Abstract
Increased angiogenesis in bone marrow (BM) is one of the characteristics of chronic myeloid leukemia (CML), a clonal myeloproliferative disorder that expresses a chimeric Bcr/Abl protein. Recently, the therapeutic strategy in CML has been totally modified with the development of a new drug: imatinib mesylate (STI571), a specific inhibitor of Bcr/Abl tyrosine kinase activity. The aim of our study was to determine, in patients with CML, the capacity of imatinib mesylate to modulate one of the most potent regulators of angiogenesis, the vascular endothelial growth factor (VEGF). In newly diagnosed CML, we observed significantly increased VEGF secretion by CML BM cells and significantly increased VEGF plasma concentrations. We showed that low plasma VEGF concentrations could be one of the characteristics of complete cytogenetic remission. To understand the molecular mechanisms leading to the inhibition of VEGF production by imatinib, we focused our experiments on the human cell line K562, which is Bcr/Abl positive. We demonstrated that imatinib inhibits VEGF gene transcription by targeting the Sp1 and Sp3 transcription factors. Taken together, our results highlight the potential prognostic value of VEGF concentrations in evaluating the evolution of CML patients treated with imatinib. (Blood. 2004;104: 495-501)
Introduction
Vascular endothelial growth factor (VEGF) principally targets endothelial cells and regulates several of their functions, including mitogenesis, permeability, and migration. VEGF is one of the most potent and specific regulators of angiogenesis1 and, as a consequence, is required for viability and growth of solid tumors. Recently, an increase in the microvessel density was reported in the bone marrow of patients with chronic myeloid leukemia (CML),2,3 suggesting a role for angiogenesis in the pathophysiology of hematologic malignancies. CML is a clonal myeloproliferative disorder of pluripotent hematopoietic stem cells with a specific cytogenetic abnormality, a balanced translocation between chromosomes 9 and 22. This translocation results in a Bcr/Abl chimeric gene that expresses an abnormal fusion protein showing constitutive tyrosine kinase activity. In the murine cell line Ba/F3, overexpression of Bcr/Abl induces VEGF expression.4 Previously, VEGF secretion was shown to be increased in myeloid cells derived from day-6 granulocyte-macrophage colony-forming unit (GM-CFU) colonies isolated from patients with CML.5 These data are supported by solid-phase radioimmunoassay (RIA) analysis of the VEGF protein in the bone marrow of patients with CML.6 Furthermore, VEGF plasma concentrations are significantly increased in CML.2 Recently, the therapeutic strategy in CML has been totally modified with the development of an effective new drug: imatinib mesylate (STI571), a specific inhibitor of Bcr/Abl protein tyrosine kinase activity. The aim of our study was to monitor the effect of imatinib on VEGF plasma concentrations in patients with CML. We also analyzed the effects of imatinib on the molecular mechanisms leading to VEGF expression in the human Bcr/Abl-positive cell line K562.
Patients, materials, and methods
Patients
All samples were obtained from patients, healthy donors of allogeneic bone marrow, or healthy volunteers as excess material after obtaining written, informed consent. Bone marrow specimens from 8 adult patients with newly diagnosed, untreated CML were studied. In addition, mononuclear cells (MNCs) were isolated from nonaffected bone marrow obtained from 4 patients (blood lymphocytosis, blood macrocytosis, bone marrow donors) who served as control subjects. Concurrently, we studied plasma samples of 14 patients with CML at presentation, of which 9 patients treated with imatinib were in complete cytogenetic response (CCR) for at least 6 months. Plasma of 10 healthy volunteers serves as control.
Materials
Restriction and DNA-modifying enzymes were obtained from New England Biolabs (Beverly, MA) or from Eurogentec (Liège, Belgium). Synthetic oligonucleotides were from Eurogentec. [α32P] deoxycytidine triphosphate (dCTP) was from Amersham Biosciences (Freiburg, Germany).
Cell culture and ELISA
Bone marrow MNCs were separated by density gradient centrifugation using Ficoll-Paque (Biochrom, Berlin, Germany) and were cultured for 48 hours in RPMI 1640 medium (Gibco, Paisley, United Kingdom) supplemented with 10% fetal calf serum (FCS; Invitrogen, Belgium) and 2 mM l-glutamine at 37°C and 5% CO2. Conditioned media were tested for VEGF-A concentrations by using the sandwich enzyme-linked immunosorbent assay (ELISA) (Quantikine Human VEGF-A; R&D System, Minneapolis, MN) according to the manufacturer's instructions. For conditioned media, VEGF concentrations were normalized to cell number. The sensitivity of the kit is 5 pg/mL. K562 cells, a Bcr/Abl-positive human cell line, were cultured at 500 000 cells/mL in RPMI 1640 medium supplemented with penicillin, streptomycin, l-glutamine, and 10% FCS. Imatinib mesylate (STI571) (kindly provided by Novartis, Basel, Switzerland) was added directly to RPMI medium at different concentrations from a 10-mmol/L stock solution in distilled water. For inhibition experiments, cells were incubated with the phosphatidylinositol-3 (PI3) kinase inhibitor LY294002, the MAPK/ERK kinase (MEK) inhibitor UO-126, and phorbol 12-myristate 13-acetate (PMA) (all from Calbiochem, San Diego, CA). In typical experiments, 5 μM LY294002, 5 μM UO-126, and 10 ng/mL PMA were applied for up to 24 hours.
