Abstract
Acute leukemias are remarkably heterogeneous as evidenced by the increasing number of recurring genetic and epigenetic abnormalities, especially the presence or absence of balanced translocations, which have been shown to be of independent prognostic significance. Thus, In a recent single center study we analyzed 250 AML patients for a series a genetic alteration and found that balanced translocations (identified by a multiplex PCR reaction; Pallisgaard et al, BLOOD, 1998 and Olesen et al, Brit. J. Hem. in press) were present in 17% of the patients, FLT3 internal tandem duplication in 24%, and MLL partial tandem duplication in 4%. In addition, we delineated the presence of promoter hypermethylation for the p15 (71%), MDR1 (4%), E-cadherin (CDH1) (64%), and Estrogen receptor (ER) (40%) genes by bisulfite DGGE (Aggerholm, Cancer Res. 1999) as well as the abnormal expression by RQ-PCR of genes related to increased chemotherapy resistance (MDR1 and MRP1) as well as resistance to undergo apoptosis (FAS, Bcl2, BAX and CASPASE3). Here we have performed gene expression profiling focusing on patients negative for all these molecular lesions. Out of the 250 patients, 8 were determined to be molecularly negative. Global gene expression analysis (Affymetrix human genome chip (U133A)) was performed on the 6 samples, from which high-quality RNA could be harvested.
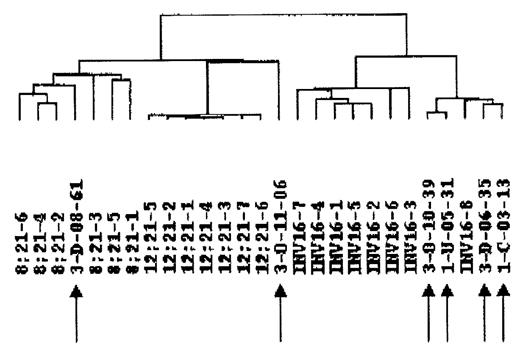
As will be seen from the Figure, all samples could be easily distinguished from TEL/AML pre-B ALL samples, and also in 4/6 cases clustered differently from AML/ETO+ and CBF/MYH11+ AML groups. Interestingly, as seen from the dendrogram, the 6 samples separated in three clusters, one also containing a CBF/MYH11+ patient (4 cases), one consisting of one single patient, while the last patient clustered together with the AML/ETO cases. While these data suggest that the gene expression in the molecularly negative patients can be different from that in CBF leukemias, it gives no clues for the leukemogenetic events in these patients. To that end, we analyzed the gene expression data and found 24 genes to be more than 5-fold overexpressed and 26 genes to be underexpressed compared to CBF AMLs. Of special notice was the increased usage of the prostaglandin/leukotriene metabolism pathway in two patients with strikingly similar global gene expression (arrows 3 and 4 from right on Fig.) including the prostaglandin I2 (prostacyclin) synthase, and the prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) genes, which were more than 10-fold increased. Also noteworthy was the upregulation of LR8 protein IQ motif containing GTPase activating protein 1 in the remaining 4 cases. Applied together with novel global cytogenetic techniques, these data will form a platform for the identification of leukemogenesis in AML patients with no demonstrable genetic aberrations.ßßß
Author notes
Corresponding author