Abstract
The pathogenesis of Hodgkin lymphoma (HL) is still largely unknown. Based on a search for footprints of pathogenetic mechanisms in global RNA expression data of Hodgkin/Reed-Sternberg (HRS) cell lines, we analyzed the expression and activation of 6 receptor tyrosine kinases (RTKs) in classic HL. Immunohistochemistry revealed that the RTKs platelet-derived growth factor receptor A (PDGFRA), DDR2, EPHB1, RON, TRKB, and TRKA were each expressed in HRS cells in 30% to 75% of patients. These RTKs were not expressed in normal B cells, the origin of HRS cells, or in most B-cell non-Hodgkin lymphoma (NHL). In the majority of patients at least one RTK was expressed, and in most patients several RTKs were coexpressed, most prominently in Hodgkin lymphoma of the nodular sclerosis subtype. Phosphotyrosine-specific antibodies revealed exemplarily the activation of PDGFRA and TRKA/B and an elevation of cellular phosphotyrosine content. Immunohistochemistry for RTK ligands indicated that DDR2 and TRKA are likely activated in a paracrine fashion, whereas PDGFRA and EPHB1 seem to be activated by autocrine loops. Activating mutations were not detected in cDNA encoding the RTKs in HRS cell lines. These findings show the unprecedented coexpression of multiple RTKs in a tumor and indicate that aberrant RTK signaling is an important factor in HL pathogenesis and that it may be a novel therapeutic target.
Introduction
Hodgkin lymphoma (HL) is one of the most common lymphomas in the Western world. It is characterized by few tumor cells, Hodgkin/Reed-Sternberg (HRS) cells, and nonneoplastic infiltrate composed of T lymphocytes, histiocytes, eosinophilic granulocytes, plasma cells, and other cells.1 HRS cells are derived from mature B cells, and their rarity (they usually represent approximately 1% of the infiltrate) has greatly impeded analysis of the pathogenesis of this lymphoma.2 In addition to the potential oncogenic Epstein-Barr virus (EBV), which is found in the HRS cells of approximately 40% of patients in the Western world,3,4 the constitutive activation of nuclear factor κB (NFκB) is observed in the HRS cells of most patients, and it may be attributed to different genetic alterations.5-10
Tyrosine kinases (TKs) are frequently involved in neoplastic transformation.11 They are important regulators of intercellular and intracellular signaling and are involved in the regulation of fundamental cellular processes such as proliferation, differentiation, survival, and cell motility.12 In normal cells, the activity of TKs is tightly regulated. Extracellular ligands binding to receptor tyrosine kinases (RTKs) induce the dimerization and subsequent phosphorylation of intracellular domains and signaling molecules. RTK activation is usually quickly terminated by phosphatases or by degradation. Aberrant TK activity caused by various genetic alterations—for example, activating point mutations, translocations, or amplifications—frequently causes cellular transformation and has been observed in many different types of tumors.11 In recent years, TK inhibitors have been used as highly effective forms of therapy for patients with tumors with activated TKs, among them imatinib for gastrointestinal stromal tumors with activated KIT and BCR-ABL (breakpoint-cluster region/Abelson leukemia)–positive chronic myeloid leukemia or gefitinib for epithelial tumors with activated epithelial growth factor receptor (EGFR).13
In mature B cells, several intracellular TKs are essential components of the B cell receptor signalosome,14 whereas the expression of RTKs seems to be limited. Thus far, only the expressions of TRKA (tyrosine kinase receptor A), which is essential for the survival of memory B cells,15,16 and of MET in germinal center B cells, where it is involved in the regulation of adhesion,17,18 have been described.
Given the prominent role of RTKs in cellular transformation, which is often caused by aberrant expression, we screened gene expression data obtained from oligonucleotide microarrays for aberrant RTK expression in HRS cell lines. Aberrant expression of several RTKs in the HRS cell lines was observed and encouraged us—especially in light of the great therapeutic success of TK inhibitors in some tumors with activated TK—to perform further analysis of these RTKs in patients with primary HL.
Materials and methods
Cell lines and tissue samples
The cell lines analyzed were established from patients with nodular sclerosis (NS) (L428 and HDLM2) or mixed cellularity (MC) HL (KMH2 and L1236) and were grown as recommended (DSMZ, Braunschweig, Germany). SUP-B15 carries the BCR-ABL translocation and is derived from a pre–B cell acute lymphoblastic leukemia and was grown as recommended (DSMZ). All tissue samples were retrieved from the files of the Department of Pathology of the University of Frankfurt and were originally submitted for diagnostic purpose. All biopsy specimens were fixed in 5% phosphate-buffered formalin, usually overnight at room temperature. Patients were classified according to the World Health Organization (WHO) classification scheme.19 Of the patients with HL, 39 had the NS subtype and 8 of these were EBV positive, 60 were of MC subtype and 31 of these were EBV positive, and 23 had lymphocyte-predominant (LP) HL.
Western blot analysis
Approximately 105 cells were harvested, and whole-cell lysates were prepared as described.20 Proteins were separated by electrophoresis on 12.5% polyacrylamide gels containing 0.1% sodium dodecyl sulfate (SDS) at 30 mA for 90 minutes and were transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Munich, Germany) at 100 mA for 240 minutes using a semidry technique. Membranes were blocked with phosphate-buffered saline (PBS) containing 5% dry milk powder and 0.05% Tween 20. Primary antibodies were used at 0.8 μg/mL in PBS with 5% milk powder, and bound antibody was detected by a horseradish peroxidase–coupled suitable secondary antibody (DAKO, Hamburg, Germany), followed by chemiluminescence detection using the ECL Plus system (Amersham Biosciences, Freiburg, Germany).
