Abstract
The expression of tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL) and TRAIL receptors was investigated in resting and cytokine-activated purified primary human natural killer (NK) and CD8+ T cells. Resting NK and CD8+ T cells expressed the mRNA for all TRAIL receptors, but TRAIL-R4 was the only receptor clearly detectable on the surface of both cell types. NK cells were activated by interleukin 2 (IL-2) or IL-15, whereas CD8+ T cells were activated by phytohemagglutinin (PHA) + IL-2 followed by IL-2 alone for up to 10 days. On activation, both cell types rapidly expressed TRAIL-R2 and TRAIL-R3, whose expression peaked at day 10 of culture. TRAIL-R1, however, was never expressed at any time point examined, whereas the expression of TRAIL-R4, which showed a progressive increase in CD8+ T cells, remained constant in NK cells. Notwithstanding the expression of TRAIL-R2, recombinant TRAIL did not show any cytotoxic activity on either NK or CD8+ T cells. Both resting and activated NK and CD8+ T cells were found to express high levels of the 2 isoforms of c-FLIP (cellular Fas-associated death domain protein [FADD]–like IL-1–converting enzyme [FLICE]–inhibitory protein). Small interference RNA-mediated inhibition of c-FLIP expression in NK cells abrogated their resistance to the apoptotic effect of soluble TRAIL. Thus, once activated the major cytotoxic effector cells are potentially sensitive to TRAIL but are physiologically protected from its apoptotic action by intracellular level of c-FLIP.
Introduction
Natural killer cells provide a line of defense against autologous tumors and virus-infected cells that have down-modulated their major histocompatibility complex (MHC) class I expression. Such tumors can evade the MHC class I–restricted immune recognition and cytotoxic response mediated by CD8+ cytotoxic T lymphocytes (CTLs) but are not able to trigger the inhibitory receptors present on natural killer (NK) cells and necessary to prevent their activation. NK cells can use 2 different mechanisms to kill their targets, either by cytotoxic granule exocytosis or by induction of death receptor–mediated apoptosis. The secretory pathway is active against relatively rare leukemia cell lines, whereas the nonsecretory killing pathway is active against a variety of tumor cell lines. Death ligands belong to the tumor necrosis factor (TNF) family of ligands, a large family of pleiotropic, membrane-bound, and soluble molecules, that include TNF-α, FasL, CD40L, lymphotoxinα (LTα) and LTβ, receptor activator of nuclear factor-kappa B ligand (RANKL), and TNF-related apoptosis-inducing ligand (TRAIL). A family of complementary molecules, TNF family receptors, has been identified and described (reviewed in LeBlanc and Ashkenazi1 ). Most of the death ligands are also known in soluble forms that can be secreted.
Primary human NK cells express several TNF family death ligands2 and use them to induce apoptosis in various tumor cells.3-5 Moreover, expression studies in mice have demonstrated the relevant role played by TRAIL expressed by tissue-resident NK cells in the control of metastatic growth.6 However, activated NK cells have been shown to express some of the death receptors, such as CD95,7,8 that have been implicated in the negative control of NK cell function. Moreover, it has recently been reported that CD8+ NK cells undergo apoptosis by triggering of CD8 with its natural ligands through a Fas-mediated pathway.9 Interestingly, members of the NK inhibitory receptor superfamily inhibit this apoptosis, acting, therefore, as survival receptors for NK cells. Although TRAIL is known to induce apoptosis in several cancer cell lines and primary tumors, it has been reported to play, similarly to Fas/FasL,10 an active inhibitory role in the physiologic control of erythropoiesis11 and in the thymic deletion induced by T-cell receptor ligation.12 However, contrasting data have been reported by Simon et al13 that did not demonstrate a role for TRAIL in antigen-induced deletion of thymocytes.
Given the potential use of TRAIL in anticancer therapy,14 here, we investigated the expression of TRAIL and TRAIL receptors in resting and activated primary human NK and CTL cells, the major players of anticancer immunity. Moreover, we investigated the functional effects of the addition of recombinant TRAIL at different culture times on the survival of both resting and activated NK and CD8+ CTLs. These biologic effects were correlated with the expression level of c-FLIP (cellular Fas-associated death domain protein [FADD]–like interleukin 1-converting enzyme [FLICE]–inhibitory protein), a major regulator of cell death induced by TRAIL and other death ligands.15
Materials and methods
Reagents
Monoclonal antibodies (MoAbs) anti-CD3 and anti-CD16 were purchased from Becton Dickinson (San Jose, CA). Antihuman TRAIL receptors and anti–human TRAIL monoclonal antibodies were purchased from Alexis Biochemical (San Diego, CA). Goat anti–mouse immunoglobulin G (IgG) phycoerythrin (PE)–labeled was purchased from Immunotech (Beckman Coulter, Miami, FL). For double staining anti–human CD8 PE-labeled and anti–human CD4 fluorescein isothiocyanate (FITC)–labeled, anti–human CD3 FITC-labeled and antihuman CD16 and CD56 PE-labeled, anti–human CD3 PE-labeled and anti–human CD19 FITC-labeled monoclonal antibodies were purchased from Beckman Coulter. Primary cultures of CD8+ T cells were stimulated with phytohemagglutinin (PHA-M; Sigma Chemical, St Louis, MO) and grown in RPMI 1640 medium (BioWhittaker, Walkersville, MD) plus 10% fetal calf serum (FCS; BioWittaker) with human recombinant interleukin 2 (rIL-2) from Genzyme (Boston, MA). Recombinant interleukin-15 (rIL-15) was purchased from Genzyme.