Preparation of RNA
Total RNA was extracted by using the Trizol method following the manufacturer's instructions (Life Technologies, Gibco BRL, Grand Island, NY). The supernatant was cleared by centrifugation, precipitated with isopropanol, and resuspended in sterile water. A minimum of 10 μg RNA was used for Northern blot and analyzed as previously described.7 The entire cDNA for mouse VEGF (kindly provided by Dr Werner Risau) was used as a probe in Northern blot experiments.
VEGF promoter constructs
We used the entire VEGF promoter gene construct (-1176/+54) as well as the -88 construct. Both constructs have already been described.7 Construct PstI was obtained by cutting the -1176/+54 vector with PstI and KpnI, blunting with T4 DNA polymerase, and re-ligating. Construct SacII was obtained by cutting the -1176/+54 vector with SacII and KpnI, blunting with T4 DNA polymerase, and re-ligating. Construct ApaI was obtained by cutting the -1176/+54 vector with ApaI and KpnI, blunting with T4 DNA polymerase, and re-ligating. Construct -27/+54 was obtained by subcloning a DraI/NheI fragment within the SmaI/NheI sites of pGL2 basic. For the vector coding the Gal4/Sp1 fusion protein, a EcoRI/XhoI fragment corresponding to the entire Sp1 cDNA was subcloned within the same site in the pCMV/BD vector (Stratagene, La Jolla, CA). The vector was then digested with EcoRI, blunted with Klenow, and religated for reestablishment of the correct reading frame.
Transient transfections and luciferase assay
Four million K562 cells were resuspended in 500 μL RPMI medium and placed in a 0.4-cm gap cuvette with 20 μg total plasmid DNA. Electroporation was performed with a single electric shock (250 V, 9 ms) using the ElectroSquarePorator ECM 830 (BTX, San Diego, CA). After electroporation half of the cells were exposed or not to 2 or 5 μM imatinib. One day after transfection, the cells were washed with ice-cold phosphate-buffered saline (PBS), and the luciferase assay was performed as previously described.7 The protein concentration was measured by using the bicinchonic acid (BCA) protein assay kit (Pierce, Rockford, IL) with bovine serum albumin as standard. The luciferase activity was normalized to the amount of protein. Results are expressed as relative light units (RLUs).
Preparation of nuclear extracts and gel mobility shift assays
K562 cells were cultured in 10% FCS for 16 hours in the absence or in the presence of 2.5 μM imatinib. Cells were resuspended in lysis buffer (Tris (tris(hydroxymethyl)aminomethane) 10 mM pH 7.9, NaCl 1.5 mM, KCl 10 mM, NaF 50 mM, phosphatase inhibitors [β-glycerophosphate 40 mM, 200 mM p-nitrophenylphosphate], and protease inhibitors [aprotinin 5 μg/mL, leupeptine 5 μg/mL, phenylmethylsulfonylfluoride 0.1 mM]) and homogenized in a Dounce homogenizer. Nuclei were recovered by centrifugation at 2500g (4800 rpm) for 5 minutes and lysed in 20 mM Tris pH 7.9, 20% glycerol, 1.5 mM MgCl2, 500 mM KCl, phosphatase, and protease inhibitors. Chromatin was eliminated by centrifugation at 18 400g (14 000 rpm), and nuclear extracts were concentrated with an Amicon Ultra 10000 filter (Millipore, Billerica, Spain). Electromobility shift assays (EMSAs), using as a probe the region between -88 and -66 bp of the VEGF promoter, were performed as previously described.7 For supershift assays, 0.2 μg Sp1 (PEP-2) and Sp3 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) were used. Each antibody was added 30 minutes before the addition of the probe. Electrophoresis was performed on a 4% polyacrylamide (29 acrylamide, 1 bisacrylamide), in 0.5 × TBE (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA (ethylenediaminetetraacetic acid)). Gels were run for 4 to 5 hours at 280 to 300 V/10 to 12 mA.
Western blotting
Cells were incubated with different effectors for the times indicated in the figure legends and were washed twice with ice-cold PBS and immediately lysed in Laemmli sample buffer. Protein extracts were resolved on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto a polyvinylidene diflouride (PVDF) membrane (Immobilon-P; Millipore). Membranes were incubated with a monoclonal antiphospho p42/p44 mitogen-activated protein (MAP) kinase antibody (1:2000; Sigma, St Louis, MO) or antiphospho AKT (1:3000; Cell Signaling Technology, Beverly, MA) and a polyclonal anti-Hsp60 antibody (1:1000; Santa Cruz) and then with an antirabbit or antimouse horseradish peroxidase-conjugated secondary antibody. Bound antibody was detected by using an enhanced chemiluminescence (ECL) system (Amersham Biosciences).
