Abstract
The bone marrow (BM) microenvironment plays a critical role in malignant cell growth, patient survival, and response to chemotherapy in hematologic malignancies. However, mechanisms associated with this environmental influence remain unclear. In this study, we investigated the role of Notch family proteins in myeloma and other malignant lymphoid cell line growth and response to chemotherapeutic drugs. All 8 tested cell lines expressed Notch-3 and Notch-4; 7 cell lines expressed Notch-1; and 6 expressed Notch-2 proteins. Interaction with BM stroma (BMS) activated Notch signaling in tumor cells. However, activation of only Notch-1, but not Notch-2, resulted in protection of tumor cells from melphalan- and mitoxantrone-induced apoptosis. This protection was associated with up-regulation of p21WAF/Cip and growth inhibition of cells. Overexpression of Notch-1 in Notch-1- U266 myeloma cells up-regulated p21 and resulted in protection from drug-induced apoptosis. Thus, this is a first report demonstrating that Notch-1 signaling may be a primary mechanism mediating the BMS influence on hematologic malignant cell growth and survival. (Blood. 2004; 103:3503-3510)
Introduction
Resistance to chemotherapy is one of the most important problems in the treatment of the patients with multiple myeloma (MM) and other hematologic malignancies. Increasing evidence suggests that the bone marrow (BM) microenvironment inhibits drug-induced apoptosis and conveys a form of de novo drug resistance for myeloma and leukemia cells.1-6 However, the molecular mechanisms of BM stroma (BMS)-mediated survival of tumor cells are poorly understood. To address this question, we have focused on the Notch family of receptors and transcriptional regulators critically important for interaction between BMS and hematopoietic cells. Notch proteins function in hematopoietic cells as both cell surface receptors and direct regulators of gene transcription.7,8 Notch ligands in vertebrates include Jagged-1, Jagged-2, Delta-1, Delta-2, Delta-3, and Delta-4.9 On the molecular level, interaction between Notch and its ligand results in proteolytic cleavage and release of Notch-1 intracellular domain (IC), its translocation to the nucleus, and association with DNA-binding transcriptional repressor recombination signal binding protein-Jκ (RBP-Jκ), converting it from a repressor into an activator.10 The NotchIC/RBP-Jκ complex transactivates different transcriptional factors and affects downstream target genes that mediate cell fate decisions. No data are available regarding the role of Notch proteins in MM. We suggest that Notch signaling may protect MM from drug-induced apoptosis, basing this on several previously established facts:
Members of the Notch family of receptors expressed on B-cells.11,12
Notch ligands Jagged-1 and Delta-1 are abundantly expressed on bone marrow stromal cells and bone marrow macrophages.11,13,14
Accumulating evidence demonstrates that wild-type Notch is overexpressed by various tumors15-20 and transformed cell lines of many different lineages (reviewed in Miele and Osborne8 ).
Notch signaling has been implicated in initial tumor transformation.21-23
In hematopoietic progenitor cells, Notch is required for nuclear factor-κB (NF-κB) synthesis and antiapoptotic features.24-26
In the present paper, we tested the hypothesis that Notch-1 activation in MM cells results in cell cycle arrest and protection of these cells from drug-induced apoptosis.
Materials and methods
Cell cultures, vectors, and drugs
NCI H929, U266, RPMI-8226, HS-Sultan, ARH-77, IM-9, and MC/CAR cell lines isolated from patients with myeloma were obtained from the American Type Culture Collection (ATCC) (Manassas, VA); the MM1S cell line was a gift from Dr Steven Rosen (Northwestern University, Chicago, IL). HS-Sultan cells, although initially isolated from a patient with multiple myeloma, were later found to be a Burkitt lymphoma cell line,27 and ARH-77, IM9, and MC/CAR have characteristics of lymphoblastoid cell lines.28 MC/CAR cells were grown in Iscove modified Dulbecco medium (IMDM) supplemented with 20% fetal bovine serum (FBS) and antibiotics (all from Invitrogen, Carlsbad, CA). All other myeloma cell lines were grown in RPMI-1640 medium (Invitrogen) with the addition of 10% FBS, antibiotics, and 0.05 mM 2-mercaptoethanol for H929 cells. BMS cultures were established from BM aspirates of healthy donors as described previously1 and were grown in α-minimal essential medium (α-MEM) (Invitrogen) supplemented with 10% FBS and antibiotics. NIH-3T3 fibroblasts transfected with full-length human Jagged-1 cDNA were kindly provided by Dr Irwin D. Bernstein (Fred Hutchinson Cancer Research Center, Seattle, WA) and were cultured in Dulbecco MEM (DMEM) (Invitrogen) supplemented with 10% FBS, antibiotics, and 1 mg/mL G418. Generation of this cell line was described elsewhere.29 A control cell line 3T3-MSCV was generated by infecting parental NIH-3T3 cells with empty retroviral vector MSCV followed by selection with 1 mg/mL G418. Vector encoding the intracellular (IC) part of the Notch-1 gene MSCV-IC-Notch-1-GFP or control vector MSCV-internal ribosome entry site-green fluorescent protein (MSCV-IRES-GFP) were kindly provided by J. C. Aster (Brigham and Women's Hospital, Boston, MA).26 Plasmid containing the IC part of Notch-1 pcDNA3.1-IC-Notch-1 was a gift from Dr Barbara Osborne (University of Massachusetts, Amherst, MA). Empty pcDNA3.1 vector was obtained from Invitrogen. Mitoxantrone and melphalan were purchased from Sigma (St Louis, MO).
Gene transfer
Vectors were introduced into myeloma U266 cells by electroporation by means of the Bio-Rad (Hercules, CA) Gene Pulser II apparatus. Briefly, cells (107) were pulsed with 240 V at a capacitance of 1070 microfarads followed by incubation for 24 hours. After that time, dead cells were eliminated by means of gradient centrifugation. Cells were then treated with melphalan for 24 hours followed by apoptosis evaluation or incubated for 24 hours and then lyzed and analyzed by Western blotting.
Packaging cell line ampho-293T was transfected with pMSCV-IC-Notch-1-GFP or control vector pMSCV-IRES-GFP with the use of GenePORTER reagent (Gene Therapy Systems, San Diego, CA). After incubation for 48 hours, supernatant containing retrovirus was collected and used for infection of U266 cells. Cells were exposed to cytotoxic drugs 24 hours after the second round of infection.