Recombinant Histinine6-tagged (His) TRAIL was produced in bacteria and purified by chromatography on Ni++ affinity resin. The absence of endotoxin contamination in recombinant TRAIL preparation was assessed as previously described.16,17 TRAIL-R1-Fc and TRAIL-R2-Fc chimera cocktail was purchased from R&D (Minneapolis, MN).
NK and CD8+ T-cell purification
Primary NK and CD8+ T cells were isolated from peripheral blood lymphocytes (PBLs) of healthy voluntary donors by immunomagnetic negative selection by using the NK cell isolation kit and the CD8+ T-cell isolation kit (Milteny Biotech, Gladbach, Germany), according to the manufacturer's protocol. Briefly, PBLs were separated from buffy coats by Ficoll-Hypaque gradient centrifugation, then magnetically labeled with a cocktail of hapten-conjugated anti-CD3, anti-CD14, anti-CD19, anti-CD36, and anti-IgE antibodies for NK purification; anti-CD4, anti-CD11b, anti-CD16, anti-CD19, anti-CD36, and anti-CD56 antibodies for CD8 T-cell isolation, followed by antihapten antibody-coupled magnetic cell sorter (MACS) microbeads. The magnetically labeled cells were then immobilized on a depletion column in the magnetic field of a Vario-MACS apparatus (Milteny Biotech). Purity of NK cells was immediately checked by anti-CD16–PE and anti-CD3–FITC labeling, whereas purity of CD8 T cells was monitored by anti-CD8–PE and anti-CD4–FITC, anti-CD3–PE, and anti-CD19–FITC. Only samples with a purity exceeding 95% were used.
Cell cultures and treatment
Primary human NK cells were cultured up to 15 days in RPMI medium supplemented with 10% FCS and 100 U/mL human recombinant rIL-2 at an optimal cell density of 1 × 106 cells/mL. In some experiments NK cells were cultured with rIL-15 (10 ng/mL) at the same cell density. Primary human CD8+ T cells were cultured in RPMI 1640 medium supplemented with 10% FCS, 5 μg/mL purified PHA-M plus 20 U/mL human rIL-2. After 3 days of cultures, cells were washed twice and seeded again in complete medium plus 20 U/mL human rIL-2 alone, which was re-added every 4 days, as previously described,16,18 up to 15 days.
Human natural killer cell line NK 3.3 was routinely cultured in RPMI 1640 medium supplemented with 15% FCS and 100 U/mL human recombinant IL-2 at an optimal cell density of 1 × 106 cells/mL. HL-60 promyelocytic and Jurkat T human cell lines were grown in RPMI 1640 medium plus 10% FCS at a density of 1 × 106 cells/mL. Human adenocarcinoma MCF-7 cells were grown adherently and maintained in RPMI 1640 medium containing 10% FCS. Cells were passaged 2 or 3 times weekly at a ratio between 1:5 and 1:10.
In cytotoxicity experiments, 1 μg/mL recombinant TRAIL was added overnight to primary cell cultures at day 1 or day 10 of activation, and to HL-60 and Jurkat cells as controls.
For neutralization experiments, the supernatant of cells treated with recombinant TRAIL was preincubated with TRAIL-R1-Fc and TRAIL-R2-Fc chimera cocktail according to the supplier's instructions (R&D), as previously reported.17
Flow cytometric analysis
Aliquots of 0.5 × 106 cells/experimental point were single- or multiple-labeled by a panel of anti–TRAIL-Rs monoclonal antibodies (MoAbs), as described previously.16 Expression of TRAIL-R1, TRAIL-R2, TRAIL-R3, and TRAIL-R4 was analyzed by indirect staining by using 1 μg HS101 anti–human TRAIL-R1, HS201 anti–human TRAIL-R2, HS301 anti–human TRAIL-R3, and HS401 anti–human TRAIL-R4 monoclonal antibody, followed by PE-labeled goat anti–mouse IgG. Surface expression of TRAIL was analyzed by indirect staining with use of 1 μg 5D5 anti–human TRAIL (Alexis Biochemicals), followed by PE-labeled goat anti–mouse IgG. Aspecific fluorescence was assessed by using isotype-matched irrelevant mAb followed by the same secondary reagent. Analysis was performed by an Epics XL flow cytometer (Beckman Coulter) and the Expo ADC software (Beckman Coulter). Data collected from 10 000 cells are reported as either percentage of positive cells or mean fluorescence intensity (MFI) values.
Reverse transcription–polymerase chain reaction (RT-PCR)
Total RNA was isolated from each cell sample (1 × 106 cells) by using the High pure RNA isolation kit (Roche Diagnostics S.p.A, Monza, Italy). cDNA was synthesized from 50 ng total RNA by using 50 U MuLV Reverse Transcriptase (Roche) and random hexamers (2.5 μM) in a 20-μL volume. RT reaction was performed at 42° C for 60 minutes followed by 5 minutes at 95° C and 5 minutes at 4° C. PCR was performed with 20 μL cDNA in a total volume of 100 μL, using AmpliTaq DNA Polymerase (Roche) according to manufacturer's instruction. FLIPLong and FLIPShort mRNAs were detected by FLIP primers 5′-GGA GGC TTA TGT CTG CTG AAG TCA TC and 3′-GGG GAA TTC CTT CTG ATT CCT GAA TGG. In particular (1) FLIPLong forward primer 5′-CGA GGC AAG ATA AGC AAG GA-3′ and reverse primer 5′-TGA CTG GTT CTT GTT GAG CG-3′, (2) FLIPShort forward primer 5′-TCA GGA ACC CTC ACC TTG TT-3′ and reverse primer 5′-ATC AGG ACAATG GGC ATA GG-3′ were used to discriminate the long and the short form of FLIP mRNA.19
The FLIPLong (327 bp) and FLIPShort (386 bp) products were amplified in PCR buffer (500 mM KCl, 100 mM Tris (tris(hydroxymethyl)aminomethane)–HCl, pH 8.3) with 2 mM MgCl2 for 40 cycles. Optimal PCR thermal profile was 1 minute at 94° C, 1 minute at 58° C followed by 30 seconds at 72° C. The PCR products were separated by electrophoresis in a 3% agarose gel and stained in ethidium bromide for 20 minutes before visualization under UV light.