Statistical analysis
For bone marrow and plasma VEGF concentrations, data are presented as medians, interquartile ranges (low quartile-high quartile [LQ-HQ]). We compared the values between groups by using nonparametric test of Kruskal-Wallis.
Results
Increased VEGF secretion by CML bone marrow cells
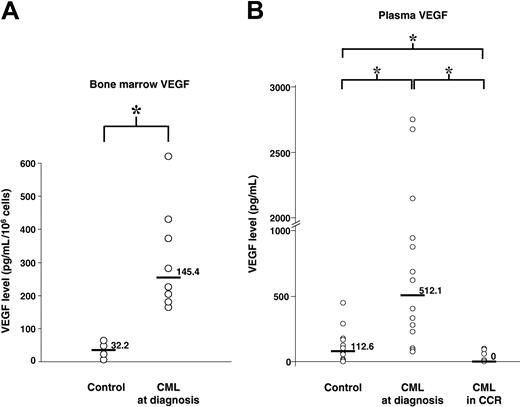
Freshly isolated bone marrow cells were incubated in RPMI medium supplemented with 10% FCS. Forty-eight hours later, conditioned medium was tested for the presence of VEGF. We found a statistically significantly higher median concentration of VEGF in CML when compared with normal bone marrow cells (P = .01) (Figure 1A). The median VEGF concentration was 145.4 pg/mL/106cells/48 hours in CML (LQ-HQ, 84.6-293.4) and 32.2 pg/mL/106/48 hours (LQ-HQ, 11.65-53.65) in normal bone marrow cells.
VEGF concentrations of patients with CML. (A) VEGF concentrations in bone marrow supernatants. The VEGF-A concentration of conditioned media was determined by ELISA and values are shown. Normal bone marrow obtained from 4 patients served as a control. Bone marrow specimens from 8 adult patients with newly diagnosed untreated CML were studied. (B) VEGF concentrations in plasma samples. Plasma samples from 14 patients with CML at diagnosis, 9 patients treated with imatinib and in complete cytogenetic response, and 10 healthy control individuals were analyzed by ELISA. Comparison between groups was performed and is represented by square brackets. Statistically significant comparisons are indicated by an asterisk (*), P < .05. Horizontal bars and numbers represent median VEGF.
VEGF concentrations of patients with CML. (A) VEGF concentrations in bone marrow supernatants. The VEGF-A concentration of conditioned media was determined by ELISA and values are shown. Normal bone marrow obtained from 4 patients served as a control. Bone marrow specimens from 8 adult patients with newly diagnosed untreated CML were studied. (B) VEGF concentrations in plasma samples. Plasma samples from 14 patients with CML at diagnosis, 9 patients treated with imatinib and in complete cytogenetic response, and 10 healthy control individuals were analyzed by ELISA. Comparison between groups was performed and is represented by square brackets. Statistically significant comparisons are indicated by an asterisk (*), P < .05. Horizontal bars and numbers represent median VEGF.
Concurrently, we measured the VEGF concentrations in plasma from 14 patients with CML at diagnosis compared with 10 healthy volunteers. We found a statistically significantly higher median concentration of VEGF in patients with CML when compared with healthy volunteers (P = .0059) (Figure 1B). The median VEGF was 512.1 pg/mL (LQ-HQ, 226.50-941.9) in CML plasma and 112.1 pg/mL (LQ-HQ, 54.45-177.18) in healthy volunteer plasma.
Decrease in plasma VEGF concentrations in patients treated with imatinib mesylate (STI571)
To analyze the effect of imatinib treatment on abnormal VEGF secretion, we compared plasma VEGF concentrations of 9 patients with CML at diagnosis and after imatinib treatment. These patients were in complete cytogenetic remission for at least 6 months. Significantly lower VEGF concentrations after treatment were observed when compared with VEGF amounts at diagnosis (P = .0002) (Figure 1B). The median VEGF concentration in patients in CCR was 0 pg/mL (LQ-HQ, 0-66.75) compared with 512.1 pg/mL at diagnosis. It is important to highlight that VEGF concentrations in the treated patients are statistically significantly lower than those of healthy volunteers (P = .0058).
