Abstract
Hypoxia is a key factor in tumor development, contributing to angiogenesis and radiotherapy resistance. Hypoxia-inducible factor-1 (HIF-1) is a major transcription factor regulating the response of cancer cells to hypoxia. However, tumors also contain areas of more severe oxygen depletion, or anoxia. Mechanisms for survival under anoxia are HIF-1α independent in Caenorhabditis elegans and, thus, differ from the hypoxic response. Here we report a differential response of cancer cells to hypoxia and anoxia by demonstrating the induction of activating transcription factor-4 (ATF-4) and growth arrest DNA damage 153 (GADD153) protein specifically in anoxia and the lack of induction in hypoxia. By applying RNAi, ATF-4 induction in anoxia was shown to be independent of HIF-1α, and desferrioxamine mesylate (DFO) and cobalt chloride induced HIF-1α but not ATF-4 or GADD153. Furthermore, the inductive response of ATF-4 and GADD153 was not related to alterations in or arrest of mitochondrial respiration and was independent of von Hippel-Lindau (VHL) disease mutations. In reoxygenated anoxic cells, ATF-4 had a half-life of less than 5 minutes; adding the proteasome inhibitor to normoxic cells up-regulated ATF-4 protein. Extracts from primary human tumors demonstrated more ATF-4 expression in tumors near necrotic areas. Thus, this study demonstrates a novel HIF-1α–independent anoxic mechanism that regulates ATF-4 induction at the protein stability level in tumor cells.
Introduction
Cells from humans to Caenorhabditis elegans have a sensing pathway for hypoxia involving the hypoxia-inducible factor-1 (HIF-1) complex.1,2 HIF-1 is a heterodimer that includes HIF-1α and the aryl hydrocarbon receptor nuclear translocator (ARNT).3 In normoxia, prolyl hydroxylase modifies HIF-1α by hydroxylation of 2 prolyl residues,4 leading to its interaction with the von Hippel-Lindau protein (pVHL) and resulting in ubiquitination and degradation in the proteasome. In hypoxia, hydroxylation is blocked, and HIF-1α is stable.5,6 Mild levels of hypoxia (5%-0.5% O2) result in the up-regulation of HIF-1,7 which activates a gene transcription program, including the switch-on of glycolysis,8 and angiogenic factors, such as vascular endothelial growth factor (VEGF),9 that serve to protect the cell from the damaging effects of hypoxia.
HIF-1α and HIF-2α have been shown to be up-regulated in many cancers compared with normal tissue and to be related to tumor hypoxia.10,11 However, in addition to milder levels of hypoxia, tumors contain areas of severe hypoxia and anoxia, which has been suggested as a functionally different state than hypoxia.12-14 In severe hypoxia (0.1% O2) or anoxia (less than 0.1% O2), cells undergo p53-dependent apoptosis,13 and HIF-1 can mediate this program of death.15 Mutations in apoptotic pathways, particularly p53, can result in the survival and, hence, the selection of tumor cells for mutations under conditions of extreme hypoxia or anoxia.16 Because HIF-1α is a major contributor to tumor cell survival pathways, it is considered a therapeutic target.17 Additionally, the use of hypoxia response elements (HREs) regulated by HIF-1α is being developed for gene therapy.18-20 HRE activity in hypoxia and anoxia is dependent on HIF-1,20 but the level of oxygen tension necessary to induce various hypoxic transcription factors seem to differ. For example, HIF-1α is induced by a broad range of hypoxic levels, including anoxia,21 whereas other factors, such as p53, apoptosis inhibitory protein IAP-2, and activating transcription factor-4 (ATF-4), are induced in severe hypoxia and anoxia.13,22,23 This differential induction of various transcription factors by hypoxia compared with anoxia suggests that cellular responses to anoxia are different from those to hypoxia. However, the potential factors that may determine the cellular response and adaptation to anoxia in an HIF-1–independent manner are not well understood. It has been shown that C elegans can survive and adapt to hypoxia through the HIF-1 signaling pathway.24 However, C elegans can also adapt to anoxia by entering a reversible state of suspended animation independent of the hypoxic HIF-1 signaling pathway.25
The basic region-leucine zipper (bZip) factor ATF-4 has been shown to be up-regulated in primary rat fibroblasts under anoxia,23 but whether this is HIF-1 regulated remains unknown. ATF-4, which generally binds the cyclic adenosine monophosphate (cAMP)–responsive element (CRE),26 showed increased DNA binding activity to a variant site under anoxia.23 Binding of ATF-4 to variant sites during conditions of stress can activate the expression of cognate target genes such as growth arrest DNA damage/C-EBP homologous protein (GADD153/CHOP).27,28
Posttranslational regulation by protein degradation is one crucial mechanism of the hypoxic signaling pathway leading to HIF-1 induction. Considering posttranslational regulation by protein degradation, C/ATF, the mouse homologue of human ATF-4, has been shown to be an unstable protein with a half-life of less than 30 minutes.29 Recently, it has been shown that human ATF-4 is an unstable protein targeted to proteasomal degradation through phosphorylation-dependent interaction with the SCFtrcp ubiquitin ligase.30
To investigate the differential response of human cancer cells to hypoxia and anoxia and to determine different regulatory pathways to the hypoxia/HIF-1 system, the expression of ATF-4 and its target gene GADD153 in normoxia (21% O2), severe hypoxia (0.1% O2), and anoxia (less than 0.1% O2) was studied. This report describes the findings that ATF-4 is induced by anoxia rather than hypoxia and that this induction does not involve HIF-1 or electron transport but occurs mainly by stabilization of the protein, which has a half-life after anoxia reoxygenation of less than 5 minutes.
