Comment on De Keersmaecker et al, page 4849
A novel fusion of ABL1 to EML1 adds to a growing list of chimeric ABL proteins in patients with acute T-cell lymphoblastic leukemia.
Until recently, our recognition of the involvement of the ABL1 gene (which encodes the nonreceptor protein tyrosine kinase c-ABL1) in the pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL) was limited to rare cases of Philadelphia chromosome-positive T-ALL, some of which likely represented lymphoid blast crisis of chronic myeloid leukemia. The first hint that ABL1 might be involved more frequently in this disease came from a United Kingdom Medical Research Council study that used fluorescent in situ hybridization (FISH) with probes from the ABL1 locus to analyze interphase nuclei from lymphoid leukemia blasts. The study was intended to detect cryptic BCR-ABL1 fusion but instead found extrachromosomal amplification of the ABL1 locus independent of BCR in 8 of 280 T-ALL samples.1 The nature of the amplification was soon revealed when researchers from the University of Leuven in Belgium demonstrated that ABL1 was fused to NUP214 via episomal amplification in 5 of 90 patients with T-ALL.2 This resulted in fusion of N-terminal sequences of the NUP214 protein, a ubiquitously expressed component of the nuclear pore complex, with the same 1104 C-terminal amino acids of c-ABL1 found in the BCR-ABL1 fusion protein. Like BCR-ABL1, the NUP214-ABL fusion is a constitutively active tyrosine kinase with transforming activity in vitro. Collectively, these results suggested that ABL1 might be involved in the pathogenesis of 5% to 6% of T-ALLs.
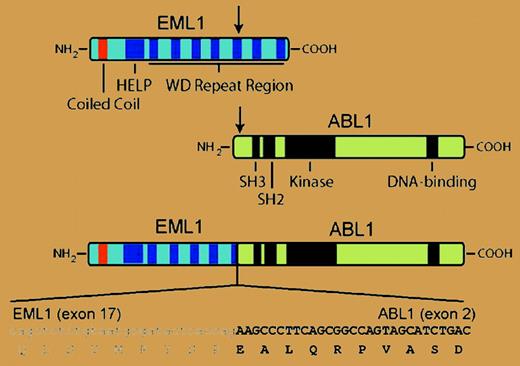
In the current issue of Blood, the Leuven group extends these results to identify a new ABL1 fusion gene in T-ALL. A patient with T-ALL whose blasts lacked ABL1 amplification but demonstrated a split in the hybridization signal between 5′ and 3′ ABL1 probes was found to have cryptic t(9;14), leading to fusion of ABL1 with EML1 on chromosome 14. EML1 encodes a protein with similarity to an echinoderm microtubule-associated protein (EML1), and the resulting 190-kDa echinoderm microtubule-associated protein-like 1-Abelson 1 (EML1-ABL1) fusion is a dysregulated tyrosine kinase that alleviates interleukin-3 dependence in Ba/F3 hematopoietic cells and constitutively activates extracellular signal-related kinase (ERK), signal transducers and activators of transcription 5 (Stat5), and Src signaling pathways. As with BCR-ABL1, EML1-ABL kinase activity was dependent on a coiled-coil domain in the N-terminus, suggesting that oligomerization and autophosphorylation are necessary to overcome the autoinhibition of the ABL kinase domain.3 The leukemic cells from this patient exhibited ectopic expression of the homeobox transcription factor TLX1 and hemizygous deletion of the CDKN2A tumor suppressor gene. These alterations were also seen frequently in patients with NUP214-ABL1 fusion2 and provide more evidence that T-ALL can be divided into subgroups with distinct molecular pathogenesis.4 Interestingly, expression of EML1 was not observed in other T-ALL blasts or cell lines, suggesting that the EML1 promoter may be activated as a consequence of the translocation.FIG1
In T-ALL with cryptic t(9;14), N-terminal sequences from the EML1 polypeptide, including a coiled-coil domain, are joined to the same C-terminal ABL sequences present in the more common BCR-ABL1 fusion protein. The resulting 190-kDa EML1-ABL1 fusion protein is a dysregulated tyrosine kinase that transforms Ba/F3 cells. For details, see the article beginning on page 4849.
In T-ALL with cryptic t(9;14), N-terminal sequences from the EML1 polypeptide, including a coiled-coil domain, are joined to the same C-terminal ABL sequences present in the more common BCR-ABL1 fusion protein. The resulting 190-kDa EML1-ABL1 fusion protein is a dysregulated tyrosine kinase that transforms Ba/F3 cells. For details, see the article beginning on page 4849.
These findings have therapeutic implications, of course. The EML1-ABL1 fusion protein was inhibited by imatinib mesylate, the small molecule inhibitor of ABL kinase activity, with approximately the same sensitivity as BCR-ABL1, raising the possibility that patients with T-ALL with NUP214-ABL1 and EML1-ABL1 might respond clinically to imatinib treatment. In the previous study, imatinib was found to inhibit the growth of a T-ALL cell line with the NUP214-ABL1 fusion,2 but the clinical utility of imatinib in these patients might be limited by rapid selection for resistance, as is observed in Philadelphia chromosome-positive patients with B-ALL with amplification of BCR-ABL1.5 Patients with ABL1 fusions from balanced translocations might fare better. Finally, the search for activated ABL1 genes in T-ALL is not over. Although the FISH studies1,2 suggest that there are no other frequent ABL1 amplifications in acute lymphoid leukemias, additional cryptic ABL1 translocations will no doubt continue to pop up. In addition, point mutations in ABL1 can also dysregulate c-ABL1 kinase activity and cause lymphoid leukemia in mouse models,6 but finding these cases will require direct exon sequencing of genomic DNA. ▪