Decrease in VEGF secretion in K562 cells cultured in the presence of imatinib mesylate (STI571)
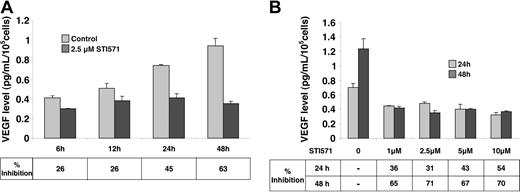
To decipher the molecular mechanisms implicated in imatinib-mediated inhibition of VEGF expression, we chose to examine the K562 Bcr/Abl-positive human cell line. The amount of VEGF protein in conditioned medium of K562 cells, cultured with 10% FCS in the presence or in the absence of 2.5 μM imatinib, was determined by ELISA (Figure 2A). In control cells, VEGF accumulated in the medium in a time-dependent manner. In contrast, in the presence of imatinib, VEGF does not accumulate in the medium (26% inhibition after 6 hours and 63% after 48 hours).
VEGF production in K562 cells treated with imatinib mesylate (STI571) in vitro. K562 cells (100 000 cells/mL) were maintained in normal medium supplemented with 10% serum for 48 hours in the absence (control) or in the presence of the indicated imatinib mesylate (STI571). VEGF concentrations in conditioned medium were measured by ELISA (median of 3 independent experiments ± SE). (A) Time course of VEGF production in K562 cells treated with imatinib. (B) VEGF production in K562 cells treated with different concentrations of imatinib during 24 or 48 hours.
VEGF production in K562 cells treated with imatinib mesylate (STI571) in vitro. K562 cells (100 000 cells/mL) were maintained in normal medium supplemented with 10% serum for 48 hours in the absence (control) or in the presence of the indicated imatinib mesylate (STI571). VEGF concentrations in conditioned medium were measured by ELISA (median of 3 independent experiments ± SE). (A) Time course of VEGF production in K562 cells treated with imatinib. (B) VEGF production in K562 cells treated with different concentrations of imatinib during 24 or 48 hours.
We then investigated the amount of VEGF in conditioned medium of K562 cells cultured with increasing concentrations of imatinib in the presence of 10% FCS (Figure 2B). Whatever the concentration (from 1 μM to 10 μM), imatinib inhibited VEGF accumulation by approximately 54% after 24 hours and by 70% after 48 hours. Analysis of [3H] leucine incorporation shows that protein synthesis is not affected for concentrations of imatinib varying from 1 to 10 μM (data not shown). This result is probably due to the maintenance of 10% serum in our culture conditions and is different from the conditions described by Ebos et al8 who performed experiments after serum removal. As a control, we performed experiments with HL60, a promyelocytic leukemia cell line known to secrete VEGF.9 We were not able to show any difference in VEGF secretion without or with imatinib mesylate (data not shown).
Inhibition of VEGF production by imatinib mesylate (STI571) is a consequence of inhibition of both the p42/p44 MAP kinase and PI3 kinase pathways
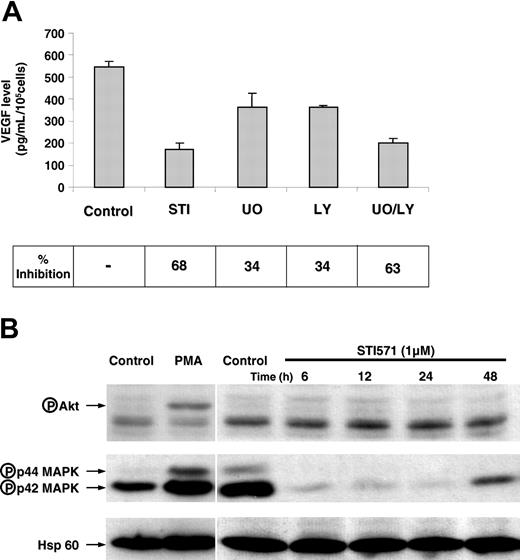
Our goal was to identify the signaling pathway(s) targeted by imatinib. To this aim, we examined VEGF concentration in the K562-conditioned medium after treatment of cells with either imatinib mesylate (STI571) or an inhibitor of p42/p44 MAP kinase pathway (UO-126) and/or an inhibitor of PI3 kinase pathway (LY294002). Figure 3A shows a 34% inhibition of VEGF secretion in the presence of U0-126 or in the presence of LY294002 and a 63% inhibition with both agents. Thus, there is an additive effect of UO-126 and LY294002 on VEGF production. To further analyze the effect of imatinib on both pathways, we determined the activity of p42/p44 MAP kinase and AKT by Western blotting by using antibodies directed against the phosphorylated form of these kinases. Figure 3B shows a high basal p42/p44 MAP kinase activity, strongly inhibited by imatinib under conditions in which the Bcr/Abl-phosphorylated protein was no longer detected (data not shown). Only a few minutes of treatment with the drug10 were required to observe inhibition. The use of phospho-specific AKT antibodies (against serine 473) has shown that the basal AKT activity was low, and, even if AKT can be stimulated by PMA, the evaluation of a modest down-regulation obtained by using such a tool is quite difficult. As we are very confident in the VEGF ELISA tests, we suspect that the phospho-AKT antibodies are not sensitive enough to detect low modifications in the AKT basal concentrations. Moreover, a recent publication describes inhibition of the AKT activity in K562 cells.11 These results suggest that imatinib-induced VEGF down-regulation is a consequence of inhibition of both p42/p44 MAP kinase and PI3 kinase pathways.