TRAIL receptor mRNA transcripts were amplified by RT-PCR with the following specific primers: 5′-CTG AGC AAC GCA GAC TCG CTG TCC AC-3′ and 5′-TCC AAG GAC ACG GCA GAG CCT GTG CCA T-3′ for TRAIL-R1, 5′-GCC TCA TGG ACAATG AGA TAAAGG TGG CT-3′ and 5′-CCA AAT CTC AAA GTA CGC ACA AAC GG-3′ for TRAIL-R2, 5′-GAA GAA TTT GGT GCC AAT GCCACT G-3′ and 5′-CTC TTG GAC TTG GCT GGG AGA TGT G-3′ for TRAIL-R3, and 5′-CTT TTC CGG CGG CGT TCATGT CCT TC-3′ and 5′-GTT TCT TCC AGG CTG CTT CCC TTT GTA G-3′ for TRAIL-R4. As control, we amplified β-actin mRNA with the following primers: 5′-TGA CGG GGT CAC CCA CAC TGT GCC CAT CTA-3′ and 5′-CTA GAA GCA TTT GCG GTG GAC GAT GGA GGG-3′. Resultant PCR products were resolved in 2% agarose gels and visualized by ethidium bromide under UV light.17,18
Semiquantitative RT-PCR
As described in “Reverse transcription–polymerase chain reaction (RT-PCR),” 1 μg total RNA was reverse transcribed, and progressive dilutions (1/8, 1/20, 1/100, 1/500, 1/2500, 1/12.500) were subjected to PCR amplification to generate β-actin, TRAIL-R1, TRAIL-R2, TRAIL-R3, TRAIL-R4, FLIPLong, and FLIPShort specific sequences.
Small interference RNA
Double-strand siRNAs (dsRNA) were designed to target sequences corresponding to nt 472 to 492 (siRNA-F1) and 908 to 928 (siRNA-F2) of human c-FLIP mRNA (U97074). The target sequences, which have been designed starting from siRNA sequences previously developed by Siegmund at al,20 were screened against all known human genes by using a BLAST (Basic Local Alignment Search Tool) search to confirm that only human c-FLIP mRNA would be targeted. The respective sense and antisense RNA sequences were synthesized by Silencer siRNA Construction Kit (Ambion, Austin, TX). To achieve an efficient transfection of human primary NK cells, siRNA-F1 and siRNA-F2 (300 nM each) were delivered using the Amaxa nucleofection technology (Amaxa, Koeln, Germany) according to manufacturer's protocols. Nucleofection efficiency was assayed by transfecting a green fluorescence protein (GFP) expression vector, obtaining a number of GFP+PI– (propidium iodide) cells more than 20% of total primary T cultures.
Western blot
Cultured cells were counted, and 5 × 106 cells were collected at specific time points, washed in PBS, and centrifuged at 200g for 10 minutes. Pellets were resuspended in RIPA buffer supplemented with fresh protease inhibitors and protein concentration determined by using Bradford assay (Pierce, Rockford, IL). Proteins (60 μg) from each sample were then migrated in 12% sodium dodecyl sulfate (SDS)–acrylamide gels and blotted onto nitrocellulose filters. Blotted filters were blocked for 60 minutes in a suspension of 3% dried skimmed milk and 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS). Filters were then incubated overnight at 4° C with rabbit 1:1000 polyclonal anti-FLIP antibody (Exalpha Biologicals, Boston, MA) directed against the N-terminal portion of FLIP (55 KDa) in a suspension of 3% dried skimmed milk in PBS–0.05% Tween 20. Filters were washed and further incubated for 1.5 hours at room temperature with 1:10 000 peroxidase-conjugated anti–rabbit IgG (Pierce, Rockford, IL) in a suspension of 3% dried skimmed milk in PBS–0.05% Tween 20 at room temperature. Specific reactions were revealed with the enhanced chemiluminescence (ECL) Supersignal West Pico Chemiluminescent Substrate detection system (Pierce).
Assessment of apoptosis
Recombinant human TRAIL (1 μg) was added overnight to primary cell cultures at day 1 or day 10 of activation and to TRAIL-sensitive HL-60 and Jurkat cells as controls.
Viability was then assessed by trypan blue exclusion. Apoptosis was evaluated by PI staining of permeabilized cells and by staining extracellular phosphatidylserine with Annexin V in nonpermeabilized cells. Briefly, samples containing 0.5 × 106 cells were harvested by centrifugation at 200g for 10 minutes at 4° C, fixed, permeabilized with cold 70% ethanol for at least 1 hour at 4° C, and treated as previously described.21 Cells were then stained with 40 μg/mL PI in the presence of RNAse; analysis of PI fluorescence was performed by an Epics XL flow cytometer (Beckman Coulter) and the Expo ADC software (Beckman Coulter). For quantitative evaluation of apoptosis, the subdiploid (< 2n) DNA content was calculated as described21 and expressed as percentage of apoptotic versus nonapoptotic cells, regardless of the specific cell cycle phase.
