Abstract
Insulin-like growth factor binding protein-3 (IGFBP-3) can cause growth suppressive and proapoptotic effects on retinoids in many types of cancer cells. However, the expression and effects of IGFBP-3 in myeloid leukemia cells have not been elucidated. In this study, we found no IGFBP-3 expression in the human myeloid leukemia cell lines either at baseline or after stimulation with all-trans retinoic acid (ATRA). Human recombinant IGFBP-3 induced growth arrest and apoptosis of HL-60 and NB4 cells. We have previously identified RXRα as a nuclear receptor for IGFBP-3 and have proceeded to examine further the role of this interaction in leukemia cell lines. In signaling assays, IGFBP-3 potently suppressed RAR- and VDR-mediated signaling while enhancing RXR signaling. Interestingly, when IGFBP-3 was administered to these cells in combination with an RAR-selective ligand, the ability of these retinoids to induce differentiation was blunted. On the other hand, IGFBP-3 enhanced the effect of an RXR-selective ligand to induce differentiation of HL-60 and NB4 cells. Further studies showed that IGFBP-3 down-regulated (at the transcriptional level) the retinoid-induced expression of C/EBPϵ in NB4 cells. Taken together, these results indicate that IGFBP-3 has antiproliferative activity against myeloid leukemia cells; while it enhances signaling through RXR/RXR, it blunts signaling by activated RAR/RXR.
Introduction
Insulin-like growth factor-I (IGF-I) and IGF-II bind to insulin-like growth factor receptors IGF-IR and IGF-IIR and promote cell proliferation.1,2 Most of the biologic functions of IGFs are mediated by the type I receptor, which activates Ras, Raf, and the mitogen-activated protein kinase (MAPK) signaling pathway, and by the phosphatidylinositol 3 kinase (PI3K) pathway.3 IGF binding proteins (IGFBPs) modulate the biologic activity of IGF by sequestering IGFs away from IGF-Rs, thereby inhibiting the mitogenic and antiapoptotic activities of IGFs. Among the 6 IGFBPs, IGFBP-3 is the most abundant protein in the circulation, where it forms a 150-kDa complex with an acid-labile subunit and with IGFs.1,2
An association has been noted between elevated serum levels of IGFBP-3 and reduced risk for acute childhood leukemia,4 prostate cancer,5 and non–small cell lung cancer (NSCLC).6 IGFBP-3 induced the growth arrest and apoptosis of cells independently of IGFs; forced expression of IGFBP-3 in murine fibroblasts inhibited their cell growth, and this was not reversible when excess insulin was added.7 In addition, IGFBP-3 mediates the antiproliferative and proapoptotic effects of retinoids,8-10 1,25 dihydroxyvitamin D3,11-15 transforming growth factor β (TGF-β),16,17 and tumor necrosis factor α (TNF-α)18 in a variety of cancer cell lines, including lung, prostate, colon, and breast. Moreover, forced expression of IGFBP-3 induced the apoptosis of NSCLC cells through the inhibition of MAPK and AKT activities.19 Thus, IGFBP-3 appears to act as a tumor-suppressor molecule.
All-trans retinoic acid (ATRA) binds to retinoic acid (RA) receptor α, which forms heterodimmers with RXR and binds to the RA response element (RARE), resulting in the activation of target genes such as the myeloid-specific transcription factor C/EBPϵ, causing growth arrest, apoptosis, and differentiation of myeloid leukemia cells.20,21 In addition, novel synthetic retinoids (rexinoids) that bind only to RXR have been synthesized and have also been shown to inhibit cancer cell growth.22 We have recently described the direct and specific binding of IGFBP-3 to RXR in the nuclei of cancer cells and have proposed that this may be the mechanism for some of IGFBP-3 actions.23 Multiple studies have suggested that retinoids inhibit the growth of breast and prostate cancer cells through IGFBP-3,8-10 which prompted us to hypothesize that IGFBP-3 may modulate the prodifferentiative and proapoptotic effects of retinoids in human myeloid leukemia cells.
This study found that human myeloid leukemia cells did not constitutively express IGFBP-3 and that retinoids did not induce its expression in these cells. However, human recombinant IGFBP-3 induced the growth arrest and apoptosis of myeloid leukemia cells on its own. Interestingly, IGFBP-3 blunted the prodifferentiative and proapoptotic effects of RARα–selective ligands but augmented the activity of RXR-selective ligands.
Materials and methods
Cell lines
Cell lines HL-60 and U937 were obtained from American Type Culture Collection (Rockville, MD). NB4 cell line was a kind gift from M. Lanotte (St Louis Hospital, Paris, France). Cells were grown in RPMI 1640 medium (Gibco Life Technologies, Grand Island, NY) with 10% heat-inactivated fetal bovine serum (FCS; Gibco).