Both the p42/p44 MAP kinase and the PI3 kinase pathways are implicated in VEGF production by K562 cells. (A) K562 cells (100 000 cells/mL) were maintained in 10% serum containing medium for 24 hours in the absence (control) or in the presence of 2 μM imatinib mesylate (STI571), 5 μM UO126, and/or 5 μM LY204002. VEGF-A production in conditioned medium following 24 hours of treatment was tested by ELISA. These results are representative of 2 independent experiments performed in triplicate ± SE. (B) K562 cells were incubated for different times with 1 μM imatinib. Cells were lysed and proteins were separated by electrophoresis on 10% polyacrylamide gels. Proteins were then blotted onto a PVDF membrane and tested for the presence of either antiphospho p42/p44 MAP kinases, antiphospho AKT, or anti-Hsp-60. A positive control for AKT activation by a PMA treatment is also shown. Arrows indicate the position of phosphorylated proteins and the loading control (Hsp-60).
Both the p42/p44 MAP kinase and the PI3 kinase pathways are implicated in VEGF production by K562 cells. (A) K562 cells (100 000 cells/mL) were maintained in 10% serum containing medium for 24 hours in the absence (control) or in the presence of 2 μM imatinib mesylate (STI571), 5 μM UO126, and/or 5 μM LY204002. VEGF-A production in conditioned medium following 24 hours of treatment was tested by ELISA. These results are representative of 2 independent experiments performed in triplicate ± SE. (B) K562 cells were incubated for different times with 1 μM imatinib. Cells were lysed and proteins were separated by electrophoresis on 10% polyacrylamide gels. Proteins were then blotted onto a PVDF membrane and tested for the presence of either antiphospho p42/p44 MAP kinases, antiphospho AKT, or anti-Hsp-60. A positive control for AKT activation by a PMA treatment is also shown. Arrows indicate the position of phosphorylated proteins and the loading control (Hsp-60).
Imatinib mesylate (STI571) inhibits VEGF mRNA expression in K562 cells
To determine at which concentration of VEGF expression imatinib exerts its effect, we next analyzed the VEGF mRNA concentrations in K562 cells cultured in 10% FCS treated or not with imatinib (Figure 4). For this purpose, K562 cells were cultured in the absence or in the presence of imatinib at 2.5 μM for different times (Figure 4A). In the control, VEGF mRNAs accumulate to reach a maximum after 48 hours of culture. In contrast, as shown for the VEGF protein in the conditioned medium, imatinib prevents VEGF mRNAs accumulation. To determine the dose of imatinib required to down-regulate VEGF mRNA expression, we examined increasing concentrations of imatinib on the mRNA concentration. Figure 4B shows an already total inhibition at 2.5 μM imatinib. As for VEGF secretion, we did not find an inhibition in VEGF mRNA expression in the presence of imatinib in HL60 cells (data not shown).
mRNA expression in K 562 cells treated by imatinib mesylate (STI571) in vitro. K562 cells (100 000 cells/mL) were maintained in 10% serum containing medium for 48 hours in the absence (control) or in the presence of the indicated imatinib concentrations. Total RNA (10 μg) isolated for the indicated periods of time were analyzed by Northern blotting. Ribosomal RNAs are shown as a loading control. A representative of 3 independent experiments is shown. (A) Time course of VEGF mRNA concentrations in K562 cells treated with or without 2.5 μM imatinib. (B) VEGF mRNA in K562 cells treated with different concentrations of imatinib for 48 hours.
mRNA expression in K 562 cells treated by imatinib mesylate (STI571) in vitro. K562 cells (100 000 cells/mL) were maintained in 10% serum containing medium for 48 hours in the absence (control) or in the presence of the indicated imatinib concentrations. Total RNA (10 μg) isolated for the indicated periods of time were analyzed by Northern blotting. Ribosomal RNAs are shown as a loading control. A representative of 3 independent experiments is shown. (A) Time course of VEGF mRNA concentrations in K562 cells treated with or without 2.5 μM imatinib. (B) VEGF mRNA in K562 cells treated with different concentrations of imatinib for 48 hours.