Interaction between myeloma cells and stroma and detection of apoptosis
Myeloma cells were cultured on a pre-established monolayer of irradiated (25 Gy) fibroblasts or BMS. After different periods of time, myeloma cells were carefully removed with the monolayer of fibroblasts kept intact. The purity of myeloma cell population was greater than 95%. In some experiments, soluble peptide representing a part of Notch-1 ligand Jagged-1 was used to activate Notch protein in myeloma cells. This peptide has been shown to mimic the function of the ligand.13 H929 or U266 myeloma cells were incubated with 50 μM Jagged-1 or scrambled peptide for 24 hours prior to treatment with cytotoxic drugs. Apoptosis of myeloma cells was analyzed by 2 different methods: annexin V binding or transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) assays. For annexin V-binding assay, myeloma cells were washed twice with ice-cold phosphate-buffered saline (PBS), washed once with binding buffer (BD Pharmingen, San Diego, CA), and then stained with annexin V and 7-aminoactinomycin D (7-AAD) (BD Pharmingen). We analyzed 10 000 events by flow cytometry using CellQuest software (Becton Dickinson, San Jose, CA).
For TUNEL assay, myeloma cells were fixed in 1% paraformaldehyde in PBS for 15 minutes on ice, washed with ice-cold PBS, and resuspended in 70% ethanol. TUNEL staining was performed by means of the Apo-BrdU kit (BD Pharmingen) according to the manufacturer's protocol. We acquired 10 000 events using CellQuest software.
Cell proliferation and cell cycle distribution
H929, RPMI-8226, HS-Sultan, and U266 cells were either kept in suspension for 48 hours or incubated on irradiated (25 Gy) 3T3-Jagged and 3T3-MSCV fibroblasts or BMS. For cell cycle analysis, myeloma cells were collected, fixed, and stained with propidium iodide. We acquired 10 000 events by flow cytometry. Cell cycle distribution was analyzed on flow cytometer FACSCalibur by means of CellQuest software (Becton Dickinson). Cell proliferation was measured by adding 1 μCi (0.37 MBq) [3H]-thymidine per well of a 96-well plate 16 hours prior to harvesting the cells. The [3H]-thymidine incorporation was measured by liquid scintillation counting.
Luciferase assay
To evaluate Notch activation in myeloma cells following coculturing with human BMS, we used Notch reporter construct containing c prompter binding factor-1 (CBF-1) responsive element. The retroviral reporter plasmid p6 × CBF-1-interleukin-2-luciferase (p6 × CBF-1-IL-2-Luc) was constructed from pKA9 plasmid (IL-2-Luc) containing the luciferase gene under the control of minimal IL-2 promoter (a gift from T. J. Murphy, Emory University, Atlanta, GA). Briefly, 2 oligonucleotides, L-1 (5′-ATGTCGACCCGACTCGTGGGAAAATGGGCGGAAGGGCACCGTGGGAAAATAGTAGCGGAGAAGCCAGCCGTGGGAAAAAGAATTCAG-3′), containing SalI at the 5′end and EcoRI at the 3′ end; and L-2 (5′-ACGAATTCCCGACTCGTGGGAA-AATGGGCGGAAGGGCACCGTGGGAAAATAGTAGCGGAGAAGCCAGCCGTGGGAAAAAGGATCCGA-3′), containing EcoRI at the 5′ end and BamHI at the 3′ end; were annealed with the respective reverse complementary strands, digested with EcoRI, and then ligated together. The resulting fragment, approximately 154 base pairs (bp) containing 6 CBF-1-binding sites, was digested with SalI and BamHI and subsequently inserted into pKA9 in front of minimal IL-2 promoter. This construct was confirmed by sequence analysis.
H929 cells were transfected with Notch reporter construct containing CBF-1 responsive element (p6 × CBF-1-IL-2-Luc) or control (IL-2-Luc) vector by electroporation. Stable transfected cell lines were established by G418 selection (0.8 mg/mL). The presence of luciferase reporter construct was confirmed by evaluation of luciferase activity in H929 cells incubated on 3T3-Jagged or 3T3-MSCV stroma. After incubation, myeloma cells were collected and lyzed in luciferase reporter buffer. Luciferase activity (LA) was measured by means of the Luciferase assay system kit (Promega, Madison, WI) and then normalized to total protein concentration. Protein concentration was evaluated in all lysates by means of the Bradford protein assay (Bio-Rad).
Electromobility shift assay (EMSA)
H929, HS-Sultan, U266, RPMI-8226, or MM1S cells were starved for one hour in serum-free RPMI-1640 medium followed by incubation on the monolayer of irradiated 3T3-Jagged or 3T3-MSCV cells for 8 hours. Cells were then collected and washed with ice-cold PBS, and nuclear extracts were prepared as described earlier.30 Double-stranded oligonucleotides containing the specific binding site for CBF-1 were made by annealing the appropriate single-stranded oligonucleotides at 65°C for 10 minutes. The probes were labeled with γ-adenosine triphosphate-[32P]deoxyadenosine triphosphate (γATP-[32P]dATP) (6000 μCi/mM [222 MBq/mM]) (Amersham Life Sciences, Arlington Heights, IL) with the use of Klenow DNA polymerase. Two probes were used: wild type (5′-TGGTGTAAACACGCCGTGGGAAAAAATTTA-3′) and mutant (5′-TGGTGTAAACACGCCGTTGGAAAAAATTTA-3′). We incubated 10 μg nuclear extract with α32P-labeled probe in binding buffer containing 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 5% glycerol, 0.2 mM EDTA (ethylenediaminetetraacetic acid), 1 mM dithiothreitol (DTT), 5 mM MgCl2, and 4 μg poly(dI-dC) to prevent nonspecific DNA binding. Samples were resolved in 4% polyacrylamide gel. Cold inhibition was performed with a 50-fold excess of unlabeled probe.
To evaluate the involvement of the NF-κB pathway following Notch activation, H929 cells were incubated on the monolayer of 3T3-Jagged or 3T3-MSCV fibroblasts for the period of time indicated, then collected and washed, and EMSA was performed as described previously.31 Specific bands were visualized by overnight exposure to x-ray films (Fuji, Stamford, CT) at -70°C. Quantitation of band intensity was performed on Storm phosphor imager by means of ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Quantitative real-time PCR of the HES-1 gene
U266 cells were electroporated with pcDNA3.1-Notch-1IC or empty pcDNA3.1 plasmid, and dead cells were eliminated after 24 hours by gradient centrifugation. Cells were collected immediately or incubated for an additional 24 hours. H929, RPMI-8226, and U266 cells were incubated on irradiated 3T3-Jagged or 3T3-MSCV fibroblasts for 48 hours. After that time, myeloma cells were collected, and RNA was extracted by means of the RNeasy Mini kit (Qiagen, Valencia, CA). First-strand cDNA was synthesized with 1 μg RNA with the use of SuperScript II reverse transcriptase (Invitrogen) and random primers according to the manufacturer's protocol. Polymerase chain reaction (PCR) was performed with 2.5 μL cDNA template with the use of the TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) and target gene assay mix containing human HES-1 sequence-specific primers and 6-carboxyfluorescein (6-FAM) dye-labeled TaqMan minor groove binder (MGB) probe (Applied Biosystems). As a control, amplification with 18S endogenous control assay mix was used. The PCR conditions were as follows: 1 cycle at 50°C for 2 minutes and 95°C for 10 minutes, and 40 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 1 minute. PCR was carried out in triplicate for each sample. Data quantitation was performed by means of the relative standard curve method according to the manufacturer's instructions (Applied Biosystems). Cycle numbers obtained at the loglinear phase of the reaction were plotted against a standard curve prepared with serially diluted control samples. Expression level of the HES-1 gene was normalized by 18S mRNA level measured concurrently.