Staining of extracellular phosphatidylserine in nonpermeabilized cells was performed by FITC conjugate Annexin V (ACTIPLATE; Valter Occhiena, Torino, Italy) in Ca2+ and PI staining buffer. Samples containing 0.5 × 106 cells were harvested by centrifugation at 200g for 10 minutes at 4° C and incubated in Ca2+ and PI staining buffer following manufacturer's protocol. Flow cytometry analysis of the samples was performed as described in “Flow cytometric analysis.”
Results
Differential expression of TRAIL-Rs and TRAIL in resting and activated NK and CD8+ CTLs
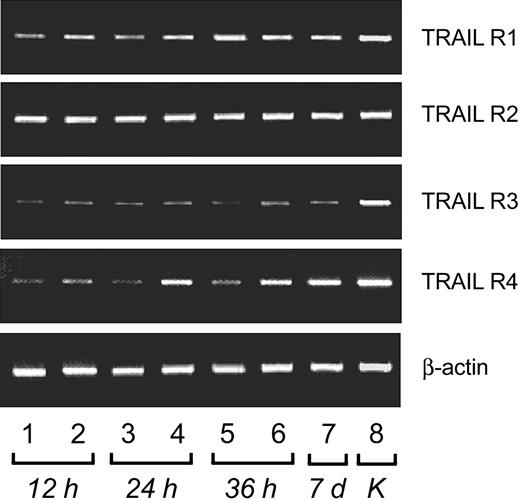
Purified NK cells were cultured for 15 days with IL-2 or with IL-15, whereas purified CD8+ CTLs were cultured for 3 days in the presence of PHA and cultured for the subsequent 12 days with IL-2. In the first group of experiments the mRNA for TRAIL receptors was examined by RT-PCR analysis both in resting and activated NK cells. As shown in Figure 1, the mRNA of all TRAIL receptors (R1, R2, R3, R4) was clearly detectable in both resting and activated NK cells. Similar data were obtained with purified CD8+ CTLs.
RT-PCR analysis of TRAIL-Receptors mRNA expression in resting and IL-2–stimulated primary human NK cells. Lanes 1, 3, and 5 show resting NK cells. Lanes 2, 4, 6, and 7 show IL-2–treated NK cells for time intervals ranging from 12 hours to 7 days. K indicates positive control (K562 cells).
RT-PCR analysis of TRAIL-Receptors mRNA expression in resting and IL-2–stimulated primary human NK cells. Lanes 1, 3, and 5 show resting NK cells. Lanes 2, 4, 6, and 7 show IL-2–treated NK cells for time intervals ranging from 12 hours to 7 days. K indicates positive control (K562 cells).
The expression profile of TRAIL receptors was next examined by flow cytometry (Figure 2A-C).
Flow cytometry analysis of the surface expression of TRAIL-Receptors and TRAIL ligand in NK and CTL cells. (A) IL-2–stimulated NK cells. (B) IL-15–stimulated NK cells. (C) CD8+ T cells, at different times of activation. (D) Surface expression of TRAIL-Rs by the NK-like cell line NK 3.3. The cell line was routinely cultured in RPMI 1640 + 15% FCS + 100 U/mL rIL-2. (E) Expression of surface TRAIL-ligand (TRAIL-L) by activated (day 10) NK and CD8+ T cells. Empty histograms indicate isotype-matched irrelevant antibody, negative controls.
Flow cytometry analysis of the surface expression of TRAIL-Receptors and TRAIL ligand in NK and CTL cells. (A) IL-2–stimulated NK cells. (B) IL-15–stimulated NK cells. (C) CD8+ T cells, at different times of activation. (D) Surface expression of TRAIL-Rs by the NK-like cell line NK 3.3. The cell line was routinely cultured in RPMI 1640 + 15% FCS + 100 U/mL rIL-2. (E) Expression of surface TRAIL-ligand (TRAIL-L) by activated (day 10) NK and CD8+ T cells. Empty histograms indicate isotype-matched irrelevant antibody, negative controls.
Despite the expression of the mRNA for all receptors, resting NK and CD8+ T cells did not show any significant surface expression of R1, R2, or R3. However, a variable proportion of freshly isolated NK and CD8+ T cells did express R4. The surface expression of TRAIL-Rs was next examined in both populations at different time points on stimulation. R1 was never expressed up to day 15 of culture, whereas R2 and, to a lower extent, R3 expression was up-regulated as soon as 36 hours after stimulation, reaching a peak around day 10. Moreover, although the expression of surface R4 remained constant in resting and activated NK cells, it showed a significant increase in activated CD8+ T cells.
The NK-like cell line NK 3.3 showed surface expression of TRAIL-R2 similar to that of primary NK cells but no expression of TRAIL-R3 or R4 (Figure 2D).
In parallel experiments, we examined the surface expression of membrane-bound TRAIL, which was undetectable in resting NK cells and CTLs (data not shown), but it became clearly detectable in activated NK cells and CTLs (Figure 2E).