Immunohistochemistry
Immunohistochemistry was performed with 5-μm sections of the tissues fixed in 5% buffered formalin and embedded in paraffin. For all patients with HL and at least half the patients with B cell non-Hodgkin lymphoma (NHL), biopsy sections of complete lymph node were used, and the remaining NHL samples were analyzed on tissue microarray. Antibodies, control experiments, and staining procedures used are given in Table 1. After antigen retrieval and blocking with normal goat serum (4 μg/mL; Santa Cruz Biotechnology, Heidelberg, Germany) and human immunoglobulin G (IgG) (4 μg/mL; Sigma, Munich, Germany), sections were incubated between 1 and 16 hours with the primary antibodies in Tris-buffered saline (TBS) at room temperature and were developed using the Envision system (DAKO, Hamburg, Germany), either with alkaline phosphatase and Fast Red or horseradish peroxidase and 3.3-diaminobenzidine (DAKO). Whenever possible, staining specificity was verified by a 4-hour preincubation of antibodies with the 20-fold quantity of the peptide used for immunization at 37°C. For platelet-derived growth factor receptor A (PDGFRA), DDR2, TRKA, PDGFA, collagen type 1, and EphrinB1, additional different primary antibodies were used at least for a fraction of the samples to confirm the staining results. The specificity of phosphotyrosine (p-Y) stainings was controlled by preincubation of the sections with T-cell phosphatase (Sigma) in the appropriate buffer (300 μL of 50 mM Tris-HCl pH 7.0, 100 mM NaCl, 5 mM dithiothreitol [DTT], and 2 mM EDTA [ethylenediaminetetraacetic acid], 20 units enzyme per section) for 3 hours at 37°C. For the p-Y-720-PDGFRA and p-Y-680/681-TRKA/B antibodies, specificity was further verified by blocking experiments with the peptides used for immunization and by immunohistochemistry with patients not expressing these RTKs. For picture acquisition, an Olympus BX40 microscope (Olympus, Hamburg, Germany) equipped with UPlanApo objective lenses (10×/0.4, 20×/0.7, and 40×/0.65) and a Coolpix 4500 digital camera (Nikon, Düsseldorf, Germany) were used. All pictures were processed with Adobe Photoshop 7.0 (Adobe, San Jose, CA).
Sequence analysis of RTK transcripts from HRS cell lines
Total RNA was isolated from all 4 HRS cell lines using a Qiagen RNA isolation kit (Qiagen, Hilden, Germany). Aliquots of the RNAs were used for cDNA synthesis with a first-strand cDNA synthesis kit (Roche, Mannheim, Germany), and aliquots of cDNA were used as templates in polymerase chain reactions (PCRs). After 2-minute denaturation at 95°C, 45 cycles of 30 seconds at 95°C, 30 seconds at 61°C, and 5 minutes at 68°C with 3.5 units Expand enzyme (Roche) in 1 × Expand PCR buffer with 2 mM MgCl2 were performed. This procedure yielded enough amplificate for visualization on ethidium bromide–stained agarose gels. For all RTKs, the entire coding region was amplified as one PCR product with RTK-specific primers from 5′- and 3′-untranslated regions (PDGFRA-US4, 5′-gcacgctctttactccatgtgtgg-3′; PDGFRA-LS1, 5′-accaagtcctcctctcaacctcc-3′; DDR2-US3, 5′-aattgaagagaagcagaggccagc-3′; DDR2-LS1, 5′-cagatggagtggcataggcatgg-3′; EPHB1-US4, 5′-agcgcgaaaggataccgagaagc-3′; EPHB1-LS1, 5′-tctctccagtacattccacagtgg-3′; RON-US1, 5′-atcctctagggtcccaggctcg-3′; RON-LS1, 5′-aaggcagctaagcaggtccagc-3′; TRKB-US1, 5′-gttaagagagccgcaagcgcagg-3′; TRKB-LS1, 5′-agtgaaggagagcagcttggtgg-3′; TRKA-US1, 5′-gcagctgggagcgcacagacg-3′; TRKA-LS1, 5′-gtcccttcccacatgctgagg-3′) and was directly sequenced with several internal RTK-specific primers.
Imatinib treatment of HL cell lines
Imatinib was prepared from 100-mg capsules solubilized in deionized water. Particulate matter was removed through 2 centrifugation steps at 2500g for 1 minute each. Activity of this preparation was tested with SUP-B15, and 50% growth reduction was observed at concentrations between 0.1 and 1 μM imatinib, which is typical for BCR/ABL–dependent cell lines.21
For imatinib treatment, growing KMH2, HDLM2, L1236, L428, and SUP-B15 cell lines were harvested and washed once in RPMI with 1% fetal calf serum (FCS) (20% for SUP-B15). Cells (3 × 105) per well were plated onto 6-well ultra-low cluster plates (Corning Incorporated, Corning, NY) in RPMI with 1% FCS (20% for SUP-B15) and were incubated for 70 hours with 0.1 to 10 μM or without imatinib. Cells were then harvested and incubated in propidium iodide/PBS (20 μg/mL) for 15 minutes, and numbers of living cells were determined using a FACScan (Becton Dickinson, Heidelberg, Germany).