Western blotting
Cells were collected at time points indicated, washed twice with ice-cold PBS, and lysed in radioimmunoprecipitation assay (RIPA) buffer. Equal amounts of protein was loaded on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. After protein transfer, membrane was blocked in 5% milk in Tris (tris(hydroxymethyl)aminomethane)-buffered saline/Tween 20 (TBS/T) for 1 hour at room temperature and probed for overnight at +4°C with the following primary antibodies: anti-Notch-1, anti-Notch-2, and anti-Jagged-1 (DSHB, University of Iowa, Iowa City, IA); anti-Notch-3, anti-Notch-4, anti-Delta-1, anti-Jagged-2, and anti-β-actin (Santa Cruz Biotechnology, Santa Cruz, CA); anti-p27 (Transduction Laboratories); anti-p21 (Lab Vision, Fremont, CA); anti-bcl-2, anti-bcl-xL, and anti-mantle cell lymphoma 1 (anti-mcl-1) (BD Pharmingen); and antisurvivin (Alpha Diagnostic, San Antonio, TX). Membrane was washed, incubated with corresponding secondary antibodies for one hour at room temperature, and then washed again with TBS/T. Specific bands were developed by means of a chemiluminescence reagent kit (Amersham Life Sciences).
Results
Notch-1 is activated in hematologic malignant cells following contact with BMS
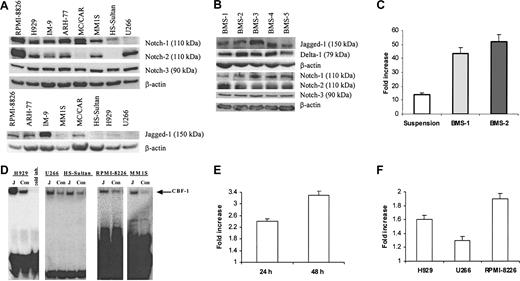
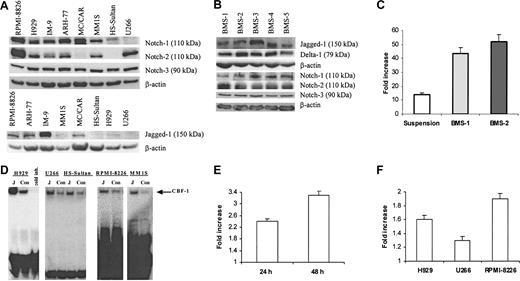
First, we evaluated the presence of the Notch family members in 8 different cell lines isolated from patients with MM. Notch-1 protein was detectable in all myeloma cell lines tested except U266. HS-Sultan and MC/CAR cells did not express Notch-2. All 8 tested cell lines expressed Notch-3 protein (Figure 1A). Multiple bands were detected on the membranes after probing with anti-Notch-4 antibodies (Santa Cruz Biotechnology). A band of approximately 70 kDa corresponding to the Notch-4 protein was detectable in all cell lines and disappeared after the use of blocking peptide (data not shown). Therefore, we considered this band specific for Notch-4 and concluded that all 8 cell lines expressed this protein. Bands of approximately 150 kDa corresponding to Jagged-1 were seen to different extents in all studied cell lines except U266. At the same time, we could not reliably detect Delta-1 protein in these cells. Notch ligands Jagged-1, and Delta-1 as well as Notch-1, Notch-2, and Notch-3 protein were detected in all tested BMS cells obtained from 5 different donors (Figure 1B). However, we could not detect an approximately 150-kDa band corresponding to Jagged-2 protein in these cells.
Activation of Notch pathway in myeloma cells following ligation to Jagged-1. (A) Expression of Notch family proteins and their ligand Jagged-1 was analyzed in 8 different human myeloma cell lines by Western blotting. (B) Expression of Notch family proteins and their ligands was evaluated by Western blotting in BMS cells obtained from 5 different BM donors. (C) Interaction between BMS and H929 cells resulted in activation of the Notch pathway in myeloma cells. H929 cells transfected with reporter or control constructs were incubated on BMS for 48 hours or kept in suspension (control). Luciferase activity (LA) was measured according to the manufacturer's protocol and normalized to 1 μg total protein. Data are presented as fold increase of the specific LA (H929 cells transfected with p6 × CBF-1-pKA9 plasmid) over control LA activity (cells transfected with pKA9 plasmid). Experiments were performed in triplicate and repeated twice with similar results. (D) Jagged-1 ligation activates Notch pathway in myeloma cells. H929, U266, RPMI-8226, or MM1S human myeloma cells and Burkitt lymphoma HS-Sultan cells were starved for one hour in serum-free RPMI-1640 medium and then incubated on the monolayer of irradiated 3T3-Jagged (J) or control 3T3-MSCV (Con) cells for 8 hours. Cells were then collected and nuclear extract was prepared. EMSA with CBF-1-specific probe was performed. “Cold inh” indicates cold inhibition control. (E-F) Notch activation in myeloma cells results in an increase of HES-1 mRNA espression. The pcDNA3.1-Notch-1IC or pcDNA3.1 plasmids were introduced in U266 cells by electroporation and cultured for 24 hours and 48 hours (E). H929, RPMI-8226, or U266 myeloma cells were cultured on 3T3-Jagged or 3T3-MSCV cells for 48 hours (F). Real-time PCR was performed as described in “Materials and methods.” The mRNA expression for each sample was normalized to 18S mRNA expression. Data shown are fold increase of HES-1 expression in myeloma cells cultured on 3T3-Jagged over cells cultured on 3T3-MSCV (F) or U266 cells transfected with Notch-1IC-carrying plasmid over control plasmid (E). Differences were statistically significant for all cell lines studied. Error bars (C, E, F) indicate standard deviation.