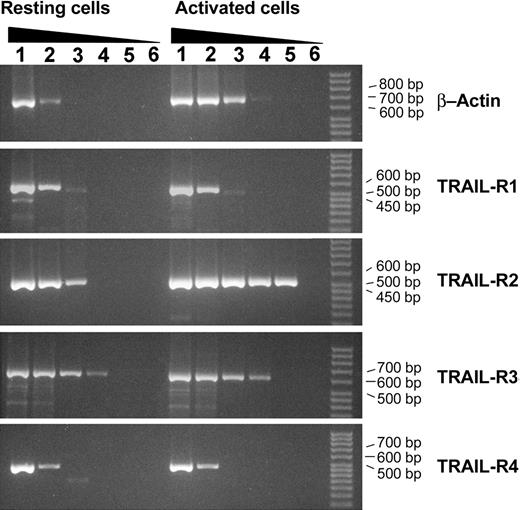
To quantify the effect of cytokine induction on TRAIL receptor mRNA expression, we performed a semiquantitative RT-PCR analysis (Figure 3). In NK cells, IL-2 promoted β-actin transcription, raising the amount of intracellular mRNA up to 26.2- ± 5.6-fold; similarly TRAIL-R2 gene transcription was induced up to 26.1- ± 3.1-fold. No induction of TRAIL-R1, TRAIL-R3, or TRAIL-R4 transcription was observed.
Quantification of gene transcription by semiquantitative RT-PCR in resting and Il-2–stimulated primary human NK cells. RT-PCR of β-actin, TRAIL-R1, TRAIL-R3, and TRAIL-R4 was performed by using the following cDNA dilutions: 1/8, 1/20, 1/100, 1/500, 1/2500 (lanes 1-5, respectively). RT-PCR of TRAIL-R2 was performed by using the following cDNA dilutions: 1/20, 1/100, 1/500, 1/2500, 1/12 500 (lanes 1-5, respectively). Lane 6 is RT-PCR negative control.
Quantification of gene transcription by semiquantitative RT-PCR in resting and Il-2–stimulated primary human NK cells. RT-PCR of β-actin, TRAIL-R1, TRAIL-R3, and TRAIL-R4 was performed by using the following cDNA dilutions: 1/8, 1/20, 1/100, 1/500, 1/2500 (lanes 1-5, respectively). RT-PCR of TRAIL-R2 was performed by using the following cDNA dilutions: 1/20, 1/100, 1/500, 1/2500, 1/12 500 (lanes 1-5, respectively). Lane 6 is RT-PCR negative control.
The increased amount of surface TRAIL-R2 protein, observed by flow cytometry on cytokine stimulation of NK cells, correlated with its increased gene expression, whereas TRAIL-R3 phenotypic up-regulation did not, suggesting a posttranscriptional regulation of TRAIL-R3 expression.
Both resting and activated NK cells and CTLs are resistant to TRAIL-induced apoptosis
The simultaneous expression of TRAIL and TRAIL-R2, 1 of the 2 death receptors, in activated NK cells and CTLs prompted us to investigate the susceptibility to TRAIL-induced cytotoxicity of both resting and activated lymphocytes. In this group of experiments, we used TRAIL-sensitive Jurkat and HL-60 cell lines as positive controls (Figure 4). The specificity of TRAIL-induced cell death was ascertained by using a TRAIL-R1-Fc and TRAIL-R2-Fc chimera cocktail, which was administered to the cell cultures at the same time as TRAIL. Figure 4A-B shows that the chimera cocktail, by sequestering soluble TRAIL, completely inhibited TRAIL-induced apoptosis on Jurkat and HL-60 cells. TRAIL did not induce apoptosis in resting NK or CD8+ T cells. This was an expected finding on the basis of the lack of surface TRAIL-R1 and TRAIL-R2 in primary resting lymphoid cells. Notwithstanding the expression of death receptor R2, however, TRAIL did not induce apoptosis either in activated NK or CD8+ cells. Similarly, NK 3.3 cells, routinely cultured in the presence of IL-2, were resistant to TRAIL-induced apoptosis (Figure 4B). These findings clearly indicate that the expression of R2 is not sufficient to sensitize NK or CD8+ cells to the apoptogenic potential of TRAIL. Given the absence of decoy receptors on NK 3.3 cells, they also suggest that intracellular mechanisms might be responsible for the resistance of activated NK cells to TRAIL-induced apoptosis.
TRAIL-induced apoptosis in NK and CD8+ T cells. NK and CD8+ T cells were stimulated for 1 day (A) or 10 days (B) as reported in “Materials and methods.” Cells were incubated overnight with 1 μg recombinant human TRAIL. TRAIL sensitivity of NK 3.3 cell line is also reported (B). TRAIL chimera indicates TRAIL-R1-Fc + TRAIL-R2-Fc chimera cocktail administered to the cells at the same time of recombinant TRAIL. TRAIL-sensitive Jurkat and HL-60 cell lines served as positive controls.
TRAIL-induced apoptosis in NK and CD8+ T cells. NK and CD8+ T cells were stimulated for 1 day (A) or 10 days (B) as reported in “Materials and methods.” Cells were incubated overnight with 1 μg recombinant human TRAIL. TRAIL sensitivity of NK 3.3 cell line is also reported (B). TRAIL chimera indicates TRAIL-R1-Fc + TRAIL-R2-Fc chimera cocktail administered to the cells at the same time of recombinant TRAIL. TRAIL-sensitive Jurkat and HL-60 cell lines served as positive controls.
