Abstract
Granzyme B (GzmB), a serine protease of cytotoxic T lymphocytes and natural killer (NK) cells, induces apoptosis by caspase activation after crossing the plasma membrane of target cells. The mechanism of this translocation during killer cell attack, however, is not understood. Killer cells release GzmB and the membrane-disturbing perforin at the contact site after target recognition. Receptor-mediated import of glycosylated GzmB and release from endosomes were suggested, but the role of the cation-independent mannose 6-phosphate receptor was recently refuted. Using recombinant nonglycosylated GzmB, we observed binding of GzmB to cellular membranes in a cell type–dependent manner. The basis and functional impact of surface binding were clarified. GzmB binding was correlated with the surface density of heparan sulfate chains, was eliminated on treatment of target cells with heparinase III or sodium chlorate, and was completely blocked by an excess of catalytically inactive GzmB or GzmK. Although heparan sulfate–bound GzmB was taken up rapidly into intracellular lysosomal compartments, neither of the treatments had an inhibitory influence on apoptosis induced by externally added streptolysin O and GzmB or by natural killer cells. We conclude that membrane receptors for GzmB on target cells are not crucial for killer cell–mediated apoptosis.
Introduction
Granzymes are a family of granular serine proteases expressed by cytotoxic T lymphocytes (CTLs) and NK cells implicated in immune defense reactions. Granzyme B (GzmB), the most prominent member of this family, induces apoptosis of target cells after cytosolic delivery by caspase-dependent and -independent pathways, resulting in the activation of effector caspases and mitochondrial depolarization, respectively.1,2 The mechanism of translocation across the plasma membrane, however, is poorly understood. In vivo, the apoptotic functions of granzymes strictly depend on the membrane-binding protein perforin. Mice without the functional perforin gene display a strong reduction of granular cytotoxicity.3-7 Although perforin can form lytic pores across membranes at high concentrations, such as the structurally related C9 component of the terminal complement complex,8 sublytic concentrations of perforin already synergize with granzymes in the absence of transmembrane pores.9,10 In vitro, the sublytic activity of perforin can be replaced by other pore-forming proteins, such as streptolysin O (SLO) and pneumolysin.9 The latter agents also cooperate in an unknown manner with GzmB at sublytic concentrations without generating pores for the delivery of proteins in the size range of granzymes.9
Although membrane-bound perforin is able to generate nonspecific calcium channels but no open pores across the membranes of nucleated target cells, it is difficult to understand how macromolecules of 25 kDa, such as nonglycosylated GzmB, can reach the cytosol. Endocytosis and redistribution of externally added GzmB inside the target cell occurs rapidly, within 15 minutes, even in the absence of perforin.11 Cells loaded with GzmB, however, do not undergo apoptosis until they receive a second signal by perforin, adenoviral particles,11 or bacterial membrane–binding toxins.9 The continuous presence of soluble GzmB in the extracellular fluid is not required to initiate the killing of target cells with the help of sublytic amounts of perforin.11,12 Because GzmB translocates to the cytosol in combination with agents such as listeriolysin O or adenovirus, which disturb the endosomal membrane after endocytosis, specific uptake and transport of GzmB to endosomal compartments9,11-13 through cell surface receptors was suggested.
Subsequent studies identified the cation-independent mannose-6-phosphate receptor (CI-MPR)14 as a GzmB-internalizing receptor that binds to terminal mannose-6-phosphates of glycosylated killer cell–derived GzmB. In an extensive study, the importance of the CI-MPR for CTL-mediated GzmB-dependent killing of target cells was recently disproved,15 and several lines of experimental evidence point toward a CI-MPR–independent translocation of GzmB to the cytosol. CTLs from patients with I-cell disease lack phosphotransferase and, therefore, cannot synthesize mannose-6-phosphate–containing GzmB, yet their CTLs kill target cells as efficiently as wild-type CTLs.16 Using dominant-negative dynamin mutants, endocytosis of the CI-MPR was prevented without suppressing target cell killing.17 Redistribution of GzmB into endosomal compartments was still reported, but the involvement and functional impact of alternative receptors mediating this uptake was not investigated.