Activation of Notch pathway in myeloma cells following ligation to Jagged-1. (A) Expression of Notch family proteins and their ligand Jagged-1 was analyzed in 8 different human myeloma cell lines by Western blotting. (B) Expression of Notch family proteins and their ligands was evaluated by Western blotting in BMS cells obtained from 5 different BM donors. (C) Interaction between BMS and H929 cells resulted in activation of the Notch pathway in myeloma cells. H929 cells transfected with reporter or control constructs were incubated on BMS for 48 hours or kept in suspension (control). Luciferase activity (LA) was measured according to the manufacturer's protocol and normalized to 1 μg total protein. Data are presented as fold increase of the specific LA (H929 cells transfected with p6 × CBF-1-pKA9 plasmid) over control LA activity (cells transfected with pKA9 plasmid). Experiments were performed in triplicate and repeated twice with similar results. (D) Jagged-1 ligation activates Notch pathway in myeloma cells. H929, U266, RPMI-8226, or MM1S human myeloma cells and Burkitt lymphoma HS-Sultan cells were starved for one hour in serum-free RPMI-1640 medium and then incubated on the monolayer of irradiated 3T3-Jagged (J) or control 3T3-MSCV (Con) cells for 8 hours. Cells were then collected and nuclear extract was prepared. EMSA with CBF-1-specific probe was performed. “Cold inh” indicates cold inhibition control. (E-F) Notch activation in myeloma cells results in an increase of HES-1 mRNA espression. The pcDNA3.1-Notch-1IC or pcDNA3.1 plasmids were introduced in U266 cells by electroporation and cultured for 24 hours and 48 hours (E). H929, RPMI-8226, or U266 myeloma cells were cultured on 3T3-Jagged or 3T3-MSCV cells for 48 hours (F). Real-time PCR was performed as described in “Materials and methods.” The mRNA expression for each sample was normalized to 18S mRNA expression. Data shown are fold increase of HES-1 expression in myeloma cells cultured on 3T3-Jagged over cells cultured on 3T3-MSCV (F) or U266 cells transfected with Notch-1IC-carrying plasmid over control plasmid (E). Differences were statistically significant for all cell lines studied. Error bars (C, E, F) indicate standard deviation.
We used the luciferase reporter-gene assay to investigate whether the interaction between BMS and myeloma cells activates the Notch pathway in myeloma cells. Myeloma cell line H929 was transfected with either CBF-1 reporter or control luciferase constructs and incubated for 2 days either in suspension or on the monolayer of BMS. Incubation of myeloma cells with BMS resulted in levels of Notch activation significantly higher (almost 3-fold) than in the cells kept in suspension (Figure 1C).
BMS express a number of different receptors and produce soluble factors that may affect survival of myeloma cells. Therefore, to elucidate the specific role of Notch signaling in myeloma cell resistance to apoptosis, we used fibroblasts expressing Notch ligand Jagged-1. HS-Sultan cells containing Notch-1 but not Notch-2; U266 myeloma cells containing Notch-2 but not Notch-1; and H929, RPMI-8226, and MM1S myeloma cells expressing both Notch-1 and Notch-2 proteins were incubated on the monolayer of 3T3-Jagged or 3T3-MSCV cells. These cells were then collected and analyzed by EMSA with a CBF-1-specific probe. As shown in Figure 1D, cell-bound Jagged-1 induced Notch activation in all tested myeloma cells.
Next, we asked whether the activation of Notch signaling in MM cells resulted in increased expression of Notch target gene HES-1. U266 cells were transfected with plasmid containing constitutively active Notch-1IC or empty plasmid and were incubated for 24 or 48 hours. Quantitative real-time PCR was performed to evaluate the expression of the HES-1 gene. Introduction of the active form of Notch-1 in U266 myeloma cells increased the HES-1 mRNA level (P = .003 and P = .02 in 24 hours and 48 hours after transfection) (Figure 1E). In parallel, myeloma cells were incubated on the monolayer of 3T3-Jagged or 3T3-MSCV fibroblasts. As shown in Figure 1F, Jagged-1 induced HES-1 expression in H929, U266, and 8226 cells (P = .04, P = .04, and P = .01, respectively).
Activation of Notch-1 pathway protects myeloma cells from drug-induced apoptosis
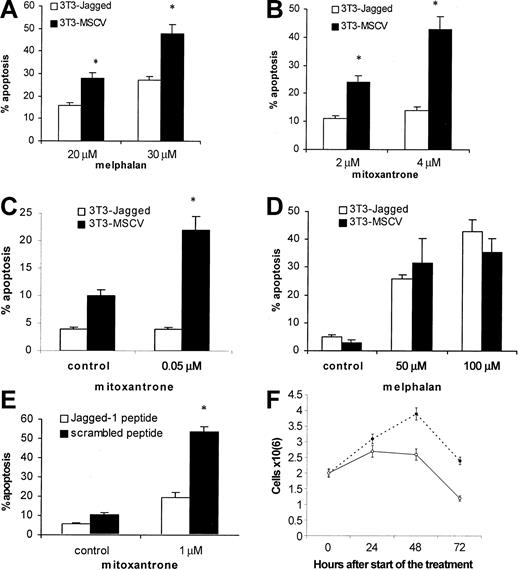
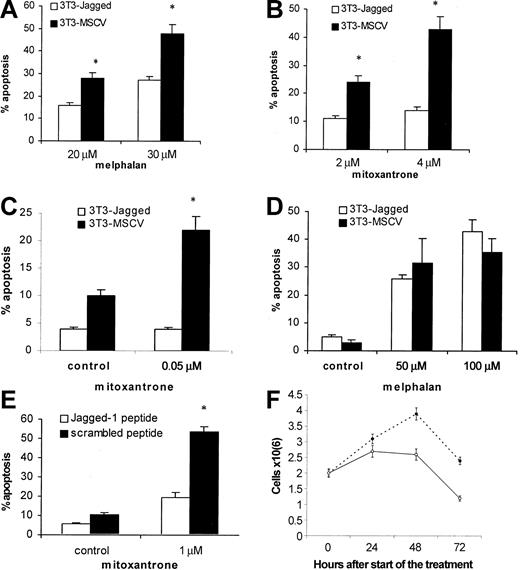
Next, we investigated whether the activation of the Notch pathway in myeloma cells could protect these cells from cytotoxic drugs. H929, U266, or HS-Sultan cells were cocultured with 3T3-Jagged or 3T3-MSCV fibroblasts followed by drug exposure. As shown in Figure 2A-B, H929 cells with activated Notch-1 were resistant to melphalan and mitoxantrone treatment. Similar results were obtained for Notch-1-expressing HS-Sultan cells (Figure 2C). However, such protection from melphalan-induced apoptosis was not observed in U266 cells, which do not express Notch-1 protein (Figure 2D).
Effect of Notch-1 activation on resistance to drug-induced apoptosis. Notch-1 activation in myeloma cells increased resistance to drug-induced apoptosis. Myeloma H929 (A-B), HS-Sultan (C), or U266 (D) cells were incubated on the monolayer of 3T3-Jagged or control fibroblasts for 24 hours. H929 cells (E) were treated with 50 μM Jagged-1 or scrambled peptide for 24 hours followed by drug or vehicle control exposure for another 24 hours. Myeloma cells were then collected, and apoptosis was measured by flow cytometry with the use of the TUNEL assay (A-B) or annexin V-binding assay (C-E). Similar results were obtained for H929 cells with the use of the annexin V-binding assay. All experiments were repeated 4 times with similar results. *Statistically significant differences from cells cultured on 3T3-Jagged stroma or with Jagged-1 peptide, P < .05. In panel F, H929 cells were treated with 50 μM Jagged-1 (○) or scrambled peptide (•) for 24, 48, or 72 hours, and the cell number was calculated in triplicates. Error bars indicate standard deviation.