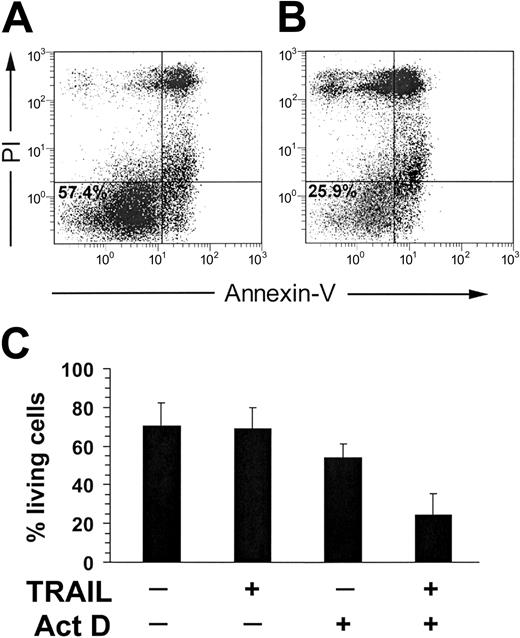
Therefore, to rule out the possible actors in the intracellular death machinery of primary lymphoid cells potentially responsible for resistance to TRAIL-induced apoptosis, we next performed experiments in which NK and CD8+ T cells were pretreated with 50 ng/mL actinomycin D before adding recombinant TRAIL. As shown in Figure 5, actinomycin D strongly sensitized activated NK cells to TRAIL-induced apoptosis, evaluated by Annexin V–FITC. Similar results were obtained for CD8+ CTLs (not shown).
TRAIL-induced apoptosis in IL-2–treated NK cells (10 days) in the presence of 50 ng/mL Actinomycin D (Act D). Act D was administered to the cells at the same time of TRAIL. (A) Negative control was IL-2–treated NK cells + Act D. The percentage of living cells (57.4%) is indicated in the Annexin V–/PI– population. (B) IL-2–treated NK cells + TRAIL + Act D. The percentage of living cells (25.9%) is indicated, as well. (C) TRAIL-induced apoptosis in IL-2–stimulated NK cells with or without Act D. Results obtained from 3 unrelated donors are reported. Values are expressed as means ± standard deviation.
TRAIL-induced apoptosis in IL-2–treated NK cells (10 days) in the presence of 50 ng/mL Actinomycin D (Act D). Act D was administered to the cells at the same time of TRAIL. (A) Negative control was IL-2–treated NK cells + Act D. The percentage of living cells (57.4%) is indicated in the Annexin V–/PI– population. (B) IL-2–treated NK cells + TRAIL + Act D. The percentage of living cells (25.9%) is indicated, as well. (C) TRAIL-induced apoptosis in IL-2–stimulated NK cells with or without Act D. Results obtained from 3 unrelated donors are reported. Values are expressed as means ± standard deviation.
Both NK cells and CTLs show a high expression of c-FLIP
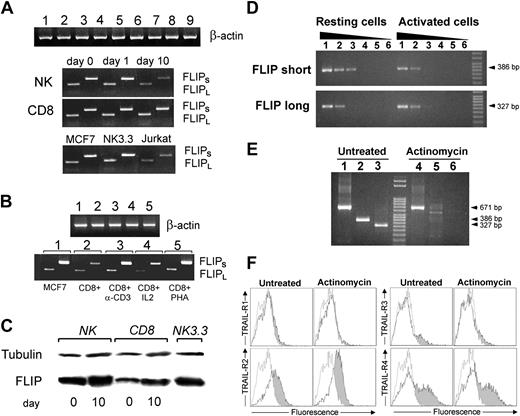
Given the well-documented inhibitory role of FLIP on the activity of apical caspase-8,15 we performed a series of experiments on purified NK and CD8+ T cells to determine the expression of FLIP at the mRNA and protein level. Figure 6A shows the RT-PCR for the long and short forms of FLIP in NK and CD8 T cells after a short (1 day) or long period (10 day) of IL-2 stimulation. Both forms are expressed at the same level by resting and activated cells. The NK cell line NK 3.3 expresses both forms of FLIP, as well. The expression of FLIP short or long mRNA in CD8+ T cells was not dependent on the type of stimulation used, as anti-CD3, IL-2, and PHA did not influence gene transcription and mRNA expression (Figure 6B).
Expression of the short and long forms of FLIP in resting and activated NK cells, CD8+ T cells, and in the NK 3.3 cell line. (A) RT-PCR of FLIPShort and FLIPLong in resting and activated (as described in “Materials and methods”) NK and CD8+ T cells for 1 day or 10 days. MCF7 cells served as positive controls. (B) RT-PCR of FLIPShort and FLIPLong in CD8+ T cells activated by anti-CD3, IL-2 alone, or PHA alone. The expression of FLIP mRNA is not dependent on the type of stimulation used to activate CD8+ CTLs. (C) Western blot analysis of FLIP protein in resting (0 day) and stimulated (10 days) NK cells, CD8+ T cells, and NK 3.3 cell line. (D) Quantification of gene transcription by semiquantitative RT-PCR in NK cells before and after IL-2 stimulation (lanes 1-5, 1/8, 1/20, 1/100, 1/500, 1/2500 cDNA dilutions were analyzed to detect FLIPShort and FLIPLong; lane 6, RT-PCR negative control). (E) RT-PCR analysis of FLIPShort and FLIPLong expression with or without actinomycin D treatment (lanes 1 and 4, β-actin; lanes 2 and 5, FLIPShort; lanes 3 and 6, FLIPLong RT-PCR products). (F) Flow cytometry analysis of the surface expression of TRAIL receptors in NK cells with or without actinomycin D treatment (50 ng/mL for 24 hours). Empty histograms indicate controls, untreated cells. Gray histograms indicate actinomycin-treated cells.