Materials and methods
Cells
The human breast cancer cell lines MCF-7, T47D, MDA-MB 231, 435, 468, the human melanoma cell line LB-4, and the VHL mutant renal cancer cell line 786-0 were provided by Cancer Research UK (London, United Kingdom). Murine neuroblastoma 2a cell line was a gift from Professor Ernst Wagner (Ludwig-Maximilian University, Munich, Germany). Cells were maintained in Dulbecco modified Eagle medium (DMEM, with 4.5 mg/mL glucose) supplemented with 10% (vol/vol) fetal calf serum, penicillin (100 U/mL) and streptomycin (100 μg/mL), and 4 mM l-glutamine (Gibco, Paisley, United Kingdom). Previously, 786-0 renal cells had been described,31 and they were supplemented with 500 μg/mL G418 for selection.
Reagents and plasmids
Rabbit polyclonal antibody to ATF-4 (sc-200) and mouse monoclonal antibody to GADD153 (sc-7351) were from Santa Cruz Biotechnology (Calne, United Kingdom). The monoclonal antibody to HIF-1α was from Signal Transduction Laboratories, and the tubulin antibody was from Sigma (Poole, United Kingdom). Tunicamycin, KCN, MG132, arsenite, desferrioxamine, and cobalt chloride were from Sigma. The ATF-4 expression plasmid pCG-ATF-4 has been described previously.26 The bluescript II KS plasmid used in RNase protection assay (RPA) was from Stratagene (Amsterdam, The Netherlands). The Hif-1 sense- and antisense-stranded siRNA oligonucleotides were designed to specifically target the Hif-1α mRNA, whereas the inverted control did not target any known gene. Details of the oligonucleotides (which were purchased from Cruachem, Glasgow, Scotland) were as follows: Hif-1α antisense, 5′-CUGAUGACCAGCAACUUGAdTdT-3′; Hif-1α sense, 5′-UCAAGUUGCUGGUCAUCAGdTdT-3′; control antisense, 5′-AGUUCAACGACCAGUAGUCdTdT-3′; and control sense, 5′-GACUACUGGUCGUUGAdTdT -3′. Duplexes were prepared by mixing 50-μM concentrations of antisense and sense oligonucleotides with annealing buffer (30 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] pH 7.0, 100 mM potassium acetate, and 2 mM magnesium acetate), heat-denaturing for 1 minute at 85°C, and annealing at 37°C for 1 hour.
Hypoxic incubation
For hypoxic incubation, cells were placed in a Heto-Holten cell house 170 (Heto-Holten, Surrey, United Kingdom) incubator set at 0.1% O2. Hypoxic conditions were achieved with 95% N2/5% CO2. The oxygen level was monitored with an in-built O2 sensor. Cells were trypsinized from a 70% to an 80% dense culture, and 1.5 to 2 × 106 cells in 100 mm tissue culture Petri dishes with 6 to 7 mL culture media were incubated immediately in hypoxia or were incubated first in a normal (normoxia) 5% CO2 incubator for 20 to 24 hours and then placed for the corresponding times in hypoxia. In all cases cells were approximately 60% to 70% confluent. The same treatment was used for cells to be incubated in normoxia (21% O2).
Anoxic incubation
A humidified gas-sorted anoxic incubator-gloved box (InVivo2 400; Ruskin, Leeds, United Kingdom) was used for anoxic experiments. The gas was sorted using a Ruskin Microaerophilic gas sorter, resulting in 5% H2, 5% CO2, and 90% N2. The humidity level was controlled by placing 4 200 × 25-mm tissue culture Petri dishes (Becton Dickinson Labware, Oxford, United Kingdom) filled with dH2O in the gloved box and using an in-built humidity sensor to ensure the humidity level was greater than 90%. Two previously unused (ie, not reheated) palladium catalysts were used to scavenge traces of oxygen. Anoxic conditions were controlled using an anaerobic indicator (Oxoid, Basingstone, United Kingdom) that demonstrated anoxic conditions within 10 to 15 minutes of gas purging and an O2 sensor installed in the gloved-box incubator that demonstrated 0% oxygen within 15 minutes of gas purging. After the indicators showed satisfactory anoxic conditions, gas was purged in the gloved box for another 45 minutes. Cells (1.5 × 106) with 6 to 7 mL media in 100-mm Petri dishes were placed in the chamber immediately or after 24 hours of normoxic incubation, with approximately 70% confluence.
RNAi treatment of cells and transfection procedures
Cells were plated onto 100-mm Petri dishes and were grown to approximately 50% confluence before transfection. HIF-1α or inverted control duplex was diluted to give a final concentration of 20 nM in Opti-Mem I (Invitrogen Life Technologies). Twenty-five microliters Oligofectamine transfection reagent (Invitrogen Life Technologies) was added, and the mixture was incubated at room temperature for 25 minutes. Cells were rinsed with Opti-Mem I to remove any residual serum and were incubated with the oligonucleotide duplexes in serum-free conditions for 4 hours at 37°C. Serum was then replenished in the cells, which were further incubated for 24 hours before anoxic pressure was applied, as described above.
To test the anti–ATF-4 antibody used in this study, MCF-7 cells were transfected with 1 μg pCG-ATF4 by using Fugene-6 according to the manufacturer's protocol (Roche Diagnostics, Sussex, United Kingdom). After 24 hours, cells were lysed by using a urea-denaturing buffer (described below) and analyzed for ATF-4 expression.