Recently, we18 and others19,20 found that recombinant nonglycosylated GzmB can be exploited to kill certain tumor cell lines in the absence of a perforin-induced second signal. Retargeting GzmB to internalizing cell surface receptors with the help of specific antibodies or ligands resulted in apoptosis induction with promising efficacy. During our previous study we encountered the unexpected binding of nonglycosylated recombinant GzmB to the plasma membranes of different cell types already at relatively low concentrations. Intriguingly, GzmB bound most strongly to monocytes and B cells, but not to NK or T cells. Because GzmB is a basic protein with a calculated isoelectric point (IEP) of 10.4, we hypothesized that ionic interactions with negatively charged glycosamino-glycans (GAGs) on cell surfaces might play a role in cellular redistribution and target cell killing. In particular, we assumed that GAGs might act as receptors for rapid internalization and accumulation of granzymes into endosomal vesicles. Here, we show that binding of recombinant GzmB on K562 or HL-60 cells is mediated exclusively by membrane-bound heparan sulfate (HS) chains. These HS structures, however, did not affect the efficacy of GzmB delivery into the cytosol of target cells by sublytic SLO concentrations or natural killer cells in killing experiments.
Materials and methods
Granzymes
All recombinant granzymes were expressed, refolded, converted, and purified as previously described.18,21,22 The concentration of all recombinant or labeled proteins was determined spectroscopically using the appropriate extinction coefficient at 280 nm. Native glycosylated GzmB purified from the NK YT-Indy cell line was a kind gift of Chris Bleackley (Edmonton, AB, Canada). Enzymatic activity of GzmB preparations was measured as described.18
Cell culture
K562, HL-60, Jurkat, and U937 cells were maintained as described.18 The human NK cell line NK92 was kindly provided by Eric Vivier (Marseille, France) and was maintained in RPMI containing GlutaMax 15% fetal bovine serum (FBS), 100 U/mL penicillin/streptomycin, and 200 U/mL interleukin-2 (IL-2).
Antibodies for FACS staining
Mouse monoclonal antibodies and the dilutions used for fluorescence-activated cell sorter (FACS) experiments and confocal immunochemistry are given in Table 1.
Preparation and differentiation of human leukocytes by flow cytometry
Peripheral blood mononuclear cells (PBMCs) and granulocytes from EDTA (ethylenediaminetetraacetic acid) blood were prepared by Lymphoprep density gradient centrifugation (Nycomed, Oslo, Norway). Erythrocyte lysis of the pellet, (granulocytes and erythrocytes) was performed in hypotonic ACK buffer (150 mM NH4Cl, 1 mM KHCO3, 0.1 mM Na2 EDTA, pH 7.4). Neutrophils were defined as CD16+ cells of the granulocyte gate. CD4+ T cells and CD8+ T cells were defined as those lymphocytes expressing high levels of CD4 and CD8, respectively. B cells were defined as CD19+ and monocytes as CD14+ lymphocytes. NK cells were defined as CD16+ small lymphocytes.
Biotinylation and labeling with Alexa 633
Recombinant GzmB, GzmK, and human iron-saturated holo-transferrin (Sigma-Aldrich, Deisenhofen, Germany) were biotinylated with Biotin X-NHS (Calbiochem, La Jolla, CA) at room temperature and were dialyzed against phosphate-buffered saline (PBS). Enzymatic activity of biotinylated GzmB against the peptide substrate Ac-IEPD-pNA was retained to approximately 80%. Similarly, biotinylated GzmB was able to induce apoptosis at comparative levels (data not shown). Enzymatically inactive GzmBS195A was labeled with AlexaFluor 633 (Molecular Probes, Eugene, OR), according to the supplier's protocol.
Binding of biotinylated GzmB or transferrin to cells and detection
Cells (0.5-2 × 105) were incubated for 1 hour at 4°C on 96-well plates with GzmB-Bio or TF-Bio in FACS medium (PBS, 3% FBS, 0.01% NaN3) in a volume of 25 to 50 μL at 10 μg/mL if not otherwise indicated. Thereafter cells were washed twice and subsequently stained with streptavidin-phycoerythrin (SA-PE; 3.33 μg/mL; Becton Dickinson Biosciences [BD], Heidelberg, Germany) for 15 minutes at 4°C. In double-staining experiments, the respective fluorescein isothiocyanate (FITC)–labeled antibodies were added to the SA-PE dilutions. Thereafter the cells were washed, resuspended in FACS medium containing 1 μg/mL propidium iodide (PI), and analyzed on a FACScalibur system (BD). Usually, 10 000 cells of the respective live gate were recorded. For quantitative comparisons of GzmB-Bio or TF-Bio binding, the geometric mean fluorescence intensity of PI-negative cells was determined. Dead cells were always excluded from FACS analyses.