Expression of the short and long forms of FLIP in resting and activated NK cells, CD8+ T cells, and in the NK 3.3 cell line. (A) RT-PCR of FLIPShort and FLIPLong in resting and activated (as described in “Materials and methods”) NK and CD8+ T cells for 1 day or 10 days. MCF7 cells served as positive controls. (B) RT-PCR of FLIPShort and FLIPLong in CD8+ T cells activated by anti-CD3, IL-2 alone, or PHA alone. The expression of FLIP mRNA is not dependent on the type of stimulation used to activate CD8+ CTLs. (C) Western blot analysis of FLIP protein in resting (0 day) and stimulated (10 days) NK cells, CD8+ T cells, and NK 3.3 cell line. (D) Quantification of gene transcription by semiquantitative RT-PCR in NK cells before and after IL-2 stimulation (lanes 1-5, 1/8, 1/20, 1/100, 1/500, 1/2500 cDNA dilutions were analyzed to detect FLIPShort and FLIPLong; lane 6, RT-PCR negative control). (E) RT-PCR analysis of FLIPShort and FLIPLong expression with or without actinomycin D treatment (lanes 1 and 4, β-actin; lanes 2 and 5, FLIPShort; lanes 3 and 6, FLIPLong RT-PCR products). (F) Flow cytometry analysis of the surface expression of TRAIL receptors in NK cells with or without actinomycin D treatment (50 ng/mL for 24 hours). Empty histograms indicate controls, untreated cells. Gray histograms indicate actinomycin-treated cells.
Western blot analysis showed that both resting NK and CD8+ T cells express FLIP protein, and prolonged activation with cytokines apparently slightly enhanced the total protein amount (Figure 6C).
To better quantify the modulation of FLIP mRNA transcription under cytokine stimulation, we performed a semiquantitative PCR analysis. Figure 6D shows that FLIPLong messenger RNA was expressed at the same level in resting and IL-2–activated NK cells, whereas FLIPShort mRNA was partially down-modulated (up to 27% ± 4% of resting cells) in IL-2–stimulated NK cells. Furthermore, actinomycin D impaired transcription and accumulation of FLIP mRNA in IL-2–stimulated NK cells (Figure 6E; compare lane 5 with lane 2 and lane 6 with lane 3). However, actinomycin D did not affect TRAIL receptors surface expression (Figure 6F).
Blockage of FLIP transcription sensitizes NK cells to TRAIL
To formally prove that FLIP levels in resting and activated NK cells are responsible for their resistance to the apoptotic effects of TRAIL, we designed siRNAs specific for FLIP mRNA and transfected them into primary, IL-2–stimulated, NK cells. Figure 7A-C shows that a significant proportion of FLIP siRNA-transfected NK cells effectively die after soluble TRAIL administration.
TRAIL-induced apoptosis of IL-2–activated NK cells (10 days) in the presence of anti-FLIP siRNAs. siRNAs were nucleofected into NK cells 24 hours before TRAIL addition. (A) Negative control was IL-2–treated NK cells + siRNA. The percentage of living cells (37.5%) was calculated by selecting the events with a PI fluorescence lower than 10° relative units. (Insert) Ten-day IL-2–stimulated control lymphocytes. (B) IL-2–treated NK cells + siRNA + TRAIL. The percentage of living cells was 15.0%. (C) TRAIL-induced apoptosis in IL-2–stimulated NK cells with or without siRNA. Results obtained from 3 unrelated donors are reported. Values are expressed as means ± standard deviation.
TRAIL-induced apoptosis of IL-2–activated NK cells (10 days) in the presence of anti-FLIP siRNAs. siRNAs were nucleofected into NK cells 24 hours before TRAIL addition. (A) Negative control was IL-2–treated NK cells + siRNA. The percentage of living cells (37.5%) was calculated by selecting the events with a PI fluorescence lower than 10° relative units. (Insert) Ten-day IL-2–stimulated control lymphocytes. (B) IL-2–treated NK cells + siRNA + TRAIL. The percentage of living cells was 15.0%. (C) TRAIL-induced apoptosis in IL-2–stimulated NK cells with or without siRNA. Results obtained from 3 unrelated donors are reported. Values are expressed as means ± standard deviation.