Effect of Notch-1 activation on resistance to drug-induced apoptosis. Notch-1 activation in myeloma cells increased resistance to drug-induced apoptosis. Myeloma H929 (A-B), HS-Sultan (C), or U266 (D) cells were incubated on the monolayer of 3T3-Jagged or control fibroblasts for 24 hours. H929 cells (E) were treated with 50 μM Jagged-1 or scrambled peptide for 24 hours followed by drug or vehicle control exposure for another 24 hours. Myeloma cells were then collected, and apoptosis was measured by flow cytometry with the use of the TUNEL assay (A-B) or annexin V-binding assay (C-E). Similar results were obtained for H929 cells with the use of the annexin V-binding assay. All experiments were repeated 4 times with similar results. *Statistically significant differences from cells cultured on 3T3-Jagged stroma or with Jagged-1 peptide, P < .05. In panel F, H929 cells were treated with 50 μM Jagged-1 (○) or scrambled peptide (•) for 24, 48, or 72 hours, and the cell number was calculated in triplicates. Error bars indicate standard deviation.
To exclude possible nonspecific effects of Jagged-expressing stroma on myeloma cells after their contact, we used Jagged-1-derived soluble peptide. This peptide directly activates Notch signaling.13 H929 myeloma cells treated with this peptide were protected from mitoxantrone-induced apoptosis (Figure 2E). It was possible that this peptide just induced proliferation of myeloma cells and thus decreased the percentage of apoptotic cells in these experiments. To test this possibility, we counted the number of myeloma cells after the treatment with soluble Notch ligand. Jagged-1-derived peptide not only did not stimulate cell division but, on the contrary, prevented accumulation of myeloma cells (Figure 2F). Similar results were obtained with RPMI 8226 cells (data not shown). These data suggest that activation of Notch-1, but not Notch-2, is important for de novo resistance of myeloma cells to cytotoxic drugs.
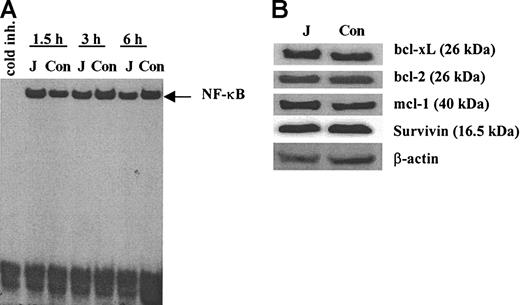
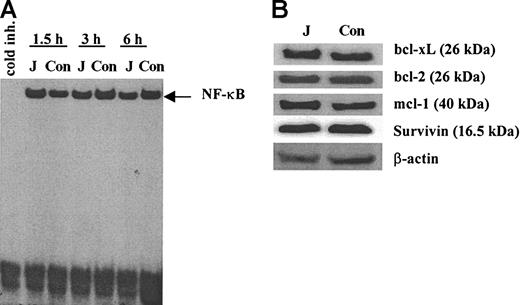
We investigated the possible mechanisms of Notch-mediated protection of myeloma cells from drug-induced apoptosis. One possible mechanism could be associated with activation of transcription factor NF-κB in these cells. NF-κB is a known antiapoptotic factor32,33 and there are data indicating that NF-κB is activated following Notch ligation.31,34,35 However, we were not able to detect any activation of NF-κB in H929 myeloma cells following ligation to cell-bound Jagged-1 (Figure 3A). We also evaluated whether the Notch activation affects the levels of antiapoptotic proteins. H929 cells were incubated on the monolayer of 3T3-Jagged or 3T3-MSCV fibroblasts for 24 hours or 48 hours. Expression of bcl-xL, bcl-2, mcl-1, and survivin proteins were not changed following ligation to Jagged-1 (Figure 3B).
Effect of Notch activation in myeloma cells on NF-κB and antiapoptotic proteins. (A) H929 cells were incubated on the monolayer of irradiated 3T3-Jagged (J) or control (Con) fibroblasts for 1.5, 3, or 6 hours. Myeloma cells were then collected, and nuclear extract was prepared followed by EMSA with NF-κB-specific probe. Similar results were obtained after H929 cells were collected at other time points: 0.5, 1, 4, 8, 12, and 24 hours. Experiments were performed twice with the same results. (B) H929 cells were incubated on the monolayer of 3T3-Jagged (J) or 3T3-MSCV (Con) stroma for 48 hours. Protein expression was evaluated by Western blotting. Experiments were repeated twice with the same results.
Effect of Notch activation in myeloma cells on NF-κB and antiapoptotic proteins. (A) H929 cells were incubated on the monolayer of irradiated 3T3-Jagged (J) or control (Con) fibroblasts for 1.5, 3, or 6 hours. Myeloma cells were then collected, and nuclear extract was prepared followed by EMSA with NF-κB-specific probe. Similar results were obtained after H929 cells were collected at other time points: 0.5, 1, 4, 8, 12, and 24 hours. Experiments were performed twice with the same results. (B) H929 cells were incubated on the monolayer of 3T3-Jagged (J) or 3T3-MSCV (Con) stroma for 48 hours. Protein expression was evaluated by Western blotting. Experiments were repeated twice with the same results.
Activation of Notch-1 causes up-regulation of p21 and growth inhibition in myeloma cells
Previous studies have suggested that cell cycle arrest can be induced in myeloma cells following adhesion to fibronectin or BMS, which could lead to a decreased drug-induced apoptosis of myeloma cells.1,3 Here, we investigated whether Notch-1 signaling affects growth of myeloma and lymphoma cells.
RPMI-8226, H929, U266, and HS-Sultan cells were incubated on the monolayer of 3T3-Jagged or 3T3-MSCV fibroblasts for 48 hours followed by proliferation assay. Ligation to Jagged-1 induced significant growth arrest in all Notch-1-expressing myeloma cells.
Growth of cells cocultured with 3T3-Jagged fibroblasts was inhibited 13-fold in H929 cells (P = .001), 4.2-fold in RPMI-8226 cells (P = 001), and 5.1-fold in HS-Sultan cells (P = .009) as compared with coculture with 3T3-MSCV cells (Figure 4A). This was consistent with the data obtained using soluble ligand (Figure 2F). However, there was no difference in proliferation of U266 cells incubated on 3T3-Jagged or control fibroblasts (P = .39). A similar effect was observed after incubation of myeloma cells on BMS. Incubation with BMS significantly inhibited proliferation of Notch-1-expressing H929 and HS-Sultan cells, but not Notch-2-expressing U266 cells (Figure 4B).
