature form of perforin was likely the cause of the defect in Fas-independent cytotoxicity of T cells and NK cells from the patient. Mutations in the patient's perforin may have altered its conformation and inhibited proteolytic processing of the protein to its mature form. Although we wanted to examine processing of perforin encoded by each allele separately, processing of perforin has never been demonstrable in cells transfected with the perforin gene. We were unable to show processing of perforin in 293T or T cells transfected with our perforin expression vector (data not shown). Similarly, Uellner et al29 could not detect processing of the protein in rat leukemia cells transfected with the perforin gene. Since precursor (uncleaved) forms of perforin were present in cells from both of the patient's parents (Figure 5), we postulate that both mutations inhibit cleavage of perforin. In addition, since both parents were seropositive for EBV and asymptomatic, and both had a mutant and normal perforin allele, one wild-type copy of the gene may be sufficient for control of EBV infection. Some perforin mutations reported in patients with FHL induce only single amino acid missense mutations as was the case in our patient, and low-level expression of perforin in T cells and NK cells from patients with FHL has been reported using the dG9 antibody.20,52 Our data suggest that at least some missense mutations in FHL may also induce conformational changes and inhibit proteolytic cleavage of the precursor form of perforin.
Our patient with mutations in both alleles of the perforin gene died from T-cell lymphoproliferative disease. Perforin mutations may contribute to uncontrolled lymphoproliferative disease.53-55 In the absence of functional perforin, T and NK cells might be unable to contain the expansion of EBV-infected cells, resulting in a persistent active infection. This process might be similar to the massive T-cell proliferation that occurs in response to acute EBV infection during infectious mononucleosis; however, in infectious mononucleosis cell proliferation is down-regulated when virus replication is reduced. The persistent activation of T cells in our patient may have increased the likelihood of development of uncontrolled T-cell lymphoproliferative disease. Since dysregulation of cytokine expression has been reported in both CAEBV and FHL,24,25 it may be associated with the persistent activation of T cells, and might not be specific for mutations in perforin.
We have identified the genetic basis for CAEBV in one patient, and postulate how mutations in the perforin protein may have led to his disease. The observation that CTLs and NK cells in other cases of CAEBV have defective cytotoxic activity13,14,56 suggests that other proteins in the perforin or Fas pathways may be responsible for CAEBV in other patients. Thus, the diverse clinical expressions of CAEBV may be due to unique, heritable disorders with the shared inability to contain proliferation of EBV-infected cells.
Prepublished online as Blood First Edition Paper, October 23, 2003; DOI 10.1182/blood-2003-06-2171.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Susan E. Stepp, Department of Pathology, University of Texas Southwestern Medical School, Dallas, TX, for advice on PCR of the perforin gene; Dong Zhang, Center for Blood Research, Department of Pediatrics, Harvard Medical School, Boston, MA, and Gillian M. Griffiths, Sir William Dunn School of Medicine, Oxford, United Kingdom, for perforin antibody 2d4-perf; Susan F. Leitman, Department of Transfusion Medicine, NIH Blood Bank, NIH, Bethesda, MD, for assistance in obtaining cells from blood bank donors; Pierre Henkart for advice on cytotoxicity assays; Ronald Hornung, SAIC, Frederick, MD, for ELISA assay results; Warren Strober and Yo Hoshino, LCI, NIAID, NIH, for helpful discussions.