Immunoblot analysis
All cell extracts were made in a cold room (4°C). The cell lysis buffer used for preparing total cell extracts was a urea-denaturing buffer (6.7 M urea, 10 mM Tris-HCl, pH 6.8, 5 mM dithiothreitol [DTT], 1% sodium dodecyl sulfate [SDS], and 10% glycerol) supplemented with Complete mini-protease inhibitor cocktail tablets (Roche Diagnostics). Cultured cells were washed rapidly once with ice-cold phosphate-buffered saline (PBS), scraped off, and centrifuged at 13 000 rpm for 20 seconds, and 250 μL urea buffer was added directly to the cell pellet. This was then homogenized rapidly on ice for 15 seconds by using the Ultra-Turrax homogenizer at full speed (IKA, Düsseldorf, Germany). Nuclear and cytoplasmic extracts were based on the method described by Schreiber et al32 but slightly modified. Cells were washed rapidly once with ice-cold phosphate-buffered saline (PBS), scraped off, and centrifuged for 20 seconds at 13 000 rpm. Cells were resuspended in 4 packed cell volumes of ice-cold buffer A (10 mM HEPES, pH 7.5, 0.1 mM EDTA [ethylenediaminetetraacetic acid], pH 8, 0.1 mM EGTA [ethyleneglycotetraacetic acid], pH 8, 10 mM KCl) (Sigma) supplemented with Complete mini-protease inhibitor cocktail tablets (Roche Diagnostics) for cytoplasmic extracts. For nuclear extracts, the cell pellet was resuspended in buffer C (20 mM HEPES, pH 7.5, 1 mM EDTA, pH 8), 1 mM EGTA, pH 8, 0.4 M NaCl) supplemented in Complete mini-protease inhibitor cocktail tablets (Roche Diagnostics). The volume of buffer C used was the same as buffer A.
To make tumor and normal tissue extracts, tumor tissue (or paired tumor/normal tissue) from patients with invasive breast cancer was frozen in liquid nitrogen immediately after surgery and stored at –80°C until extraction. Then 100 to 1000 mg frozen tissue was pulverized on dry ice to a fine powder, and ice-cold extraction buffer (20 mM HEPES, pH 7.4, 1.5 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM benzamidine, and 10 μg/mL ovomucoid trypsin inhibitor) was added at a 0.2 g tissue/2 mL buffer ratio. The resultant suspension was homogenized on ice with an Ultra-Turrax. The mixture was then centrifuged at 13 000 rpm at 4°C for 30 seconds, and the supernatant (cytosol) was collected and centrifuged at 13 000 rpm at 4°C for 30 minutes. The pellet was then resuspended in a urea-denaturing buffer supplemented with Complete mini-protease inhibitor cocktail tablets (Roche Diagnostics) and was homogenized on ice with an Ultra-Turrax. This was then centrifuged at 4°C for 5 minutes at 13 000 rpm, and the supernatant was analyzed for ATF-4.
The detergent-compatible (DC) protein assay (Bio-Rad, Hertfordshire, United Kingdom) was used to estimate the protein concentration of extracts according to the manufacturer's protocol. Nuclear/cytoplasmic (25 μg/lane) or total cell extracts (50-60 μg/lane) were subjected to reducing (10%-12%) sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS). Resolved proteins were then electroblotted (semidry) onto a Millipore Immobilon-P transfer membrane (PVDF microporous membrane). Bound antibodies were detected using the chemiluminescent substrate ECL+Plus (Amersham Biosciences, Buckinghamshire, United Kingdom).
RNase protection assay
RNA was extracted from the cells using the tri-reagent method according to the manufacturer's protocol (Sigma). The human ATF-4 gene (316 bp) was subcloned from the pCG-ATF-4 plasmid into the KpnI and EcoRV restriction sites of the bluescript II KS (Stratagene). This ATF-4 fragment was a portion of the human ATF-4 coding region from positions 565 to 881, which served as the DNA template for generating 32P-labeled RNA probes.
To generate 32P-labeled RNA probes, the bluescript II KS containing the ATF-4 fragment was linearized by using the EcoRV restriction enzyme (New England Biolabs, Hertfordshire, United Kingdom). Constitutively expressed U6 small nuclear RNA served as an internal control. RPA was carried out as described previously.33
ATF-4 immunostaining of human tumors
Five-micrometer sections were cut from 10 ductal invasive carcinomas of human breast, obtained from the Cancer Research UK, Oxford Cancer Center. After dewaxing and rehydration, endogenous peroxidase was blocked using 3% H2O2 in methanol for 20 minutes, followed by antigen retrieval (immersion in 0.01 M trisodium citrate and microwaving on high power for 10 minutes [800-W microwave]). After washing in PBS and incubation with 5% normal horse serum (Vector Laboratories, Peterborough, United Kingdom) for 20 minutes, sections were incubated for 2 hours at room temperature in a negative control antibody (purified rabbit immunoglobulin fraction) at a concentration of 2 μg/mL (DAKO) or in a polyclonal antihuman ATF-4 antibody (Santa Cruz) at the same concentration (2 μg/mL). The reaction was then detected using a standard 3-stage immunoperoxidase technique (Vector Laboratories Elite ABC kit) with the brown chromogen, diamino benzidine (DAB). Sections were counter-stained in hematoxylin (British Drug Houses [BDH]), dehydrated, cleared in xylene, and mounted in DPX (BDH).