Chemicals
Recombinant human IGFBP-3 was provided by Protigen (Mountain View, CA) and was dissolved in PBS. ATRA and 9-cis RA were purchased from Sigma Chemical (St Louis, MO) and were dissolved in 100% ethanol at a stock concentration of 10–2 M, stored at –20° C, and protected from light. RARα–specific agonist AGN197496 and RXR-specific agonist AGN194204 were synthesized at Allergan (Irvine, CA) and kindly provided to us. Protease inhibitors aprotinin, pepstatin, leupeptin, and phenylmethysulfonyl fluoride were purchased from Boehringer Mannheim (Germany) and were dissolved in phosphate-buffered saline (PBS) at stock concentrations of 100 μg/mL, 50 μg/mL, 500 μg/mL, and 500 μg/mL, respectively.
Modulation of nuclear receptor signaling by IGFBP-3
The effects of IGFBP-3 on the transcriptional activity of RXR, RAR, and VDR were assessed by measuring luciferase activity in LAPC-424 human prostate cancer cells cotransfected with these specific receptors, whose DNA-binding domains (DBDs) were replaced by the GAL4 DBD25 (generously provided by Peter Tontonoz of the University of California at Los Angeles), a GAL4 control vector, and an IGFBP-3 expressing vector9 or control vector, with or without the specific ligand for the ligand-binding domain (LBD) of each receptor. Gal4 chimeric hybrid vectors24 (1 μg/well) were cotransfected with or without ligand (10–6 M) and either IGFBP-3 expression vector (PKG-IGFBP-3)9 or control vector (pKG1226) (1 μg/well) for 5 hours and were incubated with 10% serum for 24 hours. The cells were harvested, and luciferase activity was measured using the Luciferase Assay system kit (Promega, Madison, WI) and controlled for Gal4 activity.
Trypan blue exclusion test
Leukemia cells (105/mL) were incubated with a variety of concentrations of IGFBP-3 (0.01-10 μg/mL), ATRA (10–10-10–7 M), or their combination for 4 days in 96-well plates (Flow Laboratories, Irvine, CA). After culture, cell number and viability were evaluated by staining with trypan blue and counting using light microscopy.
Assays for differentiation
Induction of differentiation of cell lines was measured by CD11b antigen expression and nitroblue tetrazolium dye (NBT) reduction, as previously described.26 Cells were harvested after 2 days' incubation. Phycoerythrin-conjugated murine antihuman CD11b (DAKO, Carpinteria, CA) was used to detect CD11b. Control studies were performed with a nonbinding murine immunoglobulin G (IgG) antibody (DAKO). Analysis of fluorescence was performed on a FACScan flow cytometer (Becton Dickinson, Mountain View, CA).
For NBT measurements, cells (105/mL) were incubated in 24-well plates (Corning Inc, Corning, NY) for 2 days. After incubation, each cell suspension was mixed with an equal volume of RPMI 1640 containing 1 mg/mL NBT (Sigma) and 10–6 M 12-O-tetradecanoyl-13-acetate (TPA; Sigma) for 30 minutes at 37° C. Cells were washed in PBS, cytocentrifuged, fixed in methanol for 5 minutes, and stained with Gram safranin for 10 minutes at room temperature; 300 cells were analyzed for blue dye using light microscopy.
Assessing apoptosis
Apoptotic cell death was examined using the terminal deoxyribonucleotide transferase-mediated dUTP nick-end labeling (TUNEL) method using the In Situ Cell Death Detection Kit (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's instruction. For quantification, 3 different fields were counted under the microscope, and at least 300 cells were enumerated in each field. All experiments were performed twice.
RNA isolation and reverse transcription–polymerase chain reaction
Total RNA was isolated as previously described using TRIzol (Life Technologies, Grand Island, NY).26 One microgram DNase I–treated RNA was reverse transcribed by using Moloney murine leukemia virus reverse transcriptase (Life Technologies), and 50 ng of the resultant complementary DNA (cDNA) was used as a template for polymerase chain reaction (PCR). We measured the expression of 18S for normalization. Real-time PCR was carried out by using Taq DNA polymerase (Qiagen), 50 ng cDNA for IGFBP-3 and C/EBPϵ (5-500 ng in serial dilutions for standard curves), or 1 pg for 18S (10-0.1 pg for standard curve), and SYBR Green I nucleic acid gel staining solution in a 1:60 000 dilution. To enhance sensitivity, the melting temperature of each PCR product was determined according to the manufacturer's recommendations. PCR conditions for all genes were as follows: a 95° C initial activation for 15 minutes followed by 45 cycles of 95° C for 15 seconds, 60° C for 15 seconds, and 72° C for 30 seconds, and fluorescence determination at the melting temperature of the product for 20 seconds on an ICycler detection system (Bio-Rad, Hercules, CA). The threshold cycle, which is defined as the cycle at which PCR amplification reaches a significant value, was determined according to the manufacturer's protocol. Normalization of the threshold cycles was carried out as follows: the relative 18S cDNA amount was extrapolated on the 18S standard curve, as was the relative IGFBP-3 and C/EBPϵ cDNA content on the corresponding standard curves. For normalization against 18S, each relative IGFBP-3 and C/EBPϵ cDNA value was divided by the mean of 3 18S cDNA value replicates. The mean of these 3 values was taken as the relative expression level.