![Figure 2. Lymphocyte phenotype, EBV DNA levels with latent gene expression, and cytokine levels in stimulated PBMCs from the patient, his parents, and a healthy blood bank donor. (A) CD4/CD8 and (B) CD8/CD56 expression of PBMCs stimulated with PHA and IL-2 for 2 days followed by IL-2 alone for 2 weeks. (C) EBV DNA levels in stimulated PBMCs. DNA from the EBV BamHI W region DNA was quantified using real-time PCR. The EBV BamHI W fragment copy number per cell was calculated by the formula N = 2 × (W/B), where N is the EBV BamHI W copy number/cell, W is the EBV BamHI W copy number, and B is the bcl-2 copy number. Copy numbers of EBV-BamHI W gene per 1×106 cells are indicated. U indicates undetectable. (D) RT-PCR analysis of EBV latency genes in stimulated PBMCs. Southern blots of PCR products were hybridized with [32P]-labeled probes. (E) Cytokine levels were measured in culture supernatants of stimulated PBMCs from a healthy donor (1), an unrelated patient with CAEBV (2), the patient with the perforin mutations (3), the patient's mother (4), and the patient's father (5). Cells were stimulated with anti-CD3 antibody (black bars) or isotype control antibody (white bars). The experiment was performed 3 times with similar results.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/4/10.1182_blood-2003-06-2171/6/m_zh80040456890002.jpeg?Expires=1764974616&Signature=LAzGb1hY6QiNfT0vK8wNDACDk4V8bO3p18Jv8M24cnefmNMItUrIoZ5NsI6z4X0dn8kGkontpI1YeXjItQPIeLDvBvkyhjCIkLlatHR4hLoDuj9-~o1Z8LrXVwiVkrDzWw4c4eFOeCP2ZhANznT97-ush65NKOFrsc6H-D0ytvJKWP0MJuWwzhDkTLIEEo0J0nqKh3oDtS--xBbEggkzghDVgazHGmq-1IjvO9zw5Yh-zT~qv86VLkJQhlkNuurk7D9Qc~MuoIITEBx4NbdxwtuueAEe6XSoJR-DnQtuFLmVLIa6eB1vu31ciLX7GYYPl3hg09QRZbaIyVO71FDLqw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 7. Cytotoxicity and granule exocytosis assays using stimulated PBMCs from the patient, his parents, an unrelated patient with CAEBV, and healthy donors. (A) PBMCs were stimulated with PHA and IL-2, cultured in IL-2 (effector cells), and flow cytometry showed lymphocyte phenotypes described in Figure 2B. The cells were incubated with [51Cr]-labeled L1210 target cells (Fas-deficient target cells) in the presence of anti-CD3 (UCHT-1; BD Pharmingen) and anti-Fas blocking antibody (ZB4; Beckman Coulter, Fullerton, CA) in a Fas-independent CTL killing assay. Cytotoxicity in this assay system is CD3-dependent, since T-cell receptor activation is required to trigger release of granules containing perforin and granzymes.20 The effector-to-target (E/T) ratio is indicated on the x axis and the percent specific lysis is indicated on the y axis. The percent lysis was calculated as follows: % lysis = (E–S)/(M–S) × 100, where E is the release from experimental samples, S is the spontaneous release, and M is the maximum release upon lysis with 2% NP-40. The gray band indicates the normal range of cytotoxicity seen in 4 healthy donors (data not shown). Cells from a healthy blood bank donor incubated in the absence of anti-CD3 antibody were assayed as a negative control. Flow cytometry showed the following lymphocyte phenotypes: donor 1: CD8 = 49%, CD56 = 42%; donor 2: CD8 = 55%, CD56 = 42%; patient: CD8 = 54%, CD56 = 25%; mother: CD8 = 50%, CD56 = 68%; father: CD8 = 91%, CD56 = 4%; unrelated patient with CAEBV: CD8 = 55%, CD56 = 40%. (B) PBMCs were stimulated with PHA and IL-2 followed by incubation in the presence (black bars) and absence (white bars) of plate-bound anti-CD3/anti-CD28 antibody. Secretion of β-hexosaminidase in the cell culture supernatant was measured to quantify the level of granule exocytosis.33 β-hexosaminidase release was expressed as a percentage of the enzyme in the supernatant divided by the total enzyme from cells lysed in 0.1% Triton X-100. Experiments in panels A and B were performed 3 times and a representative result is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/4/10.1182_blood-2003-06-2171/6/m_zh80040456890007.jpeg?Expires=1764974616&Signature=wLK89Clz~gJd3IBUJNuRxueSZmcwE~IRqEW5wsXOKrtWIA94WFVrG6s6GfCETtTzLwGCrQAj2mXfCwBTdnyHHtIIGwahnqFHFd4bym6IJjND793tBtrPhfO4-emX2cJlRqouFO9r48VTPgcoiplL5XZvVuu342Tzg-g4ozCkXK0Anic8A9Z9VHEbW0EAfkI5J7wzkFg4V~1oTauTD~CQAUfuXV25xqlY2ZVxtxYm4ZWLo4BucO5~FRH6gjav~oXE2k9lQMYVC4f~EMLqimYuRuuqWe24Yudk8pd6Q7BJ67eC2cmVmAwuxtkAp~nn---s2OniTti-El5VjOmx2vKWLA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Lymphocyte