Results
Induction of ATF-4 by anoxia
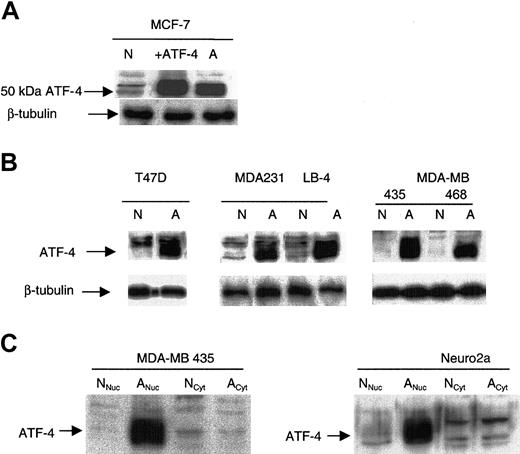
Transfection of MCF-7 cells with the pCG-ATF-4 expression plasmid resulted in a markedly increased expression of a 50-kDa protein (Figure 1A). The same 50-kDa ATF-4 protein was up-regulated in other cancer cells incubated for 16 hours in anoxia (Figure 1A-B). Anoxically induced ATF-4 was nuclear, and no differences in ATF-4 levels between normoxic and anoxic cytoplasmic extracts were detected (Figure 1C). The high expression detected in normoxia in the transfectant likely represents effective transcription and translation from the promoter and the saturation of proteasomal degradation.
ATF-4 protein is induced in cancer cells under anoxia. (A) ATF-4 overexpression in normoxia (designated as +ATF-4) in MCF-7 cells confirmed the up-regulated band in anoxia to be ATF-4 (50 kDa). (B) Various cancer cell lines, including T47D, MDA-MB 231, 435, 468, and LB-4, were incubated for 16 hours under the corresponding oxygen levels and were analyzed by immunoblot for ATF-4 protein level. N indicates normoxia (21% O2); A, anoxia. (C) MDA-MB 435 and murine N2a cells were incubated for 16 hours in either normoxia or anoxia and were analyzed by immunoblot for ATF-4 protein level in cytoplasmic and nuclear extracts. As indicated, ATF-4 induction in anoxia was only observed in nuclear extracts but not in cytoplasmic extracts. Nuc indicates nuclear; Cyt, cytoplasmic.
ATF-4 protein is induced in cancer cells under anoxia. (A) ATF-4 overexpression in normoxia (designated as +ATF-4) in MCF-7 cells confirmed the up-regulated band in anoxia to be ATF-4 (50 kDa). (B) Various cancer cell lines, including T47D, MDA-MB 231, 435, 468, and LB-4, were incubated for 16 hours under the corresponding oxygen levels and were analyzed by immunoblot for ATF-4 protein level. N indicates normoxia (21% O2); A, anoxia. (C) MDA-MB 435 and murine N2a cells were incubated for 16 hours in either normoxia or anoxia and were analyzed by immunoblot for ATF-4 protein level in cytoplasmic and nuclear extracts. As indicated, ATF-4 induction in anoxia was only observed in nuclear extracts but not in cytoplasmic extracts. Nuc indicates nuclear; Cyt, cytoplasmic.
ATF-4 mRNA level in normoxia and anoxia
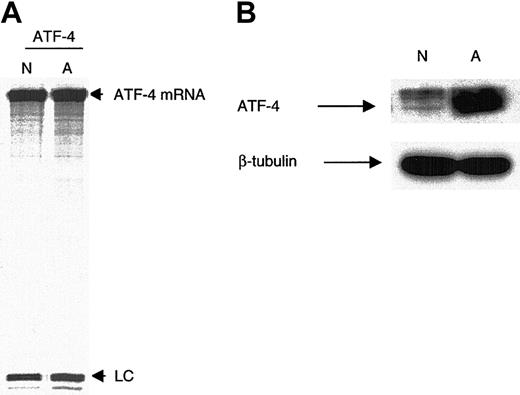
RPA demonstrated that ATF-4 mRNA was present at similar levels in normoxia and anoxia in MDA-MB 435 cells (Figure 2A). In a synchronous control, immunoblot analysis demonstrated that ATF-4 protein was induced in anoxia (Figure 2B), suggesting that ATF-4 induction in anoxia is not at the level of transcription or RNA stability.
Levels of ATF-4 mRNA between normoxia and anoxia do not vary. (A) MDA-MB 435 cells were incubated for 16 hours in either normoxia or anoxia, and ATF-4 mRNA levels were analyzed using RPA. As indicated, abundant ATF-4 mRNA was present in similar levels under normoxia and anoxia. (B) Immunoblot control for the RPA demonstrated strong anoxic induction of the ATF-4 protein. n indicates normoxia; A, anoxia; LC, loading control (small nuclear U6 RNA).
Levels of ATF-4 mRNA between normoxia and anoxia do not vary. (A) MDA-MB 435 cells were incubated for 16 hours in either normoxia or anoxia, and ATF-4 mRNA levels were analyzed using RPA. As indicated, abundant ATF-4 mRNA was present in similar levels under normoxia and anoxia. (B) Immunoblot control for the RPA demonstrated strong anoxic induction of the ATF-4 protein. n indicates normoxia; A, anoxia; LC, loading control (small nuclear U6 RNA).