Statistical analysis
Statistical analysis was performed to assess the difference between 2 groups under multiple conditions by one-way analysis of variance (ANOVA) using PRISM statistical analysis software (GraphPad Software, San Diego, CA).
Results
Modulation of nuclear receptor signaling by IGFBP-3
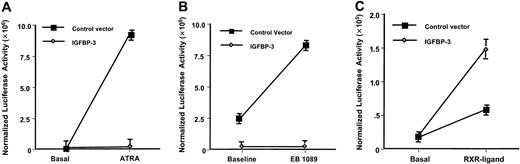
The effects of IGFBP-3 on the transcriptional activity of RXR, RAR, and VDR were assessed by measuring luciferase activity in cells cotransfected with these specific receptors whose DBDs were replaced by the GAL4 DBD and either with an IGFBP-3–expressing vector or a control vector, with or without the specific ligand for the ligand-binding domain (LBD) of each receptor. Forced expression of IGFBP-3 caused apoptosis and decreased viability of LAPC-4 cells by 15% compared with the control vector–transfected cells (data not shown). We corrected the signaling activity to a GAL4 control vector activity, which accounted for the viable cell number difference. Figure 1A shows the dramatic induction of Gal4/RARα signaling by 10–6 M ATRA (P < .0001) and the complete lack of signaling observed when IGFBP-3 was cotransfected in the presence of ATRA (P < .0001). Similarly, Figure 1B shows the effects of 10–6 M of the vitamin D analog EB1089 on Gal4/VDR signaling (P < .001). Additionally, the dramatic suppressive effects of IGFBP-3 on VDR are observed in the presence and absence of the vitamin D analog (P < .001). In contrast, Figure 1C shows the effects of 1 × 10–6 M of the RXR-selective agonist AGN194204 on Gal4/RXR signaling, which led to a 3-fold induction of signaling in the presence of the control vector (P < .01) but demonstrated a further activation of signaling when IGFBP-3 was cotransfected (P < .01).
Modulation of nuclear receptor signaling by IGFBP-3. (A) RAR signaling. Luciferase activity was measured in LAPC-4 human prostate cancer cells cultured in the presence or absence of 10–6 M ATRA and was transfected with IGFBP-3–expressing vector or control vector and with a vector expressing the RAR in which the DBD was replaced by the GAL4 DBD and with a GAL4 control vector. Each point represents the mean ± SD of 3 independent experiments performed in triplicate and normalized for GAL4 activity. (B) VDR signaling. Similar to panel A, except that cells were cultured in the presence or absence of 10–6 M of the vitamin D analog EB1089 and cotransfected with a vector expressing the VDR in which the DBD was replaced by the GAL4 DBD. (C) RXR signaling. Similar to panel A, except cells were cultured in the presence or absence of 10–6 M of the RXR-selective agonist AGN194204 and were cotransfected with a vector expressing the RXR in which the DBD was replaced by the GAL4 DBD.
Modulation of nuclear receptor signaling by IGFBP-3. (A) RAR signaling. Luciferase activity was measured in LAPC-4 human prostate cancer cells cultured in the presence or absence of 10–6 M ATRA and was transfected with IGFBP-3–expressing vector or control vector and with a vector expressing the RAR in which the DBD was replaced by the GAL4 DBD and with a GAL4 control vector. Each point represents the mean ± SD of 3 independent experiments performed in triplicate and normalized for GAL4 activity. (B) VDR signaling. Similar to panel A, except that cells were cultured in the presence or absence of 10–6 M of the vitamin D analog EB1089 and cotransfected with a vector expressing the VDR in which the DBD was replaced by the GAL4 DBD. (C) RXR signaling. Similar to panel A, except cells were cultured in the presence or absence of 10–6 M of the RXR-selective agonist AGN194204 and were cotransfected with a vector expressing the RXR in which the DBD was replaced by the GAL4 DBD.