Immunocytochemistry and confocal analysis
Binding of GzmB-Bio to PBMCs and staining was performed as described in the previous paragraph. SA-Cy3 (Caltag Laboratories, Burlingame, CA) was used at 1.33 μg/mL. Thereafter, cells were washed twice, and cytospins were prepared. Slides were dried, fixed with 4% paraformaldehyde in PBS, and rinsed 3 times with PBS. Samples were mounted using Mowiol mountain medium (Sigma-Aldrich). Confocal images were acquired using constant settings on a Leica TCS NT upright fluorescence microscope with a 63 × 1.32 oil objective using FITC and Cy3 filter sets and Leica TCS NT software version 2.585 (Leica Microsystems GmbH, Bensheim, Germany). Cells were analyzed using 10 to 20 images of planes spaced at 1 μm. Images were digitized and adjusted using Adobe Photoshop software (Adobe Systems, San Jose, CA).
Inhibition of binding
For inhibition experiments we used leukocyte elastase (Serva, Heidelberg, Germany), proteinase 3 (PR3) (Athens Research and Technology, Athens, GA) purified from human neutrophils, iC3B (Calbiochem), cationic trypsin (Sigma-Aldrich), chicken egg white lysozyme (Sigma-Aldrich), native GzmB, recombinant catalytically inactive GzmBS195A, nonlabeled recombinant GzmB, and recombinant GzmA and GzmK. Recombinant GzmK was inactivated with H-D-Phe-Pro-Arg-chlorometlyl ketone23 (FPR-CMK; Bachem, Heidelberg, Germany) at a 50-fold molar excess at room temperature for 3 hours and then dialyzed against PBS. The dialyzed GzmK-FPR-CMK was shown to be enzymatically inactive using Nα-CBZ-l-lysine thiobenzyl ester (BLT) as a substrate measured, as described21 (data not shown). Similarly, trypsin was inactivated using the irreversible inhibitor Tos-Lys-chloromethylketone (TLCK; Bachem) at a 100-fold molar excess. The integrity of trypsin-TLCK and lack of activity was confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining and by activity measurements as performed with GzmK (data not shown). Cells were preincubated with the indicated nonlabeled proteins at 4°C for 10 minutes, and then GzmB-Bio was added.
Heparinase III treatment
After washing in HS buffer (20 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] pH 7.2, 140 mM NaCl, 0.1 mg/mL bovine serum albumin [BSA], 5 mM CaCl2), cells at a density of 16 × 106 cells/mL were incubated with 10 Sigma U/mL heparinase III (EC4.4.4.8; Sigma-Aldrich) or 40 mU/mL (EC4.4.4.8; Calbiochem) for 20 minutes at 37°C in HS buffer. Then the cell suspension was diluted 2-fold by adding RPMI medium without serum and was incubated for an additional 20 minutes.
Sodium chlorate treatment
To inhibit sulfation of HS chains, HL-60 cells were treated with sodium chlorate (NaClO3; Baker, Deventer, The Netherlands) at 50 mM for 36 hours. After complete withdrawal of sodium chlorate, sulfation of carbohydrate chains reoccurred after 4 to 5 hours (data not shown). In view of this observation, we treated HL-60 cells for 36 hours and reduced the sodium chlorate concentration to 10 mM for subsequent killing experiments.
Apoptosis and DNA fragmentation (3H-thymidine release) assays
Apoptosis assays were performed as described.18 The fragmentation assay and calculation of specific 3H-thymidine release was essentially performed as reported.15 Five thousand labeled target cells were preincubated for 10 minutes at room temperature with the indicated substances. Coincubation with NK cells was performed for 3.5 hours in the presence or absence of various test substances. Spontaneous release was always lower than 7%.