Results
Aberrant RTK expression in HRS cell lines
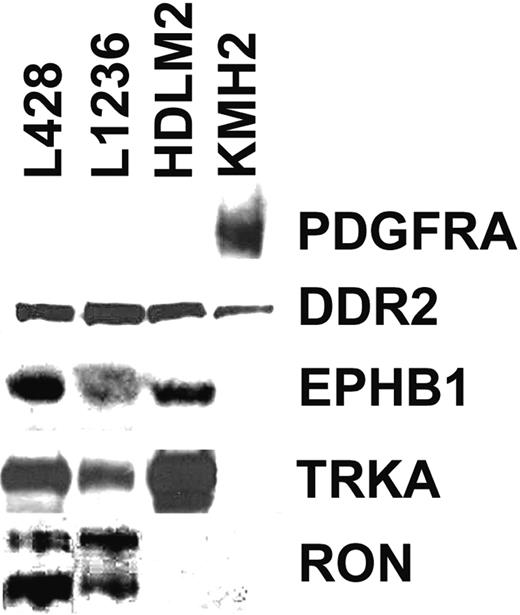
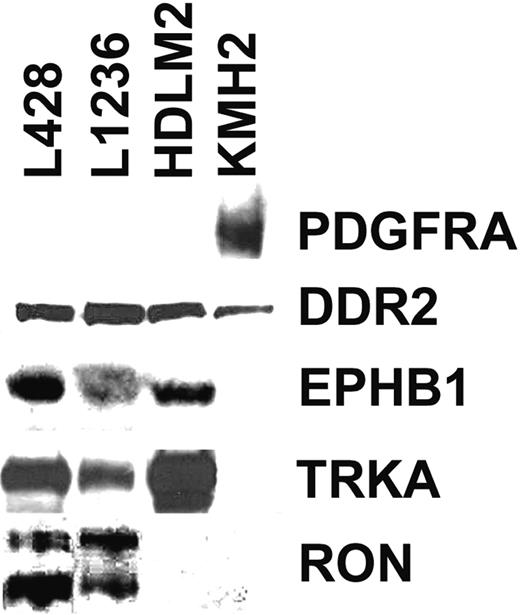
Analysis of global RNA expression data of 4 HRS cell lines, normal B cells, and several B-cell NHLs (B-NHLs) generated previously22 indicated that 8 of 50 RTKs represented by probes on the microarray were expressed in at least 2 of the 4 HRS cell lines but were not expressed or were only occasionally expressed in normal B cells or in B-NHL. Western Blot analysis with antibodies specific for the RTKs confirmed the expression of PDGFRA, DDR2, EPHB1, RON, and TRKA in HRS cell lines, whereas no expression of TRKB, TYRO3, or MER was observed (Figure 1). Expression patterns of DDR2, EPHB1, RON, and TRKA in Western blot analysis were in line with the microarray data, but we observed a discrepancy for PDGFRA. Microarray analysis indicated comparable levels of expression in all 4 cell lines, but in Western blot analysis PDGFRA expression was detected only in KMH2. However, the reason for this discrepancy became clear on sequence analysis of the cDNAs for the different RTKs from the HRS cell lines (see “No indications for constitutive activation of RTKs in HRS cell lines”).
Aberrant RTK expression in HL
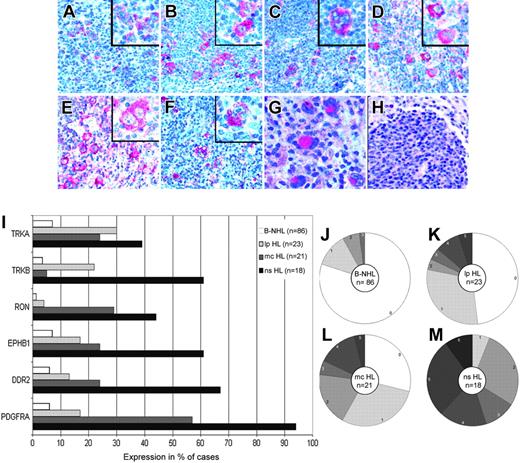
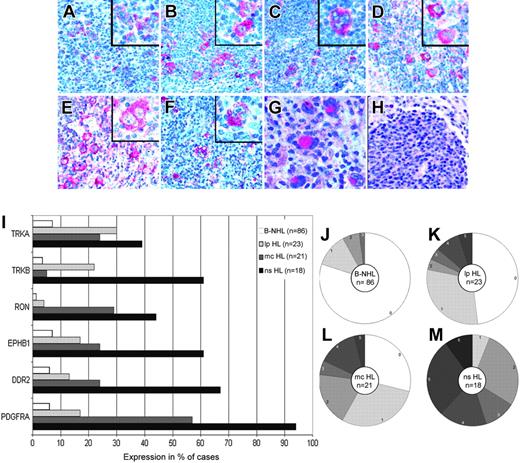
To investigate whether the RTKs expressed in HRS cell lines were also expressed in primary HL, we performed immunohistochemistry with tissue sections from normal or reactive lymphoid tissues, patients with B-NHL, and patients with primary HL with antibodies specific for PDGFRA, DDR2, EPHB1, RON, TRKA, and TRKB (Figure 2A-F). TRKB was also included in the immunohistochemical analysis despite the negativity of the HRS cell lines, because patients with primary disease showed positivity in immunostainings, and specificity of the antibody could be confirmed by other methods (Table 1). In normal or reactive lymphoid tissue (lymph nodes with unspecific lymphadenitis and with hyperplastic tonsils and spleens), these 6 RTKs were not expressed in B cells (primary and secondary follicles and marginal zones) at levels detectable by immunohistochemistry, though TRKA expression in mature B cells has previously been shown by other methods.16 In patients with B-NHL, the expression of these RTKs was only occasionally observed, with 80% of the 86 patients with B-NHL analyzed expressing none of the RTKs and with the most frequently expressed RTKs found in only 7% of patients (Figure 2I-J).