Expression of IGFBP-3 in human myeloid leukemia cells
IGFBP-3 expression in NB4, HL-60, and U937 human myeloid cells was explored by real-time PCR. No IGFBP-3 transcripts were detectable after 45 cycles of PCR using cDNA from these cells (data not shown). Exposure of these cells to ATRA (10–7 M; 18 to 48 hours) did not induce the expression of IGFBP-3 (data not shown). On the other hand, T47D breast cancer cells constitutively expressed IGFBP-3, and exposing these cells to ATRA (10–7 M; 18 hours) increased the level by 2-fold (data not shown), which was consistent with previous experiments.8
Effect of IGFBP-3 on proliferation and apoptosis in liquid culture of NB4 promyelocytic leukemia cells
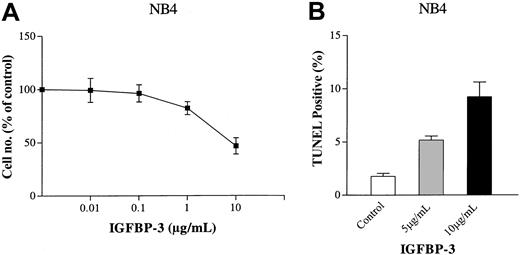
Using the trypan blue exclusion test, IGFBP-3 was examined for its effect on the growth of NB4 cells in liquid culture. IGFBP-3 inhibited the proliferation of these cells with an ED50 (effective dose that inhibited 50% cell proliferation) of 10 μg/mL (Figure 2A). IGFBP-3 caused apoptosis of NB4 cells in a dose-dependent manner, as measured by TUNEL assay (Figure 2B). On the second day of culture, 3.5 or 10 μg/mL IGFBP-3 induced a mean 5% ± 0.5% and 9% ± 2%, respectively, of NB4 cells to become apoptotic.
Dose-response activity of IGFBP-3 on proliferation of myeloid leukemia cells. (A) Trypan blue exclusion test. NB4 cells (105/mL) were cultured in the presence of various concentrations of human recombinant IGFBP-3 (0.01-10 μg/mL) for 4 days. Results are expressed as a mean percentage of the cell count in the control plates. Each point represents the mean ± SD of 3 independent experiments performed in triplicate. (B) TUNEL assay. NB4 cells (105/mL) were cultured with human recombinant IGFBP-3 (5 or 10 μg/mL) for 2 days, and apoptosis was measured by TUNEL assay. Data represent the mean ± SD of triplicate cultures.
Dose-response activity of IGFBP-3 on proliferation of myeloid leukemia cells. (A) Trypan blue exclusion test. NB4 cells (105/mL) were cultured in the presence of various concentrations of human recombinant IGFBP-3 (0.01-10 μg/mL) for 4 days. Results are expressed as a mean percentage of the cell count in the control plates. Each point represents the mean ± SD of 3 independent experiments performed in triplicate. (B) TUNEL assay. NB4 cells (105/mL) were cultured with human recombinant IGFBP-3 (5 or 10 μg/mL) for 2 days, and apoptosis was measured by TUNEL assay. Data represent the mean ± SD of triplicate cultures.
Protease inhibitors enhanced the ability of IGFBP-3 to induce growth arrest of leukemia cells
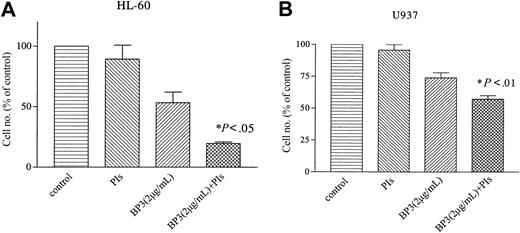
Myeloid leukemia cells secrete large amounts of various proteases that are known to cleave IGFBP-3.27 We examined whether protease inhibitors affect the biologic function of IGFBP-3. Myeloid leukemia HL-60 and U937 cells were cultured in the presence of IGFBP-3 (2 μg/mL) or protease inhibitors (5 μg/mL phenylmethysulfonyl fluoride, 1 μg/mL aprotinin, 0.5 μg/mL pepstatin, and 5 μg/mL leupeptin). IGFBP-3 alone inhibited the growth of HL-60 and U937 cells by 47% ± 12% or 26% ± 6%, respectively, on the fourth day of culture (Figure 3A-B). The effect of protease inhibitors on the growth of these cells was negligible; however, when HL-60 and U937 cells were cultured in the presence of IGFBP-3 and protease inhibitors, their growth was inhibited by approximately 80% and 43%, respectively (Figure 3A-B).
Effect of protease inhibitors on IGFBP-3-induced growth inhibition of myeloid leukemia cells. HL-60 (A) and U937 (B) cells were cultured in the presence of IGFBP-3 (2 μg/mL) or protease inhibitors (5 μg/mL phenylmethysulfonyl fluoride, 1 μg/mL aprotinin, 0.5 μg/mL pepstatin, and 5 μg/mL leupeptin). Data represent the mean ± SD of triplicate cultures; and results represent 3 independent experiments. BP-3 indicates IGFBP-3; PIs, protease inhibitors.