Effect of reoxygenation and proteasome inhibition on ATF-4 protein level
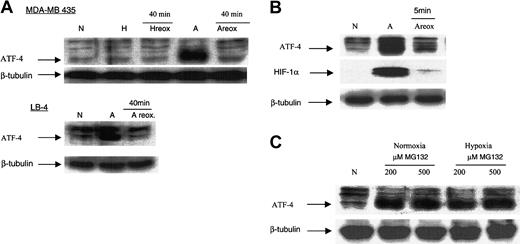
After 16 hours of exposure to anoxia, MDA-MB435 and LB-4 cells were placed for 40 minutes in an incubator with normal 5% CO2 (normoxia, 21% O2). This reoxygenation resulted in a decline in ATF-4 protein levels to those similar for normoxic cells (Figure 3A), suggesting that anoxic ATF-4 protein is unstable on reoxygenation after anoxia. This was further confirmed by reoxygenating the cells through replacing the anoxic media with normoxic media and placing the cells in a normoxic incubator for 5 minutes. Within 5 minutes, ATF-4 and HIF-1α proteins had already degraded (Figure 3B). To determine whether ATF-4 induced by anoxia was rapidly degraded in normoxia, normoxic and hypoxic cells were treated with 200 and 500 μM of the proteasome inhibitor MG132. In normoxia and hypoxia, proteasome inhibition resulted in up-regulation of the ATF-4 protein (Figure 3C). Thus, collectively these results indicated that ATF-4 induction in anoxia was caused by posttranscriptional, probably proteolytic, mechanisms.
Reoxygenation of anoxic cells results in a decrease in ATF-4 protein level, and proteasome inhibition results in an increase in ATF-4 protein level. (A) MDA-MB 435 and LB-4 cells were incubated for 16 hours in normoxia, hypoxia, or anoxia. After 16 hours, anoxic cells (or hypoxic cells, as in the case of MDA-MB 435 cells) were then immediately placed in a normoxic incubator for 40 minutes. This reoxygenation (reox.) of anoxic MDA-MB 435 and LB-4 cells resulted in the down-regulation of ATF-4 protein levels. Reoxygenation of hypoxic MDA-MB 435 cells did not affect the level of ATF-4 protein compared with hypoxic cells. (B) MDA-MB 435 cells were incubated for 16 hours in anoxia, after which the anoxic media of the cells were replaced with normal fresh media (oxygenated), and the cells were incubated in a normoxic incubator for 5 minutes. Cells were then analyzed by immunoblot for ATF-4 protein level. (C) MDA-MB 435 cells were incubated in normoxia for 16 hours, after which 200 or 500 μM (end concentration) of the proteasome inhibitor MG132 was added directly to the cells. Cells were incubated another 16 hours in normoxia or hypoxia and then analyzed using immunoblot for ATF-4 protein levels. N indicates normoxia; H, hypoxia; A, anoxia.
Reoxygenation of anoxic cells results in a decrease in ATF-4 protein level, and proteasome inhibition results in an increase in ATF-4 protein level. (A) MDA-MB 435 and LB-4 cells were incubated for 16 hours in normoxia, hypoxia, or anoxia. After 16 hours, anoxic cells (or hypoxic cells, as in the case of MDA-MB 435 cells) were then immediately placed in a normoxic incubator for 40 minutes. This reoxygenation (reox.) of anoxic MDA-MB 435 and LB-4 cells resulted in the down-regulation of ATF-4 protein levels. Reoxygenation of hypoxic MDA-MB 435 cells did not affect the level of ATF-4 protein compared with hypoxic cells. (B) MDA-MB 435 cells were incubated for 16 hours in anoxia, after which the anoxic media of the cells were replaced with normal fresh media (oxygenated), and the cells were incubated in a normoxic incubator for 5 minutes. Cells were then analyzed by immunoblot for ATF-4 protein level. (C) MDA-MB 435 cells were incubated in normoxia for 16 hours, after which 200 or 500 μM (end concentration) of the proteasome inhibitor MG132 was added directly to the cells. Cells were incubated another 16 hours in normoxia or hypoxia and then analyzed using immunoblot for ATF-4 protein levels. N indicates normoxia; H, hypoxia; A, anoxia.
Induction of ATF-4 by anoxia and relation to GADD153/CHOP10
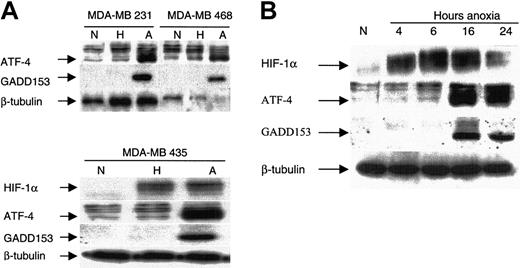
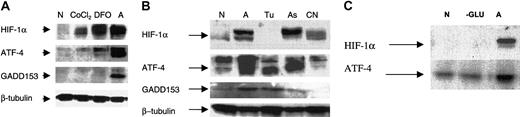
ATF-4 expression in hypoxia and anoxia and correlation with GADD153 and HIF-1α. Hypoxia (0.1% O2) did not up-regulate ATF-4 or GADD153, whereas anoxia resulted in the strong induction of both proteins in breast cancer cells (Figure 4A). In contrast, HIF-1α was up-regulated by hypoxia and anoxia (Figure 4A). This shows that ATF-4 and GADD153 induction is a response that differentiates between hypoxia and anoxia.
Induction of ATF-4 by anoxia correlates with the induction of GADD153/CHOP10 but not with that of HIF-1α. (A) Indicated MDA-MB breast cancer cells were incubated under normoxic, hypoxic, or anoxic conditions for 16 hours and were analyzed by immunoblot for the steady state levels of the indicated proteins: ATF-4, GADD153, HIF-1α, and β-tubulin (as an internal control). (B) MDA-MB435 cells were incubated under anoxia for various times, as indicated, and were analyzed by immunoblot for the indicated proteins. Cells from normoxic conditions were included as a control. N indicates normoxia; H, hypoxia; A, anoxia.