In contrast to normal B cells and cells from B-NHL patients, the RTKs analyzed were frequently expressed in patients with classic HL. Most HRS cells of most (33 of 39) patients expressed at least one of the RTKs (Figure 2I, L-M). PDGFRA was expressed in 75% of patients, and DDR2, EPHB1, RON, TRKB, and TRKA were each at least in 30% of patients. In 26 cases,26 2 different RTKs were coexpressed, with a range from 0 to 6 different RTKs per patient. Aberrant RTK expression was more pronounced in those with the NS subtype. All 18 patients with NS HL expressed at least 1 RTK, and on average nearly 4 (3.7) RTKs per patient were found. However, 6 of 21 patients with the MC subtype expressed none of the RTKs, with an average of less than 2 (1.6) RTKs expressed per patient (a highly significant statistical difference for the average number of RTKs expressed per patient; P = .001, Student t test). Analysis of 2 cases of composite follicular/HL, each with a common precursor,23,24 revealed that PDGFRA was expressed in each only in the HRS cells (Figure 2G-H) and not in the follicular lymphoma cells. In those with LP HL, expression of the RTKs analyzed was less pronounced (Figure 2I, K). Twelve of 23 patients expressed none of the RTKs; the most frequent RTK (TRKA) was expressed in 30% of patients, and an average of 1 (1.0) RTK was expressed per patient.
Western blot analysis for expression of 5 RTKs in 4 HRS cell lines. Shown are Western blots of all 4 analyzed HRS cell lines for PDGFRA, DDR2, EPHB1, TRKA, and RON. All bands shown are of expected size. In the RON Western blot, the shorter band corresponds to the processed β-chain and the longer band to the unprocessed precursor.
Western blot analysis for expression of 5 RTKs in 4 HRS cell lines. Shown are Western blots of all 4 analyzed HRS cell lines for PDGFRA, DDR2, EPHB1, TRKA, and RON. All bands shown are of expected size. In the RON Western blot, the shorter band corresponds to the processed β-chain and the longer band to the unprocessed precursor.
Activation of RTKs and aberrant tyrosine phosphorylation in primary HL
Activated RTKs are phosphorylated on several intracellular tyrosine residues.12 With an antibody specific for p-Y-720 of PDGFRA, we observed staining in 6 of 25 patients with HL expressing PDGFRA (Table 2; Figure 3). With an antibody specific for p-Y-680 and p-Y-681 of TRKA and of TRKB, we observed staining in 6 of 21 patients expressing TRKA and/or TRKB. A considerable fraction of HRS cells in positive cases were not stained, and positive and negative cells were intermingled. Taking into account technical obstacles (loss of the unstable p-Y because of an insufficient preservation of material)25,26 and biologic obstacles (not all tyrosine-residues are always phosphorylated in activated RTKs),27 it is likely that the fraction of patients with activated, phosphorylated RTKs is higher than the fraction of patients showing positivity with p-Y-RTK–specific antibodies.
Pan-p-Y antibodies detect p-Y-residues in several proteins and have been used to detect aberrant tyrosine phosphorylation in various tumors.26,28-30 With such antibodies, we observed only occasional staining of single cells in normal lymphatic tissue. Among the 86 patients with B-NHL analyzed, approximately 10% were positive for p-Y (Table 2). Staining of HRS cells was observed in 29% (11 of 38) of evaluable patients with classic HL analyzed for the expression of the 6 RTKs (Figure 3). In most patients, a fraction of HRS cells was negative, found intermingled with positive HRS cells. Pan-p-Y positivity was more pronounced in patients with NS HL (10 of 17 patients) than in those with MC HL (1 of 21 patients). The staining pattern correlated with the number of expressed RTKs because the 11 pan-p-Y-positive patients showed on average an expression of 4.4 RTKs per patient, but the 27 pan-p-Y–negative patients expressed on average only 1.9 of the RTKs analyzed. To confirm the difference between NS HL and MC HL regarding pan-p-Y positivity, we performed more staining with pan-p-Y antibodies and observed positivity in more than 50% (21 of 39) of patients with NS HL but in only 10% (6 of 60) patients with MC HL, a highly significant difference (P < .01; Pearson χ2 test with Yates continuity correction) (Table 2). Among the patients with LP HL, only 1 of 23 (4%) was found positive.
Immunohistochemical analysis of PDGFRA, DDR2, EPHB1, RON, TRKB, and TRKA expression. Immunohistochemical analysis of patients with primary HL using alkaline phosphatase and Fast Red as substrate revealed the HRS cell–specific expression of PDGFRA (A), DDR2 (B), EPHB1 (C), RON (D), TRKB (E), and TRKA (F). In positive patients, most HRS cells were stained. In a clonally related composite lymphoma, PDGFRA-specific staining was observed in the HRS cells (G) but not in the follicular lymphoma cells (H). All magnifications are 200×; those in the inserts are 400×. (I) The percentages of patients with B-NHL (n = 86; 22 mantle cell lymphomas, 15 chronic lymphocytic leukemias, 19 follicular lymphomas, 11 Burkitt lymphomas, 19 diffuse large B-cell lymphomas), LP HL (n = 23), MC HL (n = 21), and NS HL (n = 19) expressing the indicated RTKs are shown. (J-M) Pie charts showing the breakdown of patients expressing the indicated numbers of RTKs for B-NHL (J) (n = 86), LP HL (K) (n = 23), MC HL (L) (n = 21), and NS HL (M) (n = 19).
Immunohistochemical analysis of PDGFRA, DDR2, EPHB1, RON, TRKB, and TRKA expression. Immunohistochemical analysis of patients with primary HL using alkaline phosphatase and Fast Red as substrate revealed the HRS cell–specific expression of PDGFRA (A), DDR2 (B), EPHB1 (C), RON (D), TRKB (E), and TRKA (F). In positive patients, most HRS cells were stained. In a clonally related composite lymphoma, PDGFRA-specific staining was observed in the HRS cells (G) but not in the follicular lymphoma cells (H). All magnifications are 200×; those in the inserts are 400×. (I) The percentages of patients with B-NHL (n = 86; 22 mantle cell lymphomas, 15 chronic lymphocytic leukemias, 19 follicular lymphomas, 11 Burkitt lymphomas, 19 diffuse large B-cell lymphomas), LP HL (n = 23), MC HL (n = 21), and NS HL (n = 19) expressing the indicated RTKs are shown. (J-M) Pie charts showing the breakdown of patients expressing the indicated numbers of RTKs for B-NHL (J) (n = 86), LP HL (K) (n = 23), MC HL (L) (n = 21), and NS HL (M) (n = 19).