phenotype, EBV DNA levels with latent gene expression, and cytokine levels in stimulated PBMCs from the patient, his parents, and a healthy blood bank donor. (A) CD4/CD8 and (B) CD8/CD56 expression of PBMCs stimulated with PHA and IL-2 for 2 days followed by IL-2 alone for 2 weeks. (C) EBV DNA levels in stimulated PBMCs. DNA from the EBV BamHI W region DNA was quantified using real-time PCR. The EBV BamHI W fragment copy number per cell was calculated by the formula N = 2 × (W/B), where N is the EBV BamHI W copy number/cell, W is the EBV BamHI W copy number, and B is the bcl-2 copy number. Copy numbers of EBV-BamHI W gene per 1×106 cells are indicated. U indicates undetectable. (D) RT-PCR analysis of EBV latency genes in stimulated PBMCs. Southern blots of PCR products were hybridized with [32P]-labeled probes. (E) Cytokine levels were measured in culture supernatants of stimulated PBMCs from a healthy donor (1), an unrelated patient with CAEBV (2), the patient with the perforin mutations (3), the patient's mother (4), and the patient's father (5). Cells were stimulated with anti-CD3 antibody (black bars) or isotype control antibody (white bars). The experiment was performed 3 times with similar results.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/4/10.1182_blood-2003-06-2171/6/m_zh80040456890002.jpeg?Expires=1765004675&Signature=pZs4Hc4U-6XfVa1vdv-WbAqU1C8OEej4KaJTwIHWk7pUzOYXmEcOOQoh7Ea5QDjcIOfKL1zlsHEpC9Lc-2RAHeuyMBpLxrkBT9-3~075bZsrAWlVCLstJE9Ebs2Z-rALD302hrlS9tvdgbpBAJGBXXOHRT3yVYl1DtDJ-YZeLS~lN63XvHre8Yv---Hb9XUCB3DGK9HOo7uZ7R8oNPF7DkticMBMY8ITim6mGeJiavJo6IRr5aXYaIecx-04qr~qZb5fDWGny81cbw6X1zEnWqlZbSdwgueLswNaz0uEt7H7hEaQXOCDWHO1VgeQanDeSqgKjWFeLTfk~4uCI2wE4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Cytotoxicity and granule exocytosis assays using stimulated PBMCs from the patient, his parents, an unrelated patient with CAEBV, and healthy donors. (A) PBMCs were stimulated with PHA and IL-2, cultured in IL-2 (effector cells), and flow cytometry showed lymphocyte phenotypes described in Figure 2B. The cells were incubated with [51Cr]-labeled L1210 target cells (Fas-deficient target cells) in the presence of anti-CD3 (UCHT-1; BD Pharmingen) and anti-Fas blocking antibody (ZB4; Beckman Coulter, Fullerton, CA) in a Fas-independent CTL killing assay. Cytotoxicity in this assay system is CD3-dependent, since T-cell receptor activation is required to trigger release of granules containing perforin and granzymes.20 The effector-to-target (E/T) ratio is indicated on the x axis and the percent specific lysis is indicated on the y axis. The percent lysis was calculated as follows: % lysis = (E–S)/(M–S) × 100, where E is the release from experimental samples, S is the spontaneous release, and M is the maximum release upon lysis with 2% NP-40. The gray band indicates the normal range of cytotoxicity seen in 4 healthy donors (data not shown). Cells from a healthy blood bank donor incubated in the absence of anti-CD3 antibody were assayed as a negative control. Flow cytometry showed the following lymphocyte phenotypes: donor 1: CD8 = 49%, CD56 = 42%; donor 2: CD8 = 55%, CD56 = 42%; patient: CD8 = 54%, CD56 = 25%; mother: CD8 = 50%, CD56 = 68%; father: CD8 = 91%, CD56 = 4%; unrelated patient with CAEBV: CD8 = 55%, CD56 = 40%. (B) PBMCs were stimulated with PHA and IL-2 followed by incubation in the presence (black bars) and absence (white bars) of plate-bound anti-CD3/anti-CD28 antibody. Secretion of β-hexosaminidase in the cell culture supernatant was measured to quantify the level of granule exocytosis.33 β-hexosaminidase release was expressed as a percentage of the enzyme in the supernatant divided by the total enzyme from cells lysed in 0.1% Triton X-100. Experiments in panels A and B were performed 3 times and a representative result is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/4/10.1182_blood-2003-06-2171/6/m_zh80040456890007.jpeg?Expires=1765004675&Signature=h4A303qgpMCtSc8veJ9WKgVjjGgXpNtY~4eS3blbw3dZ~0EPO2fNcpKEEdAg5XEMOWOERZQCUsWPEsulM71O7HIDMIcWiAhA5XvfbYkAJtLDbLMU7AuxQYMUINIZDeWCPjJ9lCoCKcph2ck7wm0JQo~ySByDyn8yUwvWdk-466amF-momPTVhAMNzhdoMAXKzQL4m~tIpNvSGY~C~NT8Xti5wNcX1L3lb5j4sFfsRFfiUB0j9pthDL1q~MN0xpuG2Jam7eethr7v5rREsZG-YW2gFUeh6PcNHA3K3rken9W~Y8lY74K9NZ0EpPFKvRMZiglIEyjj~64CDipw90QWog__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)