Effect of Notch-1 pathway activation on p21 and cell growth. Activation of Notch-1 pathway results in up-regulation of p21 and growth arrest in myeloma cells. H929, RPMI 8226, U266, or HS-Sultan myeloma cells were incubated on the monolayer of irradiated 3T3-Jagged or 3T3-MSCV fibroblasts (A,C-D), or BMS cells (B), or kept in suspension for 48 hours. (A-B) [3H]-thymidine was added for the last 16 hours of culture. Thymidine incorporation was measured by liquid scintillation counting. Background level of thymidine incorporation into irradiated stromal cells was subtracted in all experiments. (C-D) Myeloma cells were collected, fixed, and stained with propidium iodide (PI). Cell cycle distribution was analyzed by flow cytometry. (E) H929 cells were incubated with 3T3-Jagged (J) or 3T3-MSCV (Con) cells for 24, 36, or 48 hours. Myeloma cells were collected, and p21 and p27 protein expression was analyzed by Western blotting. To confirm equal loading, membrane was reprobed with β-actin antibody. (F-G) U266 cells were cultured on 3T3-Jagged (J) or 3T3-MSCV (Con) stroma (F); H929 cells were incubated on BMS or kept in suspension for 48 hours (G). Myeloma cells were collected, and p21 expression was analyzed by Western blotting. Error bars (A-D) indicate standard deviation.
Effect of Notch-1 pathway activation on p21 and cell growth. Activation of Notch-1 pathway results in up-regulation of p21 and growth arrest in myeloma cells. H929, RPMI 8226, U266, or HS-Sultan myeloma cells were incubated on the monolayer of irradiated 3T3-Jagged or 3T3-MSCV fibroblasts (A,C-D), or BMS cells (B), or kept in suspension for 48 hours. (A-B) [3H]-thymidine was added for the last 16 hours of culture. Thymidine incorporation was measured by liquid scintillation counting. Background level of thymidine incorporation into irradiated stromal cells was subtracted in all experiments. (C-D) Myeloma cells were collected, fixed, and stained with propidium iodide (PI). Cell cycle distribution was analyzed by flow cytometry. (E) H929 cells were incubated with 3T3-Jagged (J) or 3T3-MSCV (Con) cells for 24, 36, or 48 hours. Myeloma cells were collected, and p21 and p27 protein expression was analyzed by Western blotting. To confirm equal loading, membrane was reprobed with β-actin antibody. (F-G) U266 cells were cultured on 3T3-Jagged (J) or 3T3-MSCV (Con) stroma (F); H929 cells were incubated on BMS or kept in suspension for 48 hours (G). Myeloma cells were collected, and p21 expression was analyzed by Western blotting. Error bars (A-D) indicate standard deviation.
Next, we determined the phase of cell cycle in which myeloma cells accumulated following Notch-1 activation. Cell cycle distributions were analyzed in H929 and U266 cells after coculture with 3T3-Jagged or 3T3-MSCV cells for 48 hours. As shown in Figure 4C, ligation to Jagged-1 resulted in accumulation of H929 myeloma cells in the G0/G1 phase of cell cycle. There were no differences in cell cycle distribution in U266 cells (Figure 4D).
Transition from one cell cycle phase to another is regulated by complexes of cyclins and cyclin-dependent kinases (CDKs).36 CDK inhibitors p21WAF/Cip and p27Kip1 are the 2 key regulators of this process. Therefore, we asked whether the activation of Notch-1 could up-regulate p21 or/and p27 in myeloma cells. H929 cells were cocultured with 3T3-Jagged or 3T3-MSCV fibroblasts for 24, 36, or 48 hours. Myeloma cells were then collected, and p21 and p27 protein levels were analyzed by Western blotting. As shown in Figure 4E, Notch-1 activation resulted in significant up-regulation of p21, but did not affect p27 expression. There was no difference in p21 level in U266 myeloma cells following incubation on Jagged-expressed or control stroma (Figure 4F). We investigated whether the up-regulation of p21 could represent one of the mechanisms of interaction between myeloma and BMS cells. H929 cells were incubated on BMS obtained from 3 different donors or kept in suspension. As shown in Figure 4G, interaction with BMS significantly up-regulated p21WAF/Cip level in myeloma cells.
Overexpression of Notch-1 restored drug-resistant phenotype of Notch-1-deficient myeloma cells
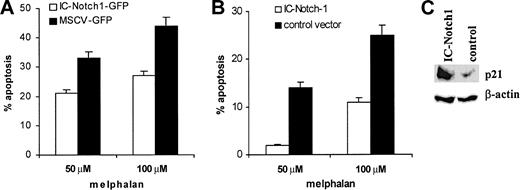
To confirm the role of Notch-1 in myeloma cell drug resistance, we overexpressed this protein in U266 myeloma cell line, which does not naturally contain Notch-1 protein. U266 cells were infected with retroviruses containing pMSCV-IC-Notch-1-GFP or control vector pMSCV-IRES-GFP. At 24 hours after infection, cells were treated with vehicle alone or melphalan for another 24 hours. Apoptosis of GFP+ cells was evaluated with the use of annexin-V. The presence of active form of Notch-1 in myeloma cells significantly increased resistance to drug-induced apoptosis (Figure 5A). These results were confirmed in experiments where constitutively active Notch-1 was expressed in U266 cells by electroporation of pcDNA3.1-IC-Notch-1 vector. Empty pcDNA3.1 vector was used as a control. At 24 hours later, dead cells were removed and myeloma cells were exposed to melphalan for another 24 hours. After that time, cells were collected and apoptosis was evaluated. As in experiments with retroviral vectors, expression of Notch-1 resulted in increased resistance of U266 cells to drug-induced apoptosis (Figure 5B).
Effect of Notch-1 overexpression on p21 accumulation and drug resistance. Overexpression of Notch-1 results in accumulation of p21 and drug resistance of myeloma cells. (A) U266 cells were infected with retroviruses containing either pMSCV-IC-Notch-1-GFP construct or control vector pMSCV-IRES-GFP. At 24 hours later, cells were exposed to melphalan or vehicle control for an additional 24 hours. Apoptosis was measured by annexin V-binding assay within the population of GFP+ cells. The level of spontaneous apoptosis in cells treated with vehicle alone was subtracted. Two experiments with the same results were performed. (B) U266 cells were electroporated with pcDNA3.1-IC-Notch-1 or control pcDNA3.1 plasmid. At 24 hours later, dead cells were eliminated by gradient centrifugation. Apoptosis was measured in the total cell population by annexin V staining. Two experiments with the same results were performed. Error bars indicate standard deviation. (C) U266 cells were transfected with IC-Notch-1 as described for panel B, dead cells were removed, and cells were cultured for an additional 24 hours. After that time, cells were collected and lyzed, and p21 expression was analyzed by Western blotting.