No indications for constitutive activation of RTKs in HRS cell lines
Aberrant RTK activity in tumors is frequently caused by activating mutations in the coding regions of RTKs.11 To search for potentially activating mutations, we amplified the open-reading frames of the 6 RTKs analyzed from all HRS cell lines expressing the RTKs by reverse transcription–polymerase chain reaction (RT-PCR) and directly sequenced the PCR products. For PDGFRA, DDR2, TRKA, TRKB, and EPHB1, only known polymorphisms and no potentially activating mutations were observed, whereas in RON transcripts from L428 and L1236, the same difference (R1321G) to all deposited sequences at the end of the kinase domain was found. That the same replacement was found in both cell lines most likely indicates an unknown polymorphism.
Analysis of RTK activation with p-Y–specific antibodies. Immunohistochemical analysis with antibodies specific for p-Y-720-PDGFRA (A) and p-Y-680/681-TRKA and -TRKB (B), using alkaline phosphatase and Fast Red as substrate, showed considerable HRS cell–specific staining in patients expressing the respective RTKs. In positive patients, only a fraction of HRS cells (approximately 30%-70%) was stained. Positive and negative cells were intermingled. Immunostainings with a pan-p-Y antibody using horseradish peroxidase and diaminobenzidine (DAB) as substrate revealed elevated levels of p-Y contents, comparable to tumors with activated RTKs, in the HRS cells of several patients (C-E). The same patient with HL is shown in panels E and F. (E) HRS cells are positive, (F) and pretreatment of the section with T cell phosphatase abolished staining completely. All magnifications except panel D are 200×, with magnifications in inserts at 400×. Magnification in panel D is 100×.
Analysis of RTK activation with p-Y–specific antibodies. Immunohistochemical analysis with antibodies specific for p-Y-720-PDGFRA (A) and p-Y-680/681-TRKA and -TRKB (B), using alkaline phosphatase and Fast Red as substrate, showed considerable HRS cell–specific staining in patients expressing the respective RTKs. In positive patients, only a fraction of HRS cells (approximately 30%-70%) was stained. Positive and negative cells were intermingled. Immunostainings with a pan-p-Y antibody using horseradish peroxidase and diaminobenzidine (DAB) as substrate revealed elevated levels of p-Y contents, comparable to tumors with activated RTKs, in the HRS cells of several patients (C-E). The same patient with HL is shown in panels E and F. (E) HRS cells are positive, (F) and pretreatment of the section with T cell phosphatase abolished staining completely. All magnifications except panel D are 200×, with magnifications in inserts at 400×. Magnification in panel D is 100×.
For PDGFRA, a second promotor in intron 12 has been described.31 A comparison of PCRs detecting transcripts originating from the conventional promotor and the internal promotor indicated that the internal promotor was predominantly used in HDLM2, L428, and L1236. This was in contrast to KMH2, in which the conventional promotor was primarily used, and it explains why Western blot analysis for only KMH2 revealed a product of expected size although the microarray data indicated expression in all 4 HRS cell lines. The relevance of the transcripts generated from the internal promotor is unclear; no bands of corresponding sizes were observed in Western blot analysis.
We amplified different splice forms of several RTKs. For TRKB, the previously described alternative splicing of exon 17 was observed.32 A previously described splice variant of RON, leading to receptor dimerization and enhanced migration of cells but not to cellular transformation, was found in each HRS cell line expressing this RTK.33 During RT-PCR for EPHB1 from HDLM2, the known EPHB1 product was obtained, but 2 previously unknown splice variants were amplified from L428 and L1236. In L428, exon 1 was spliced to exon 5, and in L1236, exon 1 was spliced to exon 10. In each transcript, the reading frame was preserved. The former protein lacked the ephrin-binding domain, the latter the complete extracellular and transmembrane domains. However, in Western blot analysis, no bands of corresponding sizes were observed.
In Western blot analysis with the 4 HRS cell lines, an anaplastic large cell lymphoma cell line with constitutive ALK activity (SuDHL1) and 2 EBV-immortalized B cell lines (WEI and J24) with the pan-p-Y–specific antibody PY99, relatively low p-Y contents were observed in HDLM2, KMH2, and L1236 compared with SuDHL1 and WEI (data not shown). Only in L428 was a more pronounced tyrosine phosphorylation found. This observation, together with the presence of several RTK transcripts that apparently do not code for functional RTKs, indicates that the HRS cell lines are, in contrast to HRS cells in most patients with primary HL, no longer under selective pressure for deregulated RTK signaling. This finding may be attributed to the fact that the HRS cell lines analyzed were established from pleural effusions or peripheral blood from patients who experienced several relapses and were thus independent of cellular interactions in the HL infiltrate, which are assumed to play an important role for HRS cells.