Imatinib mesylate (STI571) inhibits the transcriptional activity of the VEGF promoter
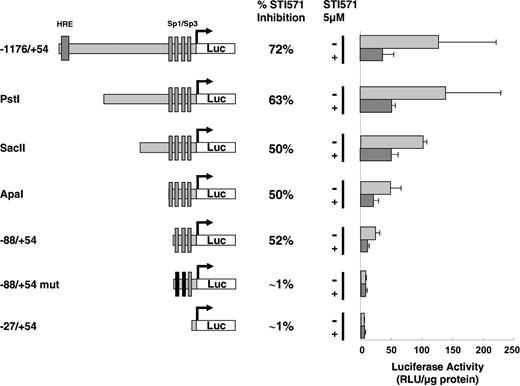
Because VEGF expression is tightly regulated at the transcriptional level by way of the p42/p44 MAP kinase and PI3 kinase pathways,4,7,12-20 we investigated the action of imatinib on the VEGF promoter. For this purpose, the entire VEGF promoter (-1176/+54) coupled to the luciferase reporter gene was used as previously reported.7 The relative VEGF promoter activity in K562 cells is strong and comparable to that determined in Ras Val12 transformed cells.7 Such activity is compatible with high VEGF mRNA and protein levels in both cell lines (data not shown). Figure 5 shows that imatinib strongly inhibits VEGF promoter activity following 24 hours of treatment (72% inhibition). To define the region of the VEGF promoter targeted by imatinib, different domains of the VEGF promoter were coupled to the luciferase reporter gene and tested for their capacity to respond to imatinib treatment (Figure 5). Successive deletions of the promoter lead to a progressive decrease in the luciferase activity to reach a minimum with the -88/+54 promoter (Figure 5). Further deletion totally abrogates VEGF promoter activity as already described7 (Figure 5). These results demonstrate that VEGF-dependent transcriptional activity depends on different regulatory sequences within the promoter. Interestingly, imatinib inhibits VEGF promoter activity by 52% even when we used the shortest -88/+54 construct (Figure 5). This activity is totally abrogated with the -88/+54 mut construct in which Sp1 binding sites are mutated. Hence, we can conclude that imatinib inhibits transcription principally by way of the proximal region (-88/+54) of the VEGF promoter.
VEGF promoter activity in K562 cells treated with imatinib mesylate (STI571). The different VEGF promoter constructs used in transient transfection assays are shown. The relative light units (RLU/μg protein) was measured 24 hours after transfection of 20 μg of the different reporter genes in the absence (-) or in the presence (+) of 5 μM imatinib is shown. The results are representative of 4 independent experiments performed in duplicate ± SE. The percentage of inhibition in the presence of imatinib is indicated.
VEGF promoter activity in K562 cells treated with imatinib mesylate (STI571). The different VEGF promoter constructs used in transient transfection assays are shown. The relative light units (RLU/μg protein) was measured 24 hours after transfection of 20 μg of the different reporter genes in the absence (-) or in the presence (+) of 5 μM imatinib is shown. The results are representative of 4 independent experiments performed in duplicate ± SE. The percentage of inhibition in the presence of imatinib is indicated.
Imatinib mesylate-dependent VEGF promoter inhibition targets Sp transcription factors
We and others have shown previously that the -88/-66 domain, which is highly GC rich, binds Sp1 as well as Sp3 transcription factors in response to growth factor stimulation, oncogenic transformation by Ras, or free radicals.21 Increased DNA binding activity as well as the transactivation capacity of Sp1 is at least mediated by direct phosphorylation of Sp1 by p42/p44 MAP kinases.20 As imatinib strongly inhibits the p42/p44 MAP kinase pathway (Figure 3), we have evaluated whether imatinib decreases the Sp1 and/or Sp3 DNA binding activity. For this purpose, we performed EMSA experiments by using the -88/-66 bp region of the VEGF promoter as a probe. Figure 6A shows 4 constitutive DNA binding complexes (a, B, c, D) with nuclear extracts from exponentially growing cells. Such complexes have already been described for nuclear extracts of fibroblastic cells stimulated with growth factors or following stimulation of the p42/p44 MAP kinase pathway.7,20 However, when extracts from imatinib-treated cells are used, a strong decrease in all 4 complexes is observed. Sp1 antibodies supershifted part of complex B (B2), and Sp3 antibodies supershifted part of complex B (B3) and complex c. In addition, the intensity of the supershifted complexes is highly reduced in the presence of imatinib, demonstrating that both Sp1 and Sp3 have a lower affinity for the probe. To test the transactivation capacity of Sp1, the major factor implicated in transactivation of the proximal region of the VEGF promoter, we used chimeric constructs in which Sp1 was fused to the DNA binding domain of the Gal4 transcription factor. The effect of imatinib was evaluated after cotransfection with a construct encoding a Gal4-dependent luciferase reporter gene. As shown in Figure 6B, imatinib treatment inhibits transcriptional activation of the Sp1/Gal4 fusion protein. Taken together, our experiments demonstrate that imatinib inhibits VEGF transcription by targeting Sp1 and Sp3 DNA binding activity and the Sp1 transactivation capacity.