Effect of Notch-1 overexpression on p21 accumulation and drug resistance. Overexpression of Notch-1 results in accumulation of p21 and drug resistance of myeloma cells. (A) U266 cells were infected with retroviruses containing either pMSCV-IC-Notch-1-GFP construct or control vector pMSCV-IRES-GFP. At 24 hours later, cells were exposed to melphalan or vehicle control for an additional 24 hours. Apoptosis was measured by annexin V-binding assay within the population of GFP+ cells. The level of spontaneous apoptosis in cells treated with vehicle alone was subtracted. Two experiments with the same results were performed. (B) U266 cells were electroporated with pcDNA3.1-IC-Notch-1 or control pcDNA3.1 plasmid. At 24 hours later, dead cells were eliminated by gradient centrifugation. Apoptosis was measured in the total cell population by annexin V staining. Two experiments with the same results were performed. Error bars indicate standard deviation. (C) U266 cells were transfected with IC-Notch-1 as described for panel B, dead cells were removed, and cells were cultured for an additional 24 hours. After that time, cells were collected and lyzed, and p21 expression was analyzed by Western blotting.
We also tested the hypothesis that overexpression of IC-Notch-1 caused up-regulation of p21. The pcDNA3.1-IC-Notch-1 or empty pcDNA3.1 vector was introduced in U266 myeloma cells by electroporation. Expression of p21WAF/Cip was evaluated by Western blotting 48 hours later. As shown in Figure 5C, overexpression of the IC domain of Notch-1 resulted in significant increase in p21 protein level in U266 cells.
Discussion
This study, for the first time, demonstrates a critical role for Notch signaling in the protection of hematopoietic cancer cells from drug-induced apoptosis. MM cells are localized primarily in BM where they come into direct contact with BMS. Previous studies have demonstrated that BMS contributed greatly to the development of MM resistance to chemotherapeutic drugs.1,37 Other groups have reported a similar effect of BMS on the protection of myelogenous leukemia cells38 and B-lineage acute lymphoblastic leukemia39 from drug-induced apoptosis. However, the precise mechanism of this effect remained unclear. In this study, we tested the hypothesis that Notch signaling may represent one of the major mechanisms associated with cell adhesion-mediated drug resistance.
As expected, BMS cells generated from all 5 tested donors expressed Notch ligands Jagged-1 and Delta-1. They also expressed Notch receptors. Importantly, BMS cells activated Notch signaling in MM cells. Since BMS cells express a multitude of surface molecules and release soluble factors influencing growth and survival of MM cells, we used the experimental system in which Notch ligand Jagged-1 was expressed on murine fibroblasts. The only difference between control fibroblasts and Jagged-1 fibroblasts was the presence of cell-bound Notch ligand. This allowed for precise identification of the role of Notch signaling in myeloma cells. In addition, cell-bound ligand more closely reflects the actual process taking place during interaction between BMS and MM cells. DNA-binding assay demonstrated that Jagged-1 activated Notch signaling in all cell lines expressing Notch-1 as well as in U266 cells that are Notch-2+ but Notch-1-. This was consistent with previous observations that Jagged-1 activated different members of the Notch family.40 Activation of Notch signaling resulted in significant protection of MM cells from melphalan- and mitoxantrone-induced apoptosis. To make sure that nonspecific factors expressed on or produced by the stromal cells are not responsible for the observed effect, in some experiments we used Notch-activating peptide from Jagged-1 instead of stromal cells.13 Treatment of MM cells with this peptide reproduced the effect of Jagged-1 stroma.
How may Notch-1 protect MM cells from drug-induced apoptosis? Notch signaling did not affect the expression of major antiapoptotic proteins such as Bcl-2, Bcl-XL, and survivin, and did not activate NF-κB, a transcription factor with established antiapoptotic function. The latter observation was consistent with our previous findings in hematopoietic progenitor cells that Notch-1 regulated transcription and synthesis of NF-κB proteins rather than NF-κB activation via inhibitory protein IκBα degradation and nuclear translocation of NF-κB subunits.25 It appears that Notch signaling protects hematopoietic cancer cells from apoptosis via another mechanism. Notch signaling inhibited cell proliferation and induced accumulation of the cells in the G0/G1 phase of cell cycle. Cell growth arrest was also observed after incubation of MM cells on BMS cells, confirming our previous observations.1 Notch signaling induced accumulation of CDK inhibitor p21WAF/Cip. The specific significance of cell growth arrest and accumulation of p21WAF/Cip for protection of MM cells from drug-induced apoptosis was evident from the analysis of cells with different expressions of Notch-1 and Notch-2. U266 cells expressed Notch-2 but not Notch-1 protein. Jagged-1 induced equal levels of Notch activation in Notch-1- and Notch-1+ cells. However, in contrast to Notch-1+ MM cells, U266 cells were not protected from drug-induced apoptosis. Similarly, activation of Notch signaling in U266 cells failed to change expression of p21WAF/Cip or inhibit cell proliferation and induce growth arrest. The causative relationship between Notch-1 signaling and protection of MM cells from drug-induced apoptosis has been confirmed in experiments in which U266 cells were transfected with an active form of Notch-1 (NotchIC). Transfected cells demonstrated accumulation of p21WAF/Cip and significant protection from melphalan-induced apoptosis. Since the levels of expression of Notch-3 and Notch-4 in all tested hematopoietic cancer cell lines were equal, we could conclude that Notch-1 signaling resulted in accumulation of p21WAF/Cip, which in turn led to accumulation of MM cells in the G0/G1 phase of the cell cycle and protection from drug-induced apoptosis.
The effect of Notch signaling on cell growth is controversial. A number of published works have demonstrated that Notch signaling increases cell proliferation.20,23,26 In contrast, a few studies have demonstrated that activation of Notch-1 in tumor cells causes growth arrest.41,42 Overexpression of the active form of Notch-1 in keratinocytes strongly suppressed DNA synthesis. It caused significant increase of the p21WAF/Cip protein, whereas p27Kip1 was not affected. The p21 null keratinocytes were completely resistant to Notch-1-mediated growth suppression, which indicated a critical role of p21WAF/Cip in Notch-1-mediated effect on cell proliferation.41 These authors have shown that RBP-Jκ can bind to the promoter region of p21WAF/Cip1 and activate transcription of this protein.41 Induction of Notch signaling induced cell cycle arrest in small cell lung cancer cells via up-regulation of both p21WAF/Cip and p27Kip1.42 The opposite effect was found in human neural stem cells. Expression of p21WAF/Cip was stimulated after inhibition of HES1, which is downstream of RBP-Jκ.43 Thus, these data support the concept that the effect of Notch signaling on cell growth and proliferation is highly dependent on cell type and lineage.20,42,44
Recent research has shown that oncogenic Ras activates Notch signaling. Oncogenic Ras increases levels and activity of the intracellular form of wild-type Notch-1 and upregulates Notch ligand Delta-1 and also presenilin-1, a protein involved in Notch processing, through a p38-mediated pathway.22 Expression of constitutively active mutated ras has been demonstrated in a number of MM cell lines.45-47 This may represent another pathway, which regulates resistance of MM cells to drug-induced apoptosis.