Autocrine and paracrine activation of RTKs in primary HL
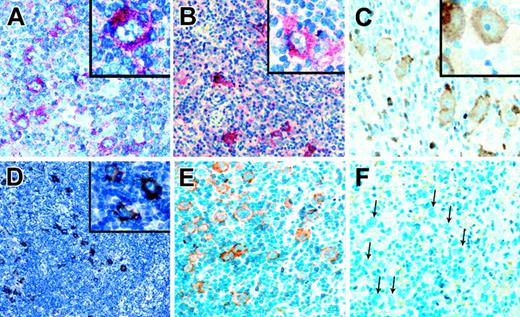
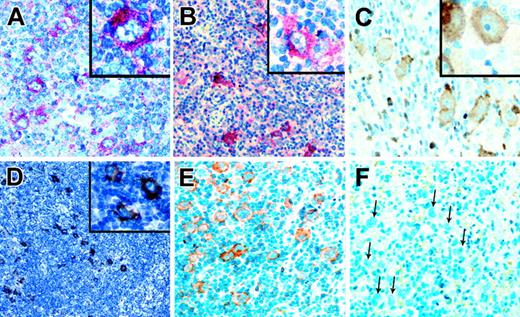
Analysis of HRS cell lines revealed no indications for constitutive RTK activation, and staining patterns with the p-Y-RTK–specific and the pan-p-Y–specific antibodies indicated that the RTKs were activated in primary HRS cells. Therefore, we tested whether the TK activity in patients with primary HL could have resulted from the presence of RTK ligands in the HL infiltrates. Ligands for DDR2 are several types of collagen, with type 1 being the most potent activator.34 Immunohistochemical analysis revealed that collagen type 1 was found in the capsule and in blood vessels of normal lymph nodes and that it was found in large amounts in the sclerotic bands of patients with classic HL. Sclerotic bands are especially abundant in NS HL (Figure 4). Collagen type 1, the high-affinity ligand for DDR2, is thus present near HRS cells in most patients with classic HL. Nerve growth factor (NGF), the ligand for TRKA, can be produced by several myeloid cell types.35 In normal lymph nodes, few cells expressing NGF were observed, whereas in classic HL, NGF-producing granulocytes were frequently found (Figure 4). Thus, sufficient amounts of ligand for TRKA activation may be available in classic HL.
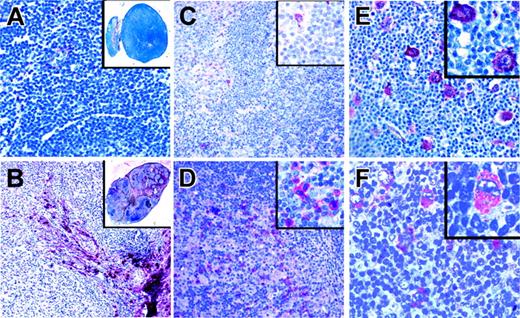
Immunostaining of classical HL for ligands of aberrantly expressed RTKs. Immunostaining of a lymph node with reactive lymphoid hyperplasia (A) and of a classical HL (B) for collagen type 1, the high-affinity ligand for DDR2, using alkaline phosphatase. Although only the capsule and occasional vessels are stained in lymphadenitis, sclerotic bands containing collagen type 1 and branching into the tumor infiltrate are numerous in HL. Immunostaining of a lymph node in a patient with reactive hyperplasia (C) and classical HL (D) for NGF, the TRKA ligand. NGF-positive cells were rarely observed in the reactive lymph node, whereas such cells were frequent in the tumor infiltrate. (E) Immunostaining of a classical HL for PDGFA, a high-affinity ligand for PDGFRA. PDGFA is expressed by HRS cells in a patient also expressing PDGFRA. (F) Immunostaining in a patient with classical HL for EphrinB1, a ligand for EPHB1, found to be expressed by HRS cells of several patients expressing EPHB1. All magnifications are 100×. Inserts in panels A and B are 0.5×; those in panels C-F are 400×.
Immunostaining of classical HL for ligands of aberrantly expressed RTKs. Immunostaining of a lymph node with reactive lymphoid hyperplasia (A) and of a classical HL (B) for collagen type 1, the high-affinity ligand for DDR2, using alkaline phosphatase. Although only the capsule and occasional vessels are stained in lymphadenitis, sclerotic bands containing collagen type 1 and branching into the tumor infiltrate are numerous in HL. Immunostaining of a lymph node in a patient with reactive hyperplasia (C) and classical HL (D) for NGF, the TRKA ligand. NGF-positive cells were rarely observed in the reactive lymph node, whereas such cells were frequent in the tumor infiltrate. (E) Immunostaining of a classical HL for PDGFA, a high-affinity ligand for PDGFRA. PDGFA is expressed by HRS cells in a patient also expressing PDGFRA. (F) Immunostaining in a patient with classical HL for EphrinB1, a ligand for EPHB1, found to be expressed by HRS cells of several patients expressing EPHB1. All magnifications are 100×. Inserts in panels A and B are 0.5×; those in panels C-F are 400×.
Immunohistochemical analysis for PDGFA and EphrinB1, the ligands for PDGFRA and EPHB1, respectively, revealed that neither ligand was detected in normal B cells but that each was expressed in HRS cells in 40% and 29% of patients, respectively (Figure 4). Coexpression of PDGFA and PDGFRA was found in 13 of 18 patients expressing PDGFRA. PDGFA was not expressed in any of 14 patients negative for PDGFRA. For EphrinB1 and EPHB1, coexpression was found in 12 of 20 patients expressing EPHB1 but in only 4 of 35 patients negative for the receptor-expressed EphrinB1. Regarding the frequencies of receptor-positive patients coexpressing the respective ligands in HRS cells, no significant differences between NS HL and MC HL were observed. Among patients coexpressing receptors and the respective ligands, most had high cellular p-Y content. Of the PDGFRA/PDGFA– and the EPHB1/EphrinB1–coexpressing patients, 14 (67%) of 21 were positive in pan-p-Y stainings compared with 27 (27%) of 99 patients with classic HL (NS and MC) and 21 (50%) of 39 patients with NS HL.
Taken together, these findings indicate that the activation of the aberrantly expressed RTKs in classic HL could be caused by the presence of high-affinity ligands in the HL infiltrate, for DDR2 and TRKA in a paracrine fashion, and for PDGFRA and EPHB1 in an autocrine fashion.