Induction of ATF-4 by anoxia correlates with the induction of GADD153/CHOP10 but not with that of HIF-1α. (A) Indicated MDA-MB breast cancer cells were incubated under normoxic, hypoxic, or anoxic conditions for 16 hours and were analyzed by immunoblot for the steady state levels of the indicated proteins: ATF-4, GADD153, HIF-1α, and β-tubulin (as an internal control). (B) MDA-MB435 cells were incubated under anoxia for various times, as indicated, and were analyzed by immunoblot for the indicated proteins. Cells from normoxic conditions were included as a control. N indicates normoxia; H, hypoxia; A, anoxia.
Time course of anoxic induction of ATF-4.Figure 4B shows that HIF-1α was already induced within 4 hours of anoxic incubation, whereas both ATF-4 and GADD153 were induced after 16 hours of anoxic incubation. Thus, as reported previously, HIF-1α induction was an early event; in contrast, ATF-4 and GADD153 induction were late responses. Furthermore, though ATF-4 and GADD153 protein levels were maintained after 24 hours of incubation, a decrease was observed in the HIF-1α protein level. These results further demonstrate that the presence of HIF-1α is not required for the maintenance of ATF-4 and GADD153 induction.
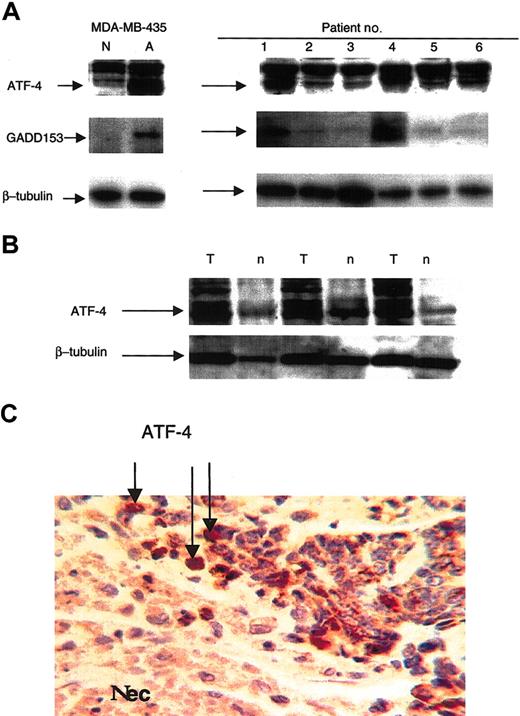
ATF-4 induction in primary tumor extracts, paired primary tumor and normal tissue extracts, and expression near necrotic areas. Extracts made from patient primary tumors demonstrated ATF-4 expression similarly to the observations made with the in vitro cell culture extracts (Figure 5A). Variations were observed among patient extracts. Some demonstrated greater levels of the 50-kDa ATF-4 protein than others, suggesting that anoxic stress may be more pronounced in some tumors than in others. Interestingly, GADD153 expression was also observed in the tumors, with the greatest levels evident in patients 1 and 4, who also showed the greatest level of ATF-4 expression (Figure 5A). In addition, paired tumor (T)/normal (n) tissue extracts demonstrated ATF-4 expression to be greater in tumors than in normal tissues (Figure 5B). ATF-4 expression was analyzed further through immunostaining of 10 ductal invasive carcinomas of the breast. Enhanced ATF-4 expression (brown) was shown by tumor cells around areas of necrosis (Nec) known to be anoxic (Figure 5C), though it should be noted that ATF-4 staining was not restricted to these areas alone because it was also seen in other tumor areas in some tumors. No such staining was seen when the primary (ATF-4) antibody was replaced with the same concentration of a purified immunoglobulin fraction from a nonspecific rabbit serum (not shown). Nuclei were highlighted in sections using hematoxylin (blue).
Expression of ATF-4 in primary human breast cancer tumors correlates with that of GADD153, and tumor-section staining indicates ATF-4 expression near necrotic areas. (A) Extracts made from primary malignant human breast tumors were analyzed by immunoblot for ATF-4 and GADD153 protein levels. MDA-MB 435 cells incubated for 16 hours in normoxia or anoxia were used as control. (B) Paired malignant tumor (T)/normal (n) tissue extracts were analyzed using immunoblot for ATF-4 expression. (C) After dewaxing and rehydration, 5-μm sections of metastatic breast tumors were analyzed for ATF-4 expression by immunostaining. Original magnification, × 120.
Expression of ATF-4 in primary human breast cancer tumors correlates with that of GADD153, and tumor-section staining indicates ATF-4 expression near necrotic areas. (A) Extracts made from primary malignant human breast tumors were analyzed by immunoblot for ATF-4 and GADD153 protein levels. MDA-MB 435 cells incubated for 16 hours in normoxia or anoxia were used as control. (B) Paired malignant tumor (T)/normal (n) tissue extracts were analyzed using immunoblot for ATF-4 expression. (C) After dewaxing and rehydration, 5-μm sections of metastatic breast tumors were analyzed for ATF-4 expression by immunostaining. Original magnification, × 120.
