Abstract
Aberrant expression of transcription factor oncogenes such as HOX11, HOX11L2, TAL1/SCL, LYL1, LMO1, and LMO2 can be detected in lymphoblasts from up to 80% of patients with acute T-cell lymphoblastic leukemia (T-ALL). Transcriptional activation of these oncogenes in leukemic cells typically results from chromosomal rearrangements that place them next to highly active cis-acting transcriptional regulatory elements. However, biallelic activation of TAL1 in some T-ALL cases has been previously proposed. We have used allele-specific mRNA analysis to show that trans-acting mechanisms leading to biallelic overexpression of TAL1 are involved in 10 (42%) of 24 TAL1+ informative T-ALL cases, 2 (17%) of 12 HOX11+ informative cases, and 7 (64%) of 11 LMO2+ informative cases. We propose that aberrant expression of oncogenic transcription factors in a significant fraction of T-ALLs may result from loss of the upstream transcriptional mechanisms that normally down-regulate the expression of these oncogenes during T-cell development.
Introduction
Many studies over the past decade have implicated the overexpression of oncogenic transcription factors, including TAL1/SCL, HOX11, and LMO2 by way of chromosomal rearrangements, in the pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL).1 However, HOX11, TAL1, and LMO2 are frequently overexpressed in T-ALL samples in the absence of detectable cytogenetic abnormalities involving the chromosomal regions that contain these genes.2,3 These observations, coupled with the demonstration of biallelic expression of TAL1 and hypomethylation of HOX11 promoter sequences in T-ALL cases, support the hypothesis that mechanisms other than cis-acting chromosomal rearrangements can contribute to the aberrant expression of transcription factor oncogenes in T-ALL.4,5
Here, we report that the T-cell ALL oncogenes TAL1, HOX11, and LMO2 are biallelically expressed in a significant fraction of primary human T-ALL samples and propose a model for the pathogenesis of T-ALL involving the disruption of transcriptional pathways, which normally maintain tight control over the expression of these transcription factors during thymocyte development.
Study design
Patients
Samples of cryopreserved lymphoblasts from 29 adults and 30 children with T-ALL were obtained from St Jude Children's Research Hospital (n = 30), the Cancer and Leukemia Group B (CALGB) (n = 25), and the Eastern Cooperative Oncology Group (ECOG) (n = 4). The cells were collected with informed consent at the time of diagnosis.
Real-time quantitative RT-PCR
Real-time quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) analysis of HOX11, TAL1, LMO2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was performed as previously described.2
Allelic expression analysis
Polymorphic markers in the 3′ untranslated regions of the TAL1, HOX11, and LMO2 genes were identified by PCR amplification and direct sequencing of genomic DNA (gDNA). Allelic expression analysis was performed by direct sequencing of RT-PCR products from heterozygous patient samples. Additional information about sample preparation, PCR and RT-PCR methods, primer sequences, and sequencing results is available on the Blood website; see the Supplemental Document link at the top of the online article.
T-cell progenitor fluorescence activated cell sorting (FACS) and RT-PCR
Single-cell suspensions from surgically removed mouse thymi were stained with conjugated antibodies against CD4, CD8, T-cell receptor β (TCRβ), CD44, CD25, and Thy1.2. Immunofluorescence analysis was performed on the Moflo flow cytometer (Cytomation, Denver, CO), and 20 000 cells were collected according to their surface markers. RNA extraction, cDNA synthesis, and RT-PCR amplification were performed as previously described.6 Primer sequences used for RT-PCR of T-ALL oncogenes and the S16 control gene are available (Supplemental Document).