Sensitivity of the PDGFRA-expressing HL cell line KMH2 to imatinib
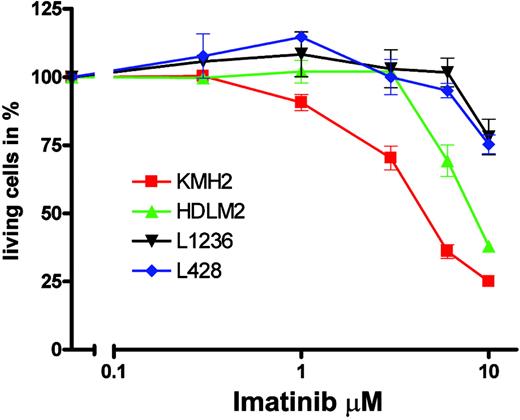
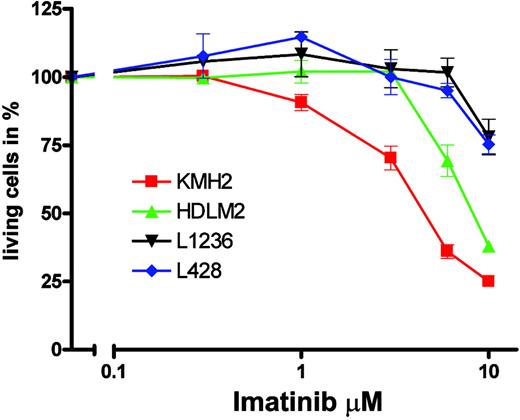
Imatinib specifically inhibits the TK activities of BCR-ABL, KIT, PDGFRA, and PDGFRB by competing with adenosine triphosphate (ATP) binding to the catalytic center of these kinases.21 KMH2 apparently expressed the full-length PDGFRA protein, and enzyme-linked immunosorbent assay (ELISA) for PDGFA showed that KMH2 cells also expressed PDGFA (data not shown), indicating that autocrine PDGFRA activation could have contributed to KMH2 survival and proliferation. Therefore, we tested whether imatinib could inhibit the proliferation of KMH2 cells. When cells were grown in media supplemented with 10% FCS, we observed no effect of imatinib on numbers of living cells with concentrations (0.1-3 μM) causing TK-specific growth inhibition in cell lines carrying BCR-ABL translocations, constitutively activated PDGFRA, or cell lines dependent on autocrine PDGF stimulation.21,36-38 This finding showed that KMH2 cell survival and proliferation is not essentially dependent on PDGFRA activation. However, autocrine PDGFRA stimulation could have a more subtle effect on KMH2 survival and proliferation recognizable only if cells were grown at low serum concentrations. We thus analyzed all 4 HL cell lines in media supplemented with 1% FCS and imatinib in the 0.3- to 10-μM range (Figure 5). Without imatinib, the 70-hour incubation in 1% FCS had no significant effect on fractions of living cells compared with incubation in 10% (KMH2, L1236, and L428) or 20% (HDLM2) FCS, and all cells proliferated. Significant effects of imatinib on L1236, L428, and HDLM2 were only observed at concentrations usually regarded to inhibit cell growth unspecifically. For KMH2, cell numbers were reduced to 50% at a concentration between 3 and 6 μM, which is above the concentration usually regarded as specific inhibition (less than 2 μM)21 but below the concentration normally regarded to cause unspecific toxicity (more than 10 μM). The finding that only KMH2, the HL cell line with functional PDGFRA expression, shows clearly the greatest sensitivity to imatinib among the HL cell lines may suggest specific inhibition.
Sensitivity of HL cell lines to imatinib. The 4 HL cell lines KMH2 (▪), HDLM2 (▴), L1236 (▾), and L428 (♦) were incubated for 70 hours with the indicated imatinib concentrations. After incubation in propidium iodide, numbers of living cells were determined in a FACScan. Experiments were repeated 3 times, and a representative experiment with 3 replicates and standard derivation is shown in a semi-logarithmic plot. Numbers of living cells are shown as fractions (in percentages) of the untreated control.
Sensitivity of HL cell lines to imatinib. The 4 HL cell lines KMH2 (▪), HDLM2 (▴), L1236 (▾), and L428 (♦) were incubated for 70 hours with the indicated imatinib concentrations. After incubation in propidium iodide, numbers of living cells were determined in a FACScan. Experiments were repeated 3 times, and a representative experiment with 3 replicates and standard derivation is shown in a semi-logarithmic plot. Numbers of living cells are shown as fractions (in percentages) of the untreated control.
Taken together, these data show that KMH2 survival and proliferation are not essentially dependent on PDGFRA signaling. Even if the imatinib effect seen at low serum concentrations is regarded as PDGFRA specific, autocrine PDGFRA stimulation alone has only a minor effect on KMH2 survival and proliferation. Given the aberrant coexpression of several RTKs in KMH2 (DDR2 and FGFR2 expression were shown by Western blot analysis, and TRKB expression was shown by microarray and RT-PCR) and the overlapping signaling pathways activated by different RTKs, the minor effect of imatinib may be attributed to the compensation of a specific inhibition of PDGFRA signaling by the activities of the other RTKs.