Results
Differential binding of GzmB to cell lines and leukocytes
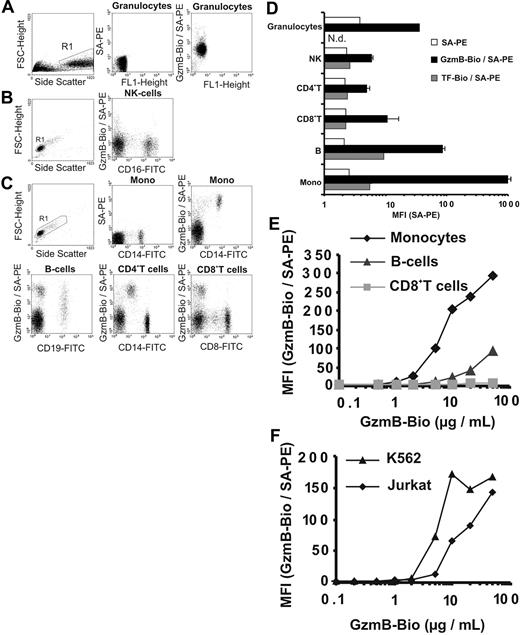
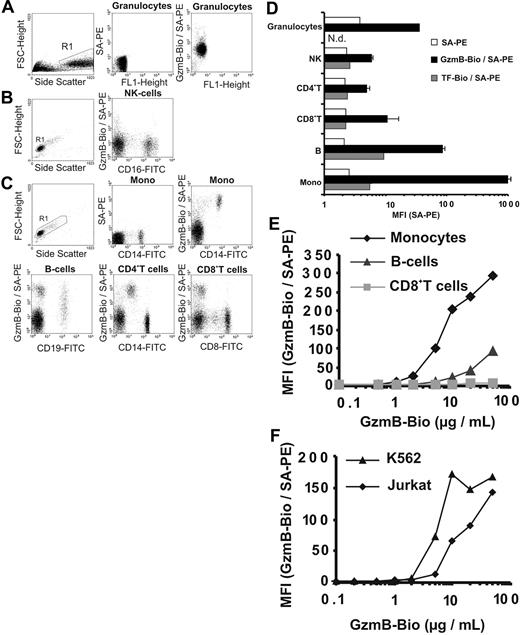
Recombinant active GzmB was generated in Escherichia coli and was prepared as described.18,22 Its enzymatic activity was similar to native glycosylated GzmB (data not shown). We investigated CI-MPR–independent binding of biotinylated recombinant GzmB (GzmB-Bio) to plasma membranes at 4°C, which was visualized by staining with streptavidin-PE (SA-PE) and was analyzed by flow cytometry (FACS). First, binding to freshly prepared human PBMCs, granulocytes (Figure 1A-E), and cell lines (Figure 1F) was examined. Granulocytes were loaded with GzmB-Bio homogeneously, but only at an intermediate level (Figure 1A, D). NK cells, CD4+ cells, and CD8+ T cells showed weak to no binding of GzmB-Bio (Figures 1B-D). In contrast, B cells and especially monocytes bound GzmB-Bio strongly. Whereas monocytes were highly homogeneous with regard to GzmB binding, B cells displayed heterogeneous binding properties (Figure 1C, left lower panel). To exclude unspecific membrane binding of biotinylated ligands, we examined the binding of biotinylated-transferrin (TF-Bio), which was low in all PBMC populations and did not follow the pattern of GzmB binding (Figure 1D, dark gray columns). In additional experiments with human PBMCs, we determined the degree of GzmB binding to monocytes, B cells, CD8+ T cells, and the Jurkat and K562 cell lines at different GzmB-Bio concentrations ranging from 0.1 μg/mL to 50 μg/mL (Figure 1E-F). Monocytes started to become positive on incubation with 1 μg/mL. Binding to B cells was comparatively low up to concentrations of 10 μg/mL. Moreover, CD8+ T cells remained negative, even at very high GzmB concentrations of 50 μg/mL. The titration curve for the 3 cell populations and the binding of GzmB to granulocytes was similar when these cells were prepared from another donor. Although the absolute mean fluorescence intensity (MFI) values varied between individual experiments, the relative signal intensities obtained for different cell populations were highly reproducible. As with monocytes, binding to Jurkat and K562 cells was observed at GzmB-Bio concentrations greater than 1 μg/mL (Figure 1F). GzmB bound slightly better to K562 cells than to Jurkat cells and reached saturation at approximately 10 μg/mL. The intriguing finding was that all subpopulations of lymphocytes, which are able to produce granzymes, had the lowest capacity to bind GzmB.
Binding of recombinant biotinylated GzmB (GzmB-Bio) to human leukocytes and cell lines analyzed by flow cytometry (FACS). (A-D) Different leukocyte subpopulations show distinct GzmB-binding patterns. (A-C) Determination of human leukocyte subpopulations and representative FACS analyses of GzmB-Bio (10 μg /mL) binding. Living cells from the chosen region (R1) were analyzed. For granulocytes and lymphocytes, background staining with SA-PE only is shown. (A) Granulocytes homogeneously bind GzmB-Bio at a low level. (B) NK cells do not bind GzmB-Bio. (C) The different lymphocyte subpopulations display strong variations in GzmB binding. Monocytes show strong and homogeneous, B cells show heterogeneous, and CD4+ and CD8+ T cells show lowest GzmB-Bio binding. Note that the cells, which are CD4low and bind GzmB-Bio strongly, are monocytes. (D) Quantitative triplicate analysis of the experiment shown in Figure 2A-C. Bars showing GzmB-Bio/SA-PE (▪) represent the average MFI with standard deviation. As specificity control, we used biotinylated transferrin (TF-Bio; ▦). SA-PE alone is indicated by □. Note the logarithmic data presentation. (E) Titration experiment with different GzmB-Bio concentrations binding to distinct human lymphocyte subpopulations.  representes monocytes;
representes monocytes;  , B cells; and ▦, CD8+ T cells, (F) Jurkat (
, B cells; and ▦, CD8+ T cells, (F) Jurkat ( ) and K562 cells (▴) bind GzmB-Bio in a concentration-dependent manner. Mono indicates monocytes.