STI571 inhibits Sp transcription factors binding and transactivation capacities. (A) EMSAs with nuclear extracts of untreated (-) or STI571 treated (+) K562 cells in the absence (Ctrl) or the presence of 0.2 μg Sp1- or Sp3-specific antibodies. Formation of specific complexes is indicated on the left (a, B, c, and D) according to the nomenclature already described.7 In the presence of the Sp1 or Sp3 antibodies, migration of specific complexes is retarded as indicated in the text, demonstrating that the complex contains Sp1 or Sp3. The position of supershifted complexes is indicated by black ovals. *Positions of the complexes that have disappeared or the intensity of which has been reduced in the presence of the antibodies. Note that the intensity of supershifted complexes decreased in the presence of STI571. (B) Relative luciferase activity measured 24 hours after the transfection of 20 μg control empty vector or Gal4/Sp1 in the absence (-) or in the presence (+) of 5 μM STI571. These results are representative of 4 independent experiments performed in duplicate ± SE.
STI571 inhibits Sp transcription factors binding and transactivation capacities. (A) EMSAs with nuclear extracts of untreated (-) or STI571 treated (+) K562 cells in the absence (Ctrl) or the presence of 0.2 μg Sp1- or Sp3-specific antibodies. Formation of specific complexes is indicated on the left (a, B, c, and D) according to the nomenclature already described.7 In the presence of the Sp1 or Sp3 antibodies, migration of specific complexes is retarded as indicated in the text, demonstrating that the complex contains Sp1 or Sp3. The position of supershifted complexes is indicated by black ovals. *Positions of the complexes that have disappeared or the intensity of which has been reduced in the presence of the antibodies. Note that the intensity of supershifted complexes decreased in the presence of STI571. (B) Relative luciferase activity measured 24 hours after the transfection of 20 μg control empty vector or Gal4/Sp1 in the absence (-) or in the presence (+) of 5 μM STI571. These results are representative of 4 independent experiments performed in duplicate ± SE.
Discussion
We have confirmed 2 previous reports2,6 that describe the capacity of bone marrow CML cells to secrete VEGF. However, the approaches we have used are different. Verstovsek et al6 used RIA to quantify the VEGF protein from total bone marrow extracts. Hence, VEGF values could originate from stromal cells. Ratajczak et al5 measured VEGF concentrations in cells following stimulation during 6 days with recombinant human growth factors to differentiate CD34 cells into GM-CFUs. This method of differentiation could induce growth factor-dependent VEGF secretion independently of the transformed status of the cells. Using our culture conditions we tested the spontaneous secretion of VEGF by CML cells in comparison to normal bone marrow cells. We showed that CML cells secreted statistically significantly more VEGF than normal bone marrow cells. Hence, VEGF concentrations in plasma of patients with CML are 4.6-fold higher than in control subjects. We then evaluated the plasma VEGF concentrations of patients treated with imatinib. All patients tested were in CCR, and their blood counts were normal with no cytopenia. We found decreased plasma VEGF concentrations in treated patients compared with concentrations reached at diagnosis. Hence, low plasma VEGF concentrations could be one of the characteristics of CCR. A previous report showed that Bcr/Abl induces VEGF expression.4 So, the decreased VEGF concentration observed with imatinib could be related to its Bcr/Abl tyrosine kinase inhibitory effect. However, if the decrease in VEGF was only related to a decrease in the number of CML cells, we should have obtained identical VEGF concentrations in patients in CCR and in healthy volunteers. In contrast, our data demonstrated that VEGF concentrations were lower than those of healthy volunteers. For 6 of 9 patients VEGF concentrations in patients were indeed under the ELISA minimum detectable amount (< 5 pg/mL). In addition to Bcr/Abl tyrosine kinase activity, imatinib also blocks platelet-derived growth factor (PDGF) and c-kit receptors normally expressed in stromal and immature hematopoietic stem cells, respectively.22 We hypothesize that imatinib could also act on the capacity of stromal cells and normal hematopoietic stem cells to secrete VEGF. Interestingly, the VEGF decrease with imatinib could be related to both Bcr/Abl, c-kit, and PDGF receptor tyrosine kinase inhibition. To our knowledge there is no study on the correlation between c-kit and PDGF receptor and VEGF. However, stimulation of NIH 3T3 cells with PDGF enhances VEGF gene expression.23 This point has to be investigated more carefully.
Acute leukemia is the natural evolution of CML. The 9 patients with CCR studied were not in molecular remission. Recent data showed that VEGF promotes the survival of leukemia cells from apoptosis by way of stimulation of the VEGF receptor-2 (VEGFR2).24 We could expect that a decrease in VEGF, by blocking clonal selection, may prevent the leukemic transformation of CML through the persistence of Bcr/Abl. Further clinical investigations will be necessary to define the effect on prognosis of the VEGF concentration, in particular on the evolution of the disease. Another unfavorable evolution of treated CML is the occurrence of imatinib resistance. Several mechanisms have been reported, including an adenosine 5′-triphosphate (ATP) binding site mutation and Lyn overexpression.25-28 For patients resistant to imatinib, determination of the VEGF concentration should be informative in evaluating the mechanism of resistance. In the case of mutations of the Bcr/Abl kinase domain, we expected a high concentration of VEGF. However, this might not be the case when Lyn overexpression is at the heart of resistance.