Discussion
Although RTKs are frequently involved in cellular transformation in several different neoplasias, analysis has been limited to a few studies of individual RTKs in HL. Reports about KIT and HER2 were controversial, and our microarray analysis indicated that neither is not expressed in HRS cell lines. The reported expression of MET in HRS cells is likely not aberrant because MET expression was also observed using similar methods in normal germinal center B cells, from which HRS cells are derived.18,39-43,60 Thus far, only aberrant PDGFRA expression has been observed in the HRS cells of 10 patients analyzed.44 Moreover, a previous analysis of several lymphomas with a pan-p-Y antibody showed HRS cell positivity in 2 of 7 patients.45
Based on a systematic analysis of global RNA expression data of 4 HRS cell lines, we show here that the RTKs PDGFRA, DDR2, EPHB1, RON, TRKB, and TRKA are aberrantly expressed and frequently coexpressed in HRS cells of most patients with primary classic HL. Although we did not recognize a specific pattern of coexpression, the expression of the 6 RTKs was largely HL specific among lymphomas derived from mature B cells. This HL-specific expression was further stressed by HRS cell–restricted expression of PDGFRA in 2 composite follicular/Hodgkin lymphomas with common tumor clone precursors. Furthermore, for PDGFRA, TRKA, and TRKB, we could demonstrate that the RTKs are activated in the tumor cells. Expression of the RTKs correlated with increased p-Y content of HRS cells, comparable to other tumors with aberrantly activated TKs such as ALK-positive anaplastic large-cell lymphoma or KIT-positive gastrointestinal stromal tumors (data not shown). Aberrant TK activity was more pronounced in NS HL than in MC HL. Interestingly, the activation of RTKs seemed largely to be ligand induced, probably in a paracrine fashion for DDR2 and TRKA and in an autocrine fashion for PDGFRA and EPHB1. We conclude from these observations that although no genetic changes in the RTKs were identified, aberrant RTK signaling is, as it is in several other tumor types, important for the pathogenesis of classic HL.
Compared with other tumors with pronounced TK activity, HL has some unique features. Thus, in most tumors with activated TKs, single aberrantly activated or expressed TKs have been identified such as HER2 in carcinoma of the breast and KIT or PDGFRA in gastrointestinal stromal tumors.46,47 In classic HL, however, as many as 6 different RTKs were aberrantly coexpressed. Another intriguing finding is that activation is likely ligand induced, at least for 4 RTKs in which analysis was possible. PDGFRA and EPHB1 are activated in an autocrine fashion, and DDR2 and TRKA are likely activated in a paracrine fashion. Autocrine and paracrine activation of different receptor types, such as cytokine or tumor necrosis factor (TNF) receptors, has previously been shown for HL,48 and autocrine and paracrine RTK activation has been previously observed in other disease settings.49,50 However, ligand-induced activation of 4 different RTKs in one entity is unprecedented.
Lp HL is different from classic HL, and this difference is reflected in the RTK expression pattern. Although overall expression of the 6 RTKs in LP HL was greater than it was in other mature B cell lymphomas, expression was less frequent than it was in classic HL. PDGFRA, the most frequently expressed RTK in classic HL, was only found in a small fraction of patients and was not the most frequently RTK found. The distinction between NS and MC classic HL subtypes is based on histologic examination. The more pronounced RTK expression and the higher TK activity in NS HL may indicate that the different histology is correlated with the preferential activation of different signaling pathways in the HRS cells of both subtypes.
The aberrant coexpression of several RTKs (up to 6 different RTKs in individual patients) and of the ligands in some patients prompts the question about the causes of these aberrant expression patterns. Common signaling pathways regulating the transcription of several RTKs and their ligands are unknown. In addition, with the exception of DDR2 and TRKA, both located on chromosome 1q12-23 and 1q21, respectively, all other RTKs are located on different arms (RON 3p21.3 and EPHB1 3q21-23) of the same chromosome or on different chromosomes (PDGFRA 4q12; TRKB 9q22.1). In addition, PDGFA and EphrinB1 are not located on the same chromosomes as their receptors (PDGFA 7p21.1; EphrinB1 Xq12). Individual genetic changes such as translocations or amplifications, which could cause deregulated expression, would thus largely affect only one of the RTKs or ligands. However, because multiple numeric imbalances are frequent in HRS cells of individual patients, several different amplifications may lead to the aberrant coexpression of different RTKs or ligands, although the above-mentioned genomic loci are not among those frequently amplified in classic HL.51 Another cause could be the disturbed regulation of gene expression in HRS cells. Aberrant expression of markers of several hematopoietic lineages is a well-known characteristic of HRS cells,2 and, interestingly, most of the aberrantly expressed RTKs are also expressed in different hematopoietic cell types.52-55
At present we can only speculate about the consequences of aberrant RTK activity in HRS cells. An intriguing question is whether the functions of the different RTKs in HL are redundant or distinct. For several of the RTKs, more or less specific functions have been described, such as a role in tissue remodeling for DDR2 or enhanced cell migration for RON, whereas TRKA and TRKB activities are essential for the differentiation and survival of neurons.56-58 However, the effects of RTK activation are often cell type dependent. Because of the largely overlapping intracellular pathways activated by the various RTKs,12 it is also conceivable that their effects in HRS cells are largely redundant and additive on coexpression. It may also be noteworthy that for patients with EPHB1/EphrinB1 coexpression, autocrine activation may not only occur through EPHB1 signaling but also through EphrinB1 because the membrane-bound B-type Ephrins also have the capacity to activate intracellular signaling pathways.59
In summary, we show here that 6 different RTKs are aberrantly expressed and activated in HRS cells of classic HL, probably in paracrine and autocrine fashions, and are most pronounced in NS HL. Although no genetic changes causing aberrant RTK activity were identified, the aberrant RTK expression and activity in patients with classic HL, highlighted by HRS cell–specific expression in 2 composite lymphomas, indicates the importance of aberrant RTK signaling for HL and implies that tyrosine kinase inhibitors may be potential therapeutic agents for classic HL, especially for NS HL.
Prepublished online as Blood First Edition Paper, January 27, 2005; DOI 10.1182/blood-2004-10-4008.
Supported by the DFG through grants KU1315/2-1, BR1238/6-1, and WI2015/1-1, and the IFORES program of the University of Essen Medical School.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Yvonne Blum and Sabine Albrecht for excellent technical assistance, John Ferebee for help with the statistical analysis, and E. Puccetti and M. Ruthardt for help with the imatinib experiments.