Discussion
Immune surveillance against tumors is mediated by natural killer cells and cytotoxic T lymphocytes. Both these cell types are equipped with receptors for MHC class I, although their molecular nature and function are different.22 Although NK cells display a spontaneous cytolytic activity against a variety of tumor cells, their antitumor activity is greatly enhanced, both in terms of efficiency and array of targets, by cytokine treatment. Antitumor CTLs physiologically act under the proliferative/activatory action of cytokines as well, and both NK cells and CTLs acquire new antitumor molecular equipment when activated by cytokines. Increasing experimental evidence suggests that TRAIL, the most recently identified of such molecules expressed on the surface of activated NK cells and CTLs, plays a key role in their antitumor activity. In fact, the expression of TRAIL has been reported on the surface of activated NK and T cells in response to several cytokines,4,23-26 and recent evidence indicates that surface TRAIL plays a major role in optimal graft-versus-tumor activity.26 Furthermore, Cretney et al27 recently generated a TRAIL-deficient mouse that was found to be more susceptible to experimental and spontaneous tumor metastasis. Studies in the mouse demonstrated that TRAIL expression is constitutive only in liver NK cells, playing a role in the control of liver metastasis. However, on cytokine activation, TRAIL expression is up-regulated also in NK cells resident in other tissues, such as spleen and lung, playing a role, as well, in the control of tumor metastasis to these organs.6 Therefore, recombinant TRAIL protein offers great promise as a cancer therapeutic,14 showing the unique property to kill several types of tumor cells, sparing most normal cells and tissues. Hence, the great expectations from the clinical trials that are about to start. Moreover, cytokine-mediated TRAIL induction on lymphoid cells has been proposed as a prognostic marker in multiple sclerosis, suggesting a role for TRAIL+ lymphocytes in patients' response to cytokine therapy.28 Our data show that both activated NK and CD8+ T cells express membrane-bound TRAIL. In transplantation tolerance, these TRAIL-expressing cells might be responsible for the “veto cell” function by killing recipient's CD8+ CTL precursors that are specific for them.29-31 Given the great importance of the anticancer effects of TRAIL, and the use of membrane-bound TRAIL made by antitumor lymphocytes, understanding whether cytokine stimulation can modulate the cellular distribution of TRAIL death receptors exposing major antitumor cytotoxic cells to the apoptogenic action of TRAIL is of great relevance. Our data essentially demonstrate that (1) resting NK and CD8+ T cells do not express TRAIL-R1, TRAIL-R2, or TRAIL-R3 but show a constitutive expression of the decoy receptor TRAIL-R4; (2) IL-2 stimulation induces the stable surface expression of TRAIL-R2 and TRAIL-R3 by both cell types; (3) both resting and activated NK cells and CTLs are insensitive to the apoptogenic effects of TRAIL; and (4) both resting and IL-2–activated NK cells and CTLs constitutively express both forms of FLIP (short and long) at the mRNA and protein level that are responsible for their resistance to the apoptotic effects of TRAIL.
With some exceptions, such as developing erythroblasts,11 hepatocytes,32 prostate cells,33 and thymic lymphocytes,12 most of the normal cells examined are resistant to TRAIL-mediated apoptosis. In the TRAIL–/– mouse model system, no specific alteration of peripheral T-cell or NK cell homeostasis was reported under physiologic conditions.27 However, controversial data have been obtained from different groups of investigators examining the response to TRAIL of primary normal T lymphocytes, as either no effect,34 cell cycle arrest without cytotoxicity,35,36 or induction of apoptosis 37 have been reported. However, several data suggest that viral infections can render lymphoid cells38-40 as well as hepatocytes41 susceptible to TRAIL-mediated cytotoxicity. Furthermore, a reciprocal expression of membrane-bound TRAIL and FasL has been recently suggested to play a role in the T helper 1 (Th1) versus Th2 cell differentiation.42
Theoretically, there are at least 2 possible, not mutually exclusive, mechanisms for NK cells and CTLs to be protected by TRAIL-induced apoptosis.43 The first mechanism is represented by the expression of TRAIL-R4 in both resting and activated cells, and TRAIL-R3 exclusively in activated cells. In fact, although TRAIL-R1 and TRAIL-R2 transduce apoptotic signals on binding of TRAIL, TRAIL-R3 and TRAIL-R4 are homologous to TRAIL-R1 and TRAIL-R2 in their cysteine-rich extracellular domain but lack intracellular death domain and apoptosis-inducing capability. Although it is not yet completely clear whether the expression of TRAIL-R3 and/or TRAIL-R4 is a key factor in determining the resistance or sensitivity to TRAIL cytotoxicity, they have been proposed to function as decoy receptors, protecting some cell types from apoptosis.44,45 However, our data show that the second possible mechanism of protection, represented by the high level of FLIP protein expression by NK cells and CTLs, is the one that accounts for their resistance to the apoptotic effect of TRAIL. A direct proof of the role for FLIP on the protection from TRAIL activity is given by the siRNA experiments, in which FLIP-siRNA transfected primary human NK cells became sensitive to TRAIL and die by apoptosis. FLIPLong, the full-length 55-kDa form of FLIP, shows overall structural homology to caspase-8, containing 2 death effector domains that interact with FADD, and an inactive caspase-like domain. FLIPShort, an alternative spliced short form of FLIP, contains only the 2 death effector domains and has lower antiapoptotic capacity.15 Our data parallel those obtained by overexpression of FLIPLong in mouse T cells, which inhibits FasL-induced apoptosis in vitro,46 whereas it is not able to block activation-induced cell death.47 Although both TRAIL-R4 and FLIP might play a role in the protection of NK and CTL cells from TRAIL-induced apoptosis, the fact that (1) selective down-modulation of FLIP exposes NK cells to TRAIL-induced apoptosis; (2) NK 3.3 cells, which are resistant to TRAIL, do express FLIP but do not express TRAIL decoy receptors; and (3) actinomycin D, which restores TRAIL sensitivity in NK cells, does not affect TRAIL receptor surface expression while decreasing FLIP expression, strongly suggest that FLIP is primarily responsible for the resistance of NK and CD8 T cells to the apoptotic effect of TRAIL.
Prepublished online as Blood First Edition Paper, June 17, 2004; DOI 10.1182/blood-2004-04-1294.
Supported by grants from Associazione Italiana per la Ricerca sul Cancro, Cassa di Risparmio di Parma, Cassa di Risparmio di Bologna, and Ministero dell'Istruzione, dell'Università e della Ricerca; programmi di ricerca cofinanziati 2002 Fondo per gli Investimenti della Ricerca di Base; Joung Researcher Grant of the Parma University, and 1% Sanità Alzheimer.
P.M. and C.P. contributed equally to this study.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.