Beyond its effect on leukemic cell survival, VEGF is an interesting cytokine because of new therapeutic possibilities. Two groups of molecules are now in clinical trials: monoclonal antibodies against VEGF and inhibitors of VEGF-R2 tyrosine kinase.29 Recent data show that CML cells express VEGF-R2.30 Indeed, these new treatments could be proposed in association with imatinib, especially in the case of resistance to the drug.
The second part of our study focused on the understanding of the mechanisms underlying the effect of imatinib on VEGF production. The K562 cell line, which is Bcr/Abl positive, is interesting because of its human origin, its VEGF production, and the absence of c-kit (data not shown). As expected, the percentage of inhibition following imatinib treatment was comparable to that determined for bone marrow cells obtained from patients. The action of imatinib mesylate (STI571) on VEGF production is specific for Bcr/Abl-positive cell line because there is no inhibition of VEGF secretion in HL60 cells, a Bcr/Abl-negative cell line. We demonstrated that imatinib inhibits VEGF transcription by targeting the Sp1 and Sp3 DNA binding activity and the Sp1 transactivation capacity. We have previously shown that p42/p44 MAP kinase directly phosphorylates Sp1.20 Such phosphorylation is implicated in Sp1 DNA binding and transactivation capacity. Because imatinib abrogates the p42/p44 MAP kinase activity, it prevents Sp1 phosphorylation, and, as a consequence, the recruitment of RNA polymerase and accessory factors for maximal transcription. EMSA experiments also show that Sp3 DNA binding is strongly inhibited on imatinib treatment. We are in the process of determining whether Sp3 is also directly phosphorylated by p42/p44 MAP kinase as it contains consensus phosphorylation sites at the same position as Sp1. The fact that Sp3 is a highly phosphorylated protein is in accordance with this hypothesis.31 Recently, Mayerhofer et al4 described increased VEGF transcriptional activity in Ba/F3 cells in which Bcr/Abl is conditionally induced by doxycycline. In this cell system, the hypoxia-inducible factor 1α (HIF-1α) and PI3 kinase pathways appear to be implicated, whereas the p42/p44 MAP kinase pathway is not. We tested the implication of both PI3 kinase and p42/p44 MAP kinase pathways on VEGF production in K562 cells. Our results show the implication of the PI3 kinase pathway that is compatible with the results of Mayerhofer et al,4 but we also show a real implication of the p42/p44 MAP kinase pathway. These results are in accordance with the results of Jacquel et al10 who showed that imatinib inhibits the p42/p44 MAP kinases. Moreover, the luciferase activity used by Mayerhofer et al4 to analyze the VEGF promoter in the presence of constitutive active members of the p42/p44 MAP kinases pathway was normalized with a CMVβgal vector. As the cytomegalovirus (CMV) promoter is strongly activated by the p42/p44 MAP kinases pathway, the luciferase values could be underestimated. In our study, the PstI construct was less sensitive to imatinib. The lack of the HIF-1 binding site (HRE), which has been deleted in this construct, could explain this result. However, we did not detect constitutive HIF-1α protein expression in K562 cells (data not shown). In this construct, a signal transducer and activator of transcription 3 (STAT-3) binding site, already shown to be implicated in VEGF gene transcription,32 was also deleted. It would be interesting to test whether imatinib could act on STAT-3 binding and transactivation capacities.
In conclusion, this study presents 2 complementary interests. First is a clinical aspect, which demonstrates, for the first time, the decrease in VEGF production in patients with CML treated with imatinib. It points out the potential prognostic value of determining the VEGF plasma concentration of patients to follow the evolution of this hematologic malignancy. Moreover, further investigations with a larger number of patients will be necessary to evaluate the potential of determining VEGF concentrations in the monitoring of patients with CML treated with imatinib, especially when patients became resistant. Second is a fundamental aspect, which allows the elucidation of the molecular mechanisms underlying the imatinib-mediated decrease of VEGF plasma concentration.
Prepublished online as Blood First Edition Paper, February 19, 2004; DOI 10.1182/blood-2003-08-2695.
Supported by grants from the CNRS, UNSA, the Association pour la Recherche Contre le Cancer, the Fondation de France, the Ligue Nationale Contre le Cancer, and the Groupe d'Etude et de Recherche Clinique, Biologique, et Thérapeutique (GERCBT).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Jean-Michel Karsenti for assistance with sternal punctures and Pr Alain Pesce. We thank Dr Patrick Berthaud and Novartis Pharma (Rueil-Malmaison, France) for supporting this work. We thank Dr Anne-Odile Hueber and Dr Christiane Brahimi-Horn for critical reading of the manuscript. We thank Dr Christian Pradier and Dr Laurent Letrilliart for statistical analysis. We thank Dr Patrick Philip and Dr Sylvia Benzaken for technical assistance.