Effect of protease inhibitors on IGFBP-3-induced growth inhibition of myeloid leukemia cells. HL-60 (A) and U937 (B) cells were cultured in the presence of IGFBP-3 (2 μg/mL) or protease inhibitors (5 μg/mL phenylmethysulfonyl fluoride, 1 μg/mL aprotinin, 0.5 μg/mL pepstatin, and 5 μg/mL leupeptin). Data represent the mean ± SD of triplicate cultures; and results represent 3 independent experiments. BP-3 indicates IGFBP-3; PIs, protease inhibitors.
IGFBP-3 inhibited the ability of retinoid to induce growth arrest of NB4 cells
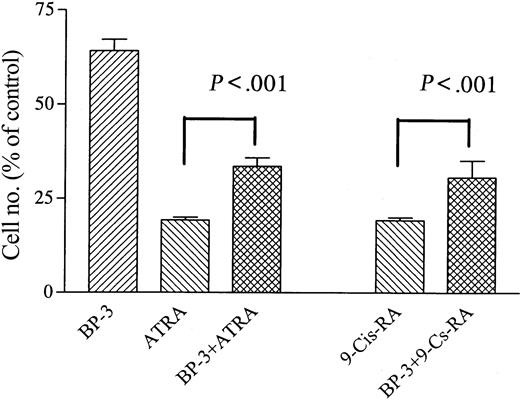
NB4 cells were cultured with ATRA (10–7 M) alone or in combination with IGFBP-3 (5 μg/mL). IGFBP-3 blunted the ability of ATRA to inhibit the proliferation of NB4 cells (Figure 4A). ATRA (10–7 M, 4 days) inhibited the growth of NB4 cells by 79% ± 2%, and IGFBP-3 (5 μg/mL) inhibited the proliferation of these cells by 36% ± 5%. The combination of ATRA (10–7 M) and IGFBP-3 (5 μg/mL) inhibited NB4 growth by 67% ± 2% (P < .001) (Figure 4). IGFBP-3 partially reversed the ability of 9-cis RA to inhibit the growth of NB4 cells, and 9-cis RA (10–7 M, 4 days) inhibited the growth of NB4 cells by 81% ± 2%. When 9-cis RA (10–7 M) was combined with IGFBP-3 (5 μg/mL), NB4 growth inhibition was 70% ± 4% (P < .001) (Figure 4).
Effect of IGFBP-3 on ATRA- or 9-cis RA–induced growth inhibition of NB4 cells. Cells were cultured with ATRA (10–7 M) or 9-cis RA (10–7 M) alone or in combination with IGFBP-3 (5 μg/mL). Data represent the mean ± SD of triplicate cultures, and results represent at least 3 independent experiments.
Effect of IGFBP-3 on ATRA- or 9-cis RA–induced growth inhibition of NB4 cells. Cells were cultured with ATRA (10–7 M) or 9-cis RA (10–7 M) alone or in combination with IGFBP-3 (5 μg/mL). Data represent the mean ± SD of triplicate cultures, and results represent at least 3 independent experiments.
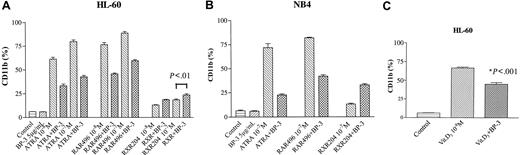
Effect of IGFBP-3 on RARα or RXR selective ligand–induced expression of CD11b on HL-60 and NB4 cells
CD11b expression increases as myeloid cells differentiate toward either macrophages or granulocytes.26 IGFBP-3 (5 μg/mL, 2 days) alone did not affect CD11b expression on HL-60 and NB4 cells (Figure 5A-B). Interestingly, when IGFBP-3 was combined with ATRA, the expression of ATRA-induced CD11b was markedly reduced; ATRA (10–8 or 10–7 M) for 2 days induced CD11b antigen expression on 60% ± 3% and 80% ± 3% of HL-60 cells, respectively. When ATRA (10–8 or 10–7 M) was combined with IGFBP-3 (5 μg/mL), 33% ± 3% and 43% ± 2% of HL-60 cells, respectively, expressed CD11b on day 2 of culture (Figure 5A). Similarly, IGFBP-3 blunted the ability of RARα–selective agonist AGN197496 to induce the expression of CD11b on HL-60 cells. AGN197496 (10–7 M, 2 days) induced CD11b antigen expression on 89% ± 2% of HL-60 cells. The combination of AGN197496 and IGFBP-3 (5 μg/mL) induced 60% ± 2% of HL-60 cells to express CD11b on day 2 of culture (Figure 5A). On the other hand, IGFBP-3 enhanced the activity of the RXR-selective agonist AGN194204 to induce CD11b expression on HL-60 cells; AGN194204 (10–7 M, 2 days) alone or in combination with IGFBP-3 induced 18% ± 1% or 24% ± 2% of HL-60 cells to express CD11b, respectively (P < .01).