Induction of ATF-4 does not involve HIF-1α, mitochondrial electron transport, or energy depletion
HIFs are another group of oxygen-sensitive transcription factors, regulated by proteolysis through the VHL protein. To investigate the relation of the ATF-4 anoxic response to the HIF proteolytic pathway, renal cell lines with mutant VHL were used. The 50-kDa ATF-4 protein was not induced constitutively in parental 786-0 cells (ie, VA, lacking pVHL) or 786-0 cells with pVHL expression vector, but it was up-regulated in both cells under anoxia only (Figure 6A). These cell lines express HIF-2α rather than HIF-1α and showed the expected constitutive up-regulation in the mutant cells and inducible expression in the cells transfected with wild-type VHL. Thus, anoxic induction of the 50-kDa ATF-4 protein is not dependent on the VHL status of cells.
Induction of ATF-4 in anoxia does not require pVHL or HIF-1α. (A) Renal cancer cell line 786-0, deficient in pVHL (VA) or expressing pVHL (+pVHL), was incubated for 16 hours in normoxia or anoxia and analyzed after 16 hours by immunoblot analysis for ATF-4 protein level and HIF-2α. VA indicates vector alone (ie, not expressing pVHL): N, normoxia; A, anoxia. (B) MDA-MB 435 cells were incubated for approximately 20 hours until they were 50% confluent. Cells were then treated with RNAi to block HIF-1α expression. Cells were incubated for 24 hours in normoxic conditions, after which they were placed in anoxia for 16 hours or left in normoxia for 16 hours. Normoxic and anoxic cells were then analyzed by immunoblot for the indicated proteins. C indicates inverted control RNAi.
Induction of ATF-4 in anoxia does not require pVHL or HIF-1α. (A) Renal cancer cell line 786-0, deficient in pVHL (VA) or expressing pVHL (+pVHL), was incubated for 16 hours in normoxia or anoxia and analyzed after 16 hours by immunoblot analysis for ATF-4 protein level and HIF-2α. VA indicates vector alone (ie, not expressing pVHL): N, normoxia; A, anoxia. (B) MDA-MB 435 cells were incubated for approximately 20 hours until they were 50% confluent. Cells were then treated with RNAi to block HIF-1α expression. Cells were incubated for 24 hours in normoxic conditions, after which they were placed in anoxia for 16 hours or left in normoxia for 16 hours. Normoxic and anoxic cells were then analyzed by immunoblot for the indicated proteins. C indicates inverted control RNAi.
Treating the cells with RNAi blocked HIF-1α induction in anoxia, whereas cells treated with sense RNA induced HIF-1α in anoxia (Figure 6B). However, RNAi- and sense RNA-treated cells responded to anoxia by inducing ATF-4. Thus, the anoxic response leading to ATF-4 induction is not dependent on the presence of HIF-1α expression in anoxia.
Desferrioxamine or cobalt chloride (200 μM) did not up-regulate ATF-4 or GADD153 but did induce HIF-1α, whereas anoxia up-regulated all 3 proteins. This also shows that DFO and CoCl2 (at 200 μM) do not imitate the anoxic conditions necessary for ATF-4 and GADD153 induction (Figure 7A).
Effect of tunicamycin, potassium cyanide, glucose deprivation, DFO, and CoCl2 on ATF-4 induction. (A) MDA-MB 435 cells were incubated for approximately 16 hours in normoxia after, which 200 μM DFO or CoCl2 (end concentration) was added directly to the cells. Cells were then analyzed after 16 hours by immunoblot for the indicated proteins: HIF-1α, ATF-4, GADD153, and β-tubulin (as an internal control). (B) MDA-MB 435 cells were incubated for 16 hours in normoxia and then stressed by either anoxia (A), tunicamycin (Tu; 1 mg/mL), arsenite (As; 2.5 μM), or cyanide (CN; 5 mM) for another 16 hours. Cells were analyzed by immunoblot for the indicated proteins. (C) MDA-MB435 cells incubated in glucose-free medium for 16 hours (–G) or in full medium and anoxia for 16 hours. Cells were then analyzed by immunoblot for the indicated proteins.
Effect of tunicamycin, potassium cyanide, glucose deprivation, DFO, and CoCl2 on ATF-4 induction. (A) MDA-MB 435 cells were incubated for approximately 16 hours in normoxia after, which 200 μM DFO or CoCl2 (end concentration) was added directly to the cells. Cells were then analyzed after 16 hours by immunoblot for the indicated proteins: HIF-1α, ATF-4, GADD153, and β-tubulin (as an internal control). (B) MDA-MB 435 cells were incubated for 16 hours in normoxia and then stressed by either anoxia (A), tunicamycin (Tu; 1 mg/mL), arsenite (As; 2.5 μM), or cyanide (CN; 5 mM) for another 16 hours. Cells were analyzed by immunoblot for the indicated proteins. (C) MDA-MB435 cells incubated in glucose-free medium for 16 hours (–G) or in full medium and anoxia for 16 hours. Cells were then analyzed by immunoblot for the indicated proteins.
Tunicamycin (1 mg/mL) induced GADD153 and the 50-kDa ATF-4 protein but failed to induce HIF-1α in MDA-MB 435 cells. Arsenite (2.5 μM) up-regulated ATF-4, GADD153, and HIF-1α; however, ATF-4 induction did not appear as a 50-kDa protein but as a slightly heavier form, suggesting that arsenite induction of ATF-4 involves signaling pathways different from those of anoxia or tunicamycin, which lead to the induction of ATF-4 (Figure 7B). Cyanide (5 mM), in the presence of glucose (4.5 mg/mL), did not induce ATF-4 or GADD153, which shows that blocking the respiratory chain is not the mechanism for the anoxic response. Similar results were also obtained with MDA-MB 468 cells and with cyanide alone (data not shown).