) and K562 cells (▴) bind GzmB-Bio in a concentration-dependent manner. Mono indicates monocytes.
Binding of recombinant biotinylated GzmB (GzmB-Bio) to human leukocytes and cell lines analyzed by flow cytometry (FACS). (A-D) Different leukocyte subpopulations show distinct GzmB-binding patterns. (A-C) Determination of human leukocyte subpopulations and representative FACS analyses of GzmB-Bio (10 μg /mL) binding. Living cells from the chosen region (R1) were analyzed. For granulocytes and lymphocytes, background staining with SA-PE only is shown. (A) Granulocytes homogeneously bind GzmB-Bio at a low level. (B) NK cells do not bind GzmB-Bio. (C) The different lymphocyte subpopulations display strong variations in GzmB binding. Monocytes show strong and homogeneous, B cells show heterogeneous, and CD4+ and CD8+ T cells show lowest GzmB-Bio binding. Note that the cells, which are CD4low and bind GzmB-Bio strongly, are monocytes. (D) Quantitative triplicate analysis of the experiment shown in Figure 2A-C. Bars showing GzmB-Bio/SA-PE (▪) represent the average MFI with standard deviation. As specificity control, we used biotinylated transferrin (TF-Bio; ▦). SA-PE alone is indicated by □. Note the logarithmic data presentation. (E) Titration experiment with different GzmB-Bio concentrations binding to distinct human lymphocyte subpopulations.  representes monocytes;
representes monocytes;  , B cells; and ▦, CD8+ T cells, (F) Jurkat (
, B cells; and ▦, CD8+ T cells, (F) Jurkat ( ) and K562 cells (▴) bind GzmB-Bio in a concentration-dependent manner. Mono indicates monocytes.
) and K562 cells (▴) bind GzmB-Bio in a concentration-dependent manner. Mono indicates monocytes.
GzmB binding analyzed by confocal laser scanning microscopy
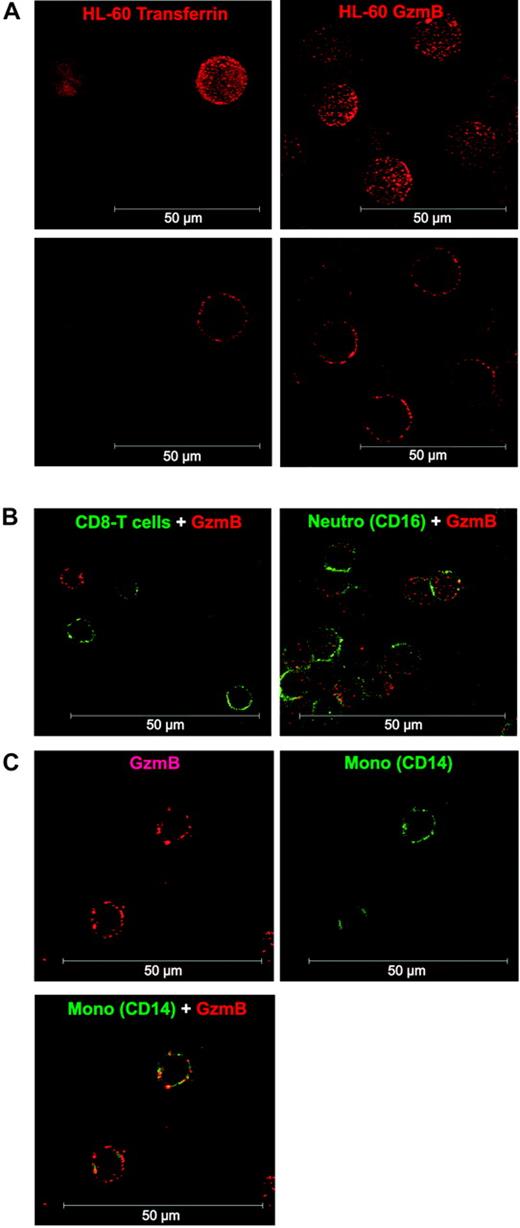