Different patterns of expression of Notch-1 and Notch-2 in different organs have been previously described.11,12,48 However, in this study we, for the first time, have described a unique function attributed exclusively to Notch-1 but not to Notch-2. Although hematopoietic malignant cell lines HS-Sultan and U266 express equal amounts of Notch-3 and Notch-4, significant differences are observed for Notch-1 and Notch-2; HS-Sultan cells are Notch-1+Notch-2-, whereas U266 cells are Notch-1-Notch-2+. Jagged-1 activates Notch signaling in both cell lines, but consequences are significantly different. Activation of Notch in HS-Sultan cells resulted in cell cycle arrest and protection from drug-induced apoptosis, whereas no such effects were observed in U266 cells. It appears that protection of MM cells from drug-induced apoptosis was mediated specifically via Notch-1 signaling. This suggests that Notch signaling in MM cells could be a potential target for therapeutic intervention. At this time, there is no information about Notch expression on myeloma cells obtained directly from patients. However, a recent study demonstrated strong Notch expression in all tested patients with Hodgkin lymphoma and anaplastic large cell lymphoma and weak expression in all patients with sporadic Burkitt lymphoma and large B-cell lymphoma.49
Our data presented here not only demonstrate a potential mechanism of BMS involvement in the protection of hematopoietic malignant cells from drug-induced apoptosis but suggest a new therapeutic approach to decrease the resistance of MM to chemotherapy. Inhibition of Notch-1 signaling with siRNAs or small molecules could provide substantial benefits in the treatment of this disease.
Prepublished online as Blood First Edition Paper, December 11, 2003; DOI 10.1182/blood-2003-07-2340.
Supported by a fellowship grant from the Multiple Myeloma Research Foundation (Y.N.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs L. Miele and B. Osborne for providing us with reagents and fruitful discussion.

![Figure 4. Effect of Notch-1 pathway activation on p21 and cell growth. Activation of Notch-1 pathway results in up-regulation of p21 and growth arrest in myeloma cells. H929, RPMI 8226, U266, or HS-Sultan myeloma cells were incubated on the monolayer of irradiated 3T3-Jagged or 3T3-MSCV fibroblasts (A,C-D), or BMS cells (B), or kept in suspension for 48 hours. (A-B) [3H]-thymidine was added for the last 16 hours of culture. Thymidine incorporation was measured by liquid scintillation counting. Background level of thymidine incorporation into irradiated stromal cells was subtracted in all experiments. (C-D) Myeloma cells were collected, fixed, and stained with propidium iodide (PI). Cell cycle distribution was analyzed by flow cytometry. (E) H929 cells were incubated with 3T3-Jagged (J) or 3T3-MSCV (Con) cells for 24, 36, or 48 hours. Myeloma cells were collected, and p21 and p27 protein expression was analyzed by Western blotting. To confirm equal loading, membrane was reprobed with β-actin antibody. (F-G) U266 cells were cultured on 3T3-Jagged (J) or 3T3-MSCV (Con) stroma (F); H929 cells were incubated on BMS or kept in suspension for 48 hours (G). Myeloma cells were collected, and p21 expression was analyzed by Western blotting. Error bars (A-D) indicate standard deviation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/9/10.1182_blood-2003-07-2340/6/m_zh80080459720004.jpeg?Expires=1766081909&Signature=VBN22bRCcDFZlHaSfJ5cWV~2KZEGZKOgc~J6DS1WmBeeDn4zJieB5s5k0W8cXEx~D2WrDGET0wZXvcwxioVuarO1jf5H7TfHrYaj3YVB8FB-yeaY-zoSGqwl01h0UT~AJ2jjeYZUI4y~oKpBe3RE1HVS98Tt5D7FaLnHeefIuyDX0BZpk2mEXHKoXzLa0DlQIIcSg~tJbs57yBAxScn8cDdcQu3Kj9JYN0-wNz7aGrCGrBI~wEa3fl3T4tktvAhJfQg5pdVss-Mdsj2AKkVeG9ZAT2UBkb4CkCb-c8NFjF1ik-NPP8Gmw23P0PCnww4BRqcSHgFlGIWibla8UAKeeA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. Effect of Notch-1 pathway activation on p21 and cell growth. Activation of Notch-1 pathway results in up-regulation of p21 and growth arrest in myeloma cells. H929, RPMI 8226, U266, or HS-Sultan myeloma cells were incubated on the monolayer of irradiated 3T3-Jagged or 3T3-MSCV fibroblasts (A,C-D), or BMS cells (B), or kept in suspension for 48 hours. (A-B) [3H]-thymidine was added for the last 16 hours of culture. Thymidine incorporation was measured by liquid scintillation counting. Background level of thymidine incorporation into irradiated stromal cells was subtracted in all experiments. (C-D) Myeloma cells were collected, fixed, and stained with propidium iodide (PI). Cell cycle distribution was analyzed by flow cytometry. (E) H929 cells were incubated with 3T3-Jagged (J) or 3T3-MSCV (Con) cells for 24, 36, or 48 hours. Myeloma cells were collected, and p21 and p27 protein expression was analyzed by Western blotting. To confirm equal loading, membrane was reprobed with β-actin antibody. (F-G) U266 cells were cultured on 3T3-Jagged (J) or 3T3-MSCV (Con) stroma (F); H929 cells were incubated on BMS or kept in suspension for 48 hours (G). Myeloma cells were collected, and p21 expression was analyzed by Western blotting. Error bars (A-D) indicate standard deviation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/9/10.1182_blood-2003-07-2340/6/m_zh80080459720004.jpeg?Expires=1766081910&Signature=hxPqotL~N4LKe1oKMR2icF3foxQUeZRYnQBIAdREKo06C69fdv9VaAYmLjhQusaoiAdQnXxp90SDS7kZ6D-ftZqe4ho0gJjlT-EUzMBhrXZTMNV~NvAe6pABcDu64~2x9XqPGuyqm17WN6TeidMaGepPzU~jNvFa-S~x7lQG0cf-fbTteYvaAdHp-3BNovlMpDJ50i0bNPXTkZa0rkxamF4Wj19I5UONOxUXfEUoXQZji4yyzKwSDC6IsBog-cmC9NlLCAo5W6BOThsJhiP5wPsNC8ukZJE7ioSf4VRhOmobf8r8c0L4MMeoTibgpM-K9Kh2A05RnYbw97kZYYtTYQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)