Effect of IGFBP-3. Retinoid (A-B) or 1,25(OH)2 vitamin D3 (C) induced the expression of CD11b on HL-60 (A, C) and NB4 (B) cells. HL-60 (A) and NB4 (B) cells were cultured for 2 days with IGFBP-3 (5 μg/mL), a retinoid (10–8 or 10–7 M), or both in combination and then were analyzed by FACscan for CD11b expression. (C) HL-60 cells were cultured for 2 days with IGFBP-3 (5 μg/mL), 1,25(OH)2 vitamin D3 (10–8 M), or both in combination and then were analyzed by FACscan for CD11b expression. Data represent the mean ± SD of triplicate cultures, and results were derived from 2 independent experiments. RAR496 indicates AGN197496; RXR204, AGN194204; BP-3, IGFBP-3.
Effect of IGFBP-3. Retinoid (A-B) or 1,25(OH)2 vitamin D3 (C) induced the expression of CD11b on HL-60 (A, C) and NB4 (B) cells. HL-60 (A) and NB4 (B) cells were cultured for 2 days with IGFBP-3 (5 μg/mL), a retinoid (10–8 or 10–7 M), or both in combination and then were analyzed by FACscan for CD11b expression. (C) HL-60 cells were cultured for 2 days with IGFBP-3 (5 μg/mL), 1,25(OH)2 vitamin D3 (10–8 M), or both in combination and then were analyzed by FACscan for CD11b expression. Data represent the mean ± SD of triplicate cultures, and results were derived from 2 independent experiments. RAR496 indicates AGN197496; RXR204, AGN194204; BP-3, IGFBP-3.
A similar effect was observed with NB4 cells. Either RAR-selective ATRA (10–7 M, 2 days) or AGN 197496 (10–7 M, 2 days) alone induced CD11b expression on 71% ± 2% and 82% ± 1% of NB4 cells, respectively; the combination of either ATRA or AGN197496 with IGFBP-3 resulted in approximately 22% ± 1% and 42% ± 2% of these cells expressing CD11b (Figure 5B). In contrast, the RXR-selective ligand AGN194204 (10–7 M, 2 days) induced 13% ± 1% of NB4 cells to express CD11b. When it was combined with IGFBP-3 (5 μg/mL), CD11b expression increased, with a mean 33% ± 2% of NB4 cells expressing CD11b on day 2 of culture.
IGFBP-3 inhibited the activity of 1,25 dihydroxyvitamin D3 to induce expression of CD11b on HL-60 cells
1,25 Dihydroxyvitamin D3 (10–8 M, 2 days) induced 67% ± 2% of HL-60 cells to express CD11b, and the combination of 1,25 dihydroxyvitamin D (10–8 M) and IGFBP-3 (5 μg/mL) resulted in a blunting of differentiation with a mean 45% ± 3% of HL-60 cells expressing CD11b on day 2 of culture (P < .001) (Figure 5C).
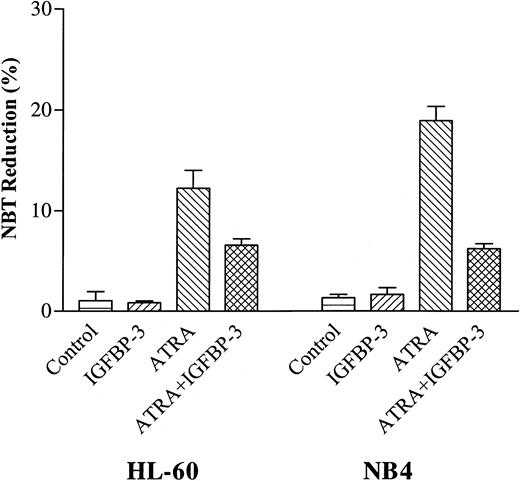
IGFBP-3 reduced the ability of ATRA to induce superoxide production in HL-60 and NB4 cells
Superoxide production is a marker of granulocyte-like differentiation, which can be measured by the ability of the cell to reduce NBT.26 Using this marker, we examined whether IGFBP-3 affected the capacity of ATRA to induce differentiation of HL-60 and NB4 cells. IGFBP-3 (5 μg/mL) alone did not reduce NBT in the HL-60 and NB4 cells. ATRA (10–7 M) induced 12% ± 3% and 19% ± 2% of HL-60 and NB4 cells to reduced NBT, respectively; however, when ATRA (10–7 M) was combined with IGFBP-3 (5 μg/mL), a mean of 7% ± 1% and 6% ± 1% of HL-60 and NB4 cells reduced NBT, respectively (Figure 6).