Finally, glucose deprivation using glucose-free DMEM for 16 hours, which deprives cells of glycolytic and oxidative phosphorylation substrates, did not induce HIF1-α or ATF-4 (Figure 7C). Thus, these results indicate that the anoxia signaling pathway does not involve HIF-1α, VHL, adenosine triphosphate (ATP) depletion, or mitochondrial electron transport in inducing the 50-kDa ATF-4 protein.
Discussion
The data presented in this paper suggest that cells can sense and discriminate between severe hypoxia (0.1% O2) and anoxia in an HIF-1–independent manner. The hypoxia mimetics DFO and CoCl2 failed to induce ATF-4. The observations that hypoxia did not induce ATF-4 but did induce HIF-1α and that blocking HIF-1α induction in anoxia did not prevent the anoxic ATF-4 response suggest that cells can respond to anoxia independently of the hypoxic HIF-1 signaling pathway. Our data indicate the anoxic ATF-4 protein to be unstable in normoxia and the protein degradation mechanism to be a major pathway controllingATF-4 induction in cancer cell lines under anoxia.ATF-4 has been shown to contain a nuclear targeting signal in the C-terminus,34 which may account for the nuclear translocation of increased levels of ATF-4 protein during anoxic stress. ATF-4 mRNA was expressed at similar levels under normoxia and anoxia in MDA-MB 435 cells. This finding is concordant with the observation that ATF-4 mRNA is abundant in human tumor cell lines in normoxia.35 It also suggests primarily that ATF-4 up-regulation is not caused by an increase in mRNA transcription but rather by posttranscriptional events. However, this may be cell type dependent because previously it had been shown that anoxia results in increased levels of ATF-4 mRNA in primary rat fibroblasts.23
One pathway that may contribute to ATF4 regulation is that induced by endoplasmic reticulum (ER) stress, which can be initiated by several mechanisms, including unfolded proteins, tunicamycin, and calcium ionophores. Whether ER stress through tunicamycin mimics anoxic signaling pathways that up-regulate ATF-4 is uncertain, but several proteins induced by ER stress, such as ORP 150 or ATF-6, are also induced in anoxic conditions.36,37 Protein disulfide isomerase (PDI), which is necessary for disulfide bond formation and, hence, protein folding, is also induced maximally in anoxia.38 In addition, the endoplasmic reticulum resident kinase PERK has been shown recently to become hyper-phosphorylated under hypoxia and anoxia, independent of HIF-1α, leading to the phosphorylation of eIF2α, which then results in hypoxia-induced translational attenuation39 of most mRNAs. Moreover, the PERK-dependent translational attenuation pathway was shown to selectively increase the translation of ATF-4 mRNA during ER stress,40 and this ER-generated signal may mediate an adaptive cellular response to hypoxia.39 Thus, anoxia may also increase the translation efficiency of ATF-4 mRNA, contributing with protein stability pathways to the increase in ATF-4 protein levels. Based on these findings, anoxia but not hypoxia may result in pronounced ER stress, leading to the induction ATF-4.
Our observations are similar in some respects to those of Dong et al,22 who showed recently the up-regulation of another factor, the antiapoptotic protein IAP-2, in anoxia through an HIF-1–independent pathway. Milder hypoxic conditions (2% O2) than those used in our experiments (0.1% O2) were shown to induce HIF-1α but not IAP-2, whereas anoxia resulted in the induction of IAP-2. DFO, CoCl2, or arrest of the mitochondrial respiratory chain also could not mimic such anoxic conditions. Dong et al22 used 1 mg/mL glucose to prevent ATP depletion, and we used 4.5 mg/mL. Therefore, in neither case was ATP depletion a likely explanation. In contrast to our results, IAP-2 induction in anoxia was shown at the transcriptional level to be mainly attributed to activation of the IAP-2 gene promoter.41
We demonstrated ATF-4 expression to be greater in tumors than in normal tissue, but the function of ATF-4 in human tumors remains unknown. It has been shown in ATF-4 knock-out transgenic mice that ATF-4 is critical for processes that require high-level proliferation, such as during fetal-liver hematopoiesis.42 Other knock-out transgenics have suggested ATF-4 to be critical for preventing p53-dependent apoptosis in anterior lens epithelial cells.43,44 Recently, transgenic mice that specifically overexpressed ATF-4 in mammary epithelial cells demonstrated ATF-4 to function as an antiproliferation and a proapoptotic factor during mammary gland development,45 roles that may be relevant to cancer when expressed in anoxic areas. ATF-4 was also shown to block differentiation, another pathway important in cancer development. Thus, the role of ATF-4 in cancer development and progression must be investigated, and the use of ATF-4 as a potential marker to predict the severity of hypoxia in perinecrotic tumor areas must be considered.
ATF-4 is a substrate for β-trcp, an F-box protein involved with degradation within the proteasome and showing some similarity to HIF regulation. Although other β-trcp substrates, such as IκBα and CREB, are degraded in hypoxia (ie, the opposite effect to ATF4),46,47 at the completion of this paper, it was reported that in conditions of severe hypoxic stress, CREB and IκBα are modified by sumo—a small, ubiquitin-like protein—and, hence, stabilized.48 A similar mechanism may be involved in ATF-4 induction under anoxia. The critical factors responsible for ATF-4 stabilization in anoxia remain to be identified, particularly posttranslational modifications, which are being investigated in our laboratory.
Prepublished online as Blood First Edition Paper, November 20, 2003; DOI 10.1182/blood-2003-06-1859.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.