Results and discussion
Allelic expression analysis of T-ALL oncogenes
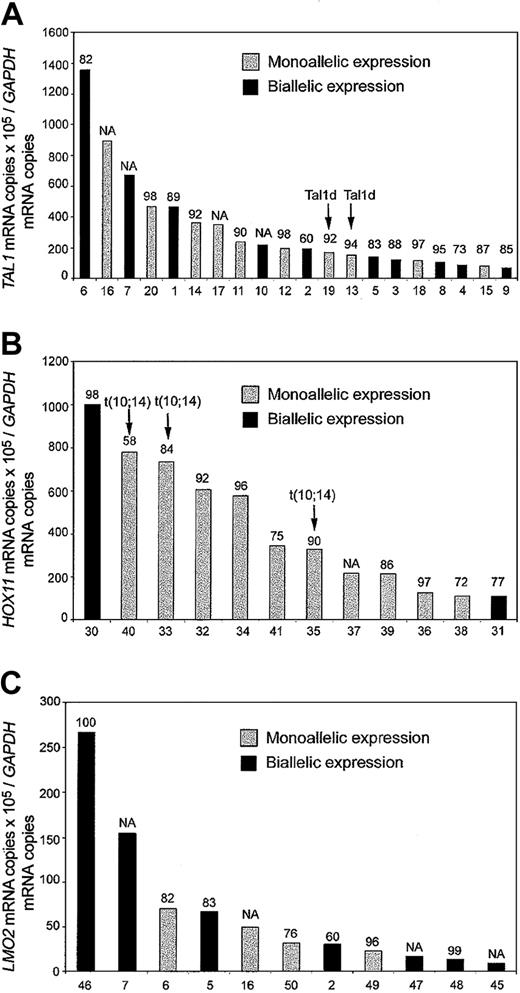
Allele-specific analysis of TAL1 mRNA was performed in 29 T-ALL cases showing aberrant expression of this oncogene (Figure 1). Analysis of genomic sequences identified 11 single nucleotide polymorphisms (SNPs) in the 3′ untranslated region (3′UTR) of TAL1. One or more of these SNPs were informative (heterozygous) in 24 of the cases (83%). Allele-specific expression analysis was performed on informative cases by direct sequencing of RT-PCR products of the TAL1 3′UTR region. Biallelic expression of TAL1 was observed in 10 (42%) of the 24 informative cases, whereas 10 cases (42%) showed monoallelic expression of this oncogene (Figure 1A). These results are in agreement with a previous report of Bash et al4 that demonstrated biallelic TAL1 activation in a significant proportion of childhood T-ALL cases.
Quantitative RT-PCR and allelic expression analysis of T-ALL transcription factor oncogenes. Black bars indicate biallelic expression, and gray bars indicate monoallelic expression. The levels of expression used as cutoff for positivity are indicated.2 Numbers at the top of each bar indicate the percentage of blast cells in the samples analyzed. Numbers at the bottom of each panel correspond to the sample identification numbers in the table in the Supplemental Document. (A) TAL1 expression in TAL1+ T-ALL allelic informative cases. Samples with a TAL1-d variant, resulting from deletion of a 90-kb genomic DNA fragment adjacent to the TAL1 locus in chromosome band 1p32, are indicated. (B) HOX11 expression in HOX11 T-ALL allelic informative cases. Samples harboring the chromosomal translocation t(10; 14)(q24;q11) that induces activation of the HOX11 locus are indicated. (C) LMO2 expression in LMO2 T-ALL allelic informative cases. NA indicates not available.
Quantitative RT-PCR and allelic expression analysis of T-ALL transcription factor oncogenes. Black bars indicate biallelic expression, and gray bars indicate monoallelic expression. The levels of expression used as cutoff for positivity are indicated.2 Numbers at the top of each bar indicate the percentage of blast cells in the samples analyzed. Numbers at the bottom of each panel correspond to the sample identification numbers in the table in the Supplemental Document. (A) TAL1 expression in TAL1+ T-ALL allelic informative cases. Samples with a TAL1-d variant, resulting from deletion of a 90-kb genomic DNA fragment adjacent to the TAL1 locus in chromosome band 1p32, are indicated. (B) HOX11 expression in HOX11 T-ALL allelic informative cases. Samples harboring the chromosomal translocation t(10; 14)(q24;q11) that induces activation of the HOX11 locus are indicated. (C) LMO2 expression in LMO2 T-ALL allelic informative cases. NA indicates not available.
Allelic expression could not be evaluated in 4 of our cases because of the presence of inconclusive sequencing results showing 2 alleles but with predominance of one of them. These cases most likely represent monoallelic TAL1-expressing leukemias with limited amounts of biallelic TAL1 expression from contaminating residual healthy bone marrow cells and show that, despite the high blast cell percentages of the T-ALL samples analyzed, some TAL1 expression may be attributed to the presence of residual healthy cells in our samples. Importantly, 2 of the 24 informative TAL1+ cases showed the TAL1d deletion, a cis-acting intrachromosomal rearrangement responsible for the activation of TAL1 in 10% to 25% of T-ALL cases,1,7,8 and lymphoblasts from each of these cases showed monoallelic expression of the TAL1 oncogene. To investigate whether the presence of biallelic activation was restricted to the TAL1 oncogene or was, in fact, a more general mechanism of oncogene activation in T-ALL, we performed allelic expression analysis of 2 additional T-ALL oncogenes, HOX11 and LMO2. PCR analysis of gDNA sequences from 15 T-ALL cases with aberrant expression of HOX11 identified 12 polymorphic SNP markers in the 3′UTR region of HOX11. Twelve (80%) of the 15 HOX11+ T-ALL cases were heterozygous for one or more SNPs. Expression of HOX11 was monoallelic in each of the 3 informative cases harboring cytogenetic abnormalities of the HOX11 locus on chromosome band 10q24. Two of the 9 informative cases lacking translocations showed biallelic expression of the HOX11 oncogene (Figure 1B). Biallelic expression of HOX11 in these cases can only be derived from T-ALL lymphoblasts and not from residual healthy cells, as HOX11 is not expressed in healthy bone marrow.9 These results agree with previous observations that show hypomethylation of HOX11 promoter sequences in T-ALL5 and demonstrate that transacting mechanisms also lead to the aberrant expression of HOX11 in this type of leukemia.
Analysis of LMO2 3′ UTR genomic sequences identified 5 polymorphic markers (4 SNPs and a 2–base pair deletion) that were informative in 11 (52%) of the 21 LMO2-overexpressing T-ALL cases. None of these cases harbored detectable cytogenetic abnormalities involving the LMO2 locus in chromosome 11p13. Allelic expression analysis demonstrated biallelic expression of LMO2 in 7 (64%) of these 11 informative LMO2+ cases (Figure 1C). All cases analyzed had a high blast cell content (Figure 1 and Supplemental Document); however, given that LMO2 is expressed by some healthy hemopoietic cells, we cannot exclude the possibility that some of the LMO2 signal could come from residual healthy bone marrow cells. Five of the cases analyzed in these series were informative for both TAL1 and LMO2 polymorphisms: (1) in 3 of them both genes were expressed biallelically; (2) one case showed monoallelic expression of both TAL1 and LMO2; and (3) one case showed biallelic expression of TAL1 with monoallelic expression of LMO2 (Figure 1A,C).
Expression of TAL1 and LMO2 in restricted populations of T-cell progenitors
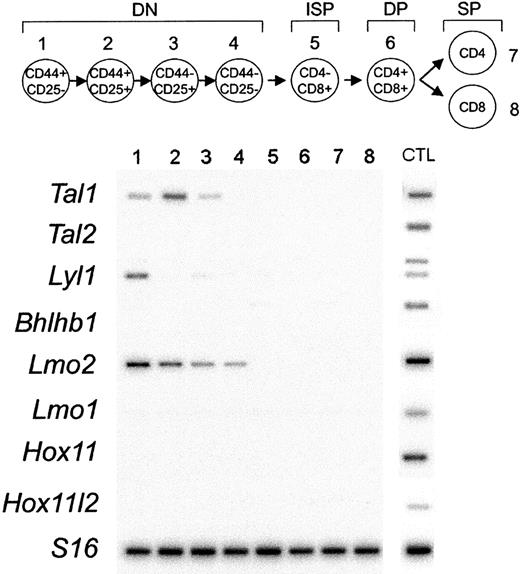
To asses the possible relationship between biallelic oncogene expression in T-ALL samples and normal transcriptional programs active in T-cell development, we analyzed the expression of T-ALL oncogenes in sorted populations of murine thymocytes. RT-PCR analysis demonstrated the expression of Tal1 in early DN1 to DN3 cells, Lmo2 in DN1 to DN4 cells, and Lyl1 in DN1 T-cell progenitors (Figure 2). By contrast, we failed to detect expression of Hox11, Hox11l2, Lmo1, Tal2, or Bhlhb1 in thymocytes of any stage. These results support a role in the pathogenesis of T-ALL for the loss of mechanisms limiting the expression of the Tal1 and Lmo2 genes to the earliest stages of T-cell development.
Expression analysis of T-ALL transcription factor oncogenes during T-cell development. Thymocyte subsets were purified by flow cytometry according to their surface markers, as indicated by the schematic diagram. The earliest T-cell precursors are characterized by the lack of expression of CD4 and CD8 surface markers (double-negative thymocytes) and can be subdivided into 4 different stages of development (DN1 to DN4). The rearrangement of the TCRβ chain at the end of the double-negative (DN) stage of development drives the production of intermediate-single-positive (ISP) cells that differentiate into CD4, CD8 double-positive (DP) cells. DP cells undergo a selection process that results in the production of functionally competent mature CD4 or CD8 single-positive (SP) cells.10 Semiquantitative RT-PCR was performed by using specific primers for Tal1, Tal2, Lyl1, Bhlhb1, Lmo2, Lmo1, Hox11, Hox11l2, and S16 as control for the amounts of cDNA. PCR products were transferred to membranes and hybridized with internal oligonucleotide probes. cDNA prepared from E11.5 or E13.5 embryonic heads or total bone marrow (Lmo2) were used as positive controls for PCR amplification (CTL).
Expression analysis of T-ALL transcription factor oncogenes during T-cell development. Thymocyte subsets were purified by flow cytometry according to their surface markers, as indicated by the schematic diagram. The earliest T-cell precursors are characterized by the lack of expression of CD4 and CD8 surface markers (double-negative thymocytes) and can be subdivided into 4 different stages of development (DN1 to DN4). The rearrangement of the TCRβ chain at the end of the double-negative (DN) stage of development drives the production of intermediate-single-positive (ISP) cells that differentiate into CD4, CD8 double-positive (DP) cells. DP cells undergo a selection process that results in the production of functionally competent mature CD4 or CD8 single-positive (SP) cells.10 Semiquantitative RT-PCR was performed by using specific primers for Tal1, Tal2, Lyl1, Bhlhb1, Lmo2, Lmo1, Hox11, Hox11l2, and S16 as control for the amounts of cDNA. PCR products were transferred to membranes and hybridized with internal oligonucleotide probes. cDNA prepared from E11.5 or E13.5 embryonic heads or total bone marrow (Lmo2) were used as positive controls for PCR amplification (CTL).
Thus, our study suggests that in addition to gene rearrangements responsible for the activation of T-ALL transcription factor oncogenes on one chromosomal allele,1 a totally different class of “trans-acting” mechanisms affect a substantial fraction of T-ALL cases. These as yet undefined mechanisms lead to high levels of expression of T-ALL oncogenes, with equal contributions from both chromosomal alleles, probably because of the disruption of gene silencing mechanisms that operate during normal T-cell development.
Prepublished online as Blood First Edition Paper, November 6, 2003; DOI 10.1182/blood-2003-07-2577.
Supported in part by grants from the National Institutes of Health (NIH) (CA68484, CA21115, CA21765, CA101140, and CA92551) and the National Cancer Institute of Canada (T.H.); Leukemia and Lymphoma Society Fellowship Award (A.A.F.); Coleman Leukemia Research Fund; Leukemia Research Fund of Canada (S.H.); and Lady Tata Memorial Trust (T.P.).
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front ot this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Michael Jaynes (St Jude Children's Research Hospital Cell Bank), Michael Caligiuri (CALGB Leukemia Tissue Bank, supported by a grant from the National Cancer Institute [CA31946] to the Cancer and Leukemia Group B, Richard L. Schilsky, MD, Chair), and Elisabeth Paietta (ECOG Cell Bank) for providing cryopreserved T-ALL samples; the CALGB Cytogenetics Committee for centrally reviewed karyotype data on CALGB cases; and John-Paul Hezel for editorial assistance.