Effect of IGFBP-3 on ATRA-induced NBT reduction of HL-60 and NB4 cells. Cells were cultured for 2 days with IGFBP-3 (5 μg/mL) alone or in combination with ATRA (10–7 M), and differentiation was determined by NBT reduction. Data represent the mean ± SD of triplicate cultures.
Effect of IGFBP-3 on ATRA-induced NBT reduction of HL-60 and NB4 cells. Cells were cultured for 2 days with IGFBP-3 (5 μg/mL) alone or in combination with ATRA (10–7 M), and differentiation was determined by NBT reduction. Data represent the mean ± SD of triplicate cultures.
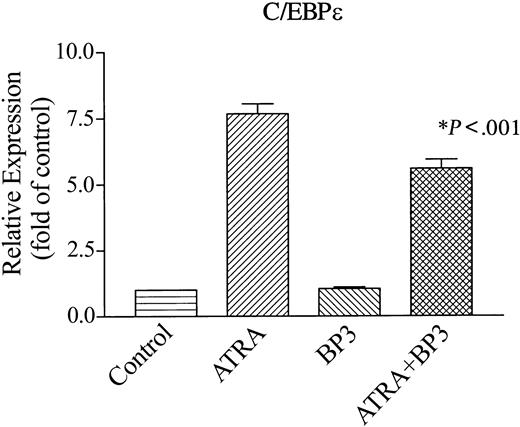
IGFBP-3 transcriptionally inhibited the ATRA induction of a myeloid transcription factor (C/EBPϵ)
C/EBPϵ is a CCAAT/enhancer-binding transcriptional factor whose expression is restricted to maturing myeloid cells.21 Forced expression of it can induce granulocytic differentiation of myeloid leukemia cells,28 and deletion or mutation of this transcription factor in mice and humans results in incomplete myeloid differentiation.21 Expression of C/EBPϵ occurs as NB4 cells differentiate to granulocytes. We examined whether IGFBP-3 affects ATRA-induced expression of C/EBPϵ mRNA in NB4 cells (Figure 7). These cells constitutively expressed C/EBPϵ mRNA; and ATRA (10–7 M, 2 days) enhanced expression of C/EBPϵ mRNA by 7.5-fold compared with untreated control cells (Figure 7). IGFBP-3 (5 μg/mL, 2 days) decreased ATRA-induced expression of C/EBPϵ mRNA by 30% compared with those treated with ATRA alone (P < .001).
Effect of IGFBP-3 on ATRA-induced levels of C/EBPϵ transcripts in NB4 cells. Cells were cultured for 2 days with IGFBP-3 (5 μg/mL), ATRA (10–7 M), or both in combination. RNA was extracted. cDNA was synthesized and subjected to real-time PCR to measure the levels of C/EBPϵ transcripts. Data represent the mean ± SD of triplicate cultures.
Effect of IGFBP-3 on ATRA-induced levels of C/EBPϵ transcripts in NB4 cells. Cells were cultured for 2 days with IGFBP-3 (5 μg/mL), ATRA (10–7 M), or both in combination. RNA was extracted. cDNA was synthesized and subjected to real-time PCR to measure the levels of C/EBPϵ transcripts. Data represent the mean ± SD of triplicate cultures.
Discussion
RAR binds RAR-selective retinoids, including ATRA, forming heterodimers with RXR that bind to RAREs, resulting in the expression of target genes.29 RXRs predominantly act as coregulators for many nuclear receptor transcription factors, including vitamin D3 receptor, thyroid hormone receptor, and peroxisome proliferator-activated receptors (PPARs). Ligand-activated heterodimer receptors bind to their response elements and transactivate their target genes.30 In addition, RXR-selective retinoids activate RXRs to form homodimers, bind to RXREs, and activate target genes, although the activation is usually through ligand-stimulated RAR/RXR heterodimers31,32 ; 9-cis RA can activate RXR homodimers and RAR/RXR heterodimers.31,32 This study has found that IGFBP-3 augmented the prodifferentiative effects of RXR-selective retinoids but inhibited the ability of RAR-selective retinoids to mediate these effects, suggesting that IGFBP-3 stimulates signaling by RXR/RXR but inhibits signaling by RAR/RXR. A possible explanation is that IGFBP-3 can bind to RXR and augment the transcriptional activity of RXR/RXR, whereas it can inactivate RAR/RXR transcriptional activity by sequestering RXR away from RAR/RXR heterodimers.
Recently, we identified RXR as a partner of IGFBP-3 by using a yeast 2-hybrid system.23 Glutathione S-transferase (GST) pull-down assay showed that the heparin-binding domain of IGFBP-3 is critical for binding IGFBP-3 and RXR.23 Immunocytochemistry showed colocalization of IGFBP-3 and RXR in the nucleus and the cytoplasm.23 With RXR-selective ligand stimulation, both proteins were more prominent in the nucleus, suggesting ligand-dependent nuclear translocation of these factors.23 Additionally, the gel retardation assay confirmed that IGFBP-3 selectively binds to RXR/RXR homodimers, not RXR/RAR heterodimers.23 Moreover, IGFBP-3 increased RXR-selective ligand-induced DR1-RXRE reporter activity and decreased RAR-selective ligand-induced reporter activity in COS-7 cells.23 These observations, together with the results obtained in this study, strongly reinforce our hypothesis that IGFBP-3 sequesters RXR away from forming RAR/RXR heterodimers.
One possible unifying hypothesis for the interactions between retinoids and IGFBP-3 is that retinoids induce IGFBP-3 in many cell types and that IGFBP-3 feeds back in a negative fashion on RAR signaling to form a closed-loop system that regulates its own expression, making it a sensitive responsive molecule for multiple stimuli. This probably is not the case in the myeloid leukemia cells because these cells do not express IGFBP-3.
In this study, we used nonglycosylated IGFBP-3. Physiologically, IGFBP-3 is posttranslationally modified by the phosphorylation and glycosylation.33 IGFBP-3 has 3 N-glycosylation sites in its nonconserved central region (Asn89, Asn109, Asn172), and each site contains 4 to 5 kDa carbohydrate.33 Other investigators have shown that glycosylated IGFBP-3 has more cell surface–binding affinity than its glycosylated form.33 Nonglycosylated IGFBP-3 might elicit effects not observed with the natural form of IGFBP-3. However, we have recently found that the transferrin-binding C-terminal peptide region of IGFBP-3 is critical for uptake and nuclear translocation of IGFBP-3; IGFBP-3 binds transferrin and forms a ternary complex with transferrin receptor 1 and is internalized into the cells.34 Therefore, the non-glycosylated IGFBP-3 was unlikely to have elicited non-physiological activities of IGFBP-3 in this study. The effect of the carbohydrate moiety of IGFBP-3 on internalization and nuclear translocation of IGFBP-3 is unknown. Differences between forms of IGFBP-3 may exist that were not addressed by us.
IGFBP-3 circulating levels are 3000 to 6000 ng/mL,35 and we have shown that secreted IGFBP-3 levels in conditioned media of prostate cancer cells in response to cytokine stimuli were approximately 1000 ng/mL.36 We used higher concentrations of IGFBP-3 (5-10 μg/mL) to observe the unique biologic effects shown in this study. Myeloid leukemia cells secrete large amounts of various proteases known to cleave IGFBP-3.27 It is possible that the effective concentration of IGFBP-3 is much lower but that much of the IGFBP-3 in vitro is cleaved. Indeed, the presence of the protease inhibitors (5 μg/mL phenylmethysulfonyl fluoride, 1 μg/mL aprotinin, 0.5 μg/mL pepstatin, and 5 μg/mL leupeptin) enhanced the antiproliferative activity of IGFBP-3 against HL-60 and U937 cells was observed (Figure 3A-B). In vivo, we believe that inhibitors that limit the degree of proteolysis regulate these proteases.
Taken together, this study found that IGFBP-3 induced the growth arrest and apoptosis of human myeloid leukemia cells. It enhanced the differentiation-inducing activity of RXR-selective ligands but inhibited the ability of RAR-selective retinoids to induce differentiation of myeloid leukemia cells. Further studies will be required to determine the molecular mechanism by which IGFBP-3 causes these effects in myeloid leukemia cells. In addition, the role IGFBP-3 plays in leukemogenesis and responsiveness to retinoid and rexinoid therapies should be investigated.
Prepublished online as Blood First Edition Paper, March 16, 2004; DOI 10.1182/blood-2003-07-2203.
Supported by in part by National Institutes of Health grants R01AG20954, P50CA9231, R01CA100938, and UO1CA84128 (P.C.) and AT00151 (H.P.K.) and by the George Harrison Fund, the Horn Trust, the Parker Hughes Fund, and the Aaron Eschman Fund. H.P.K. holds an endowed Mark Goodson Chair of Oncology Research at Cedars-Sinai Medical Center.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Patricia Lin (Flow Cytometry Core Facility, Cedars-Sinai Medical Center) for her generous technical assistance. We also thank Kim Burgin for her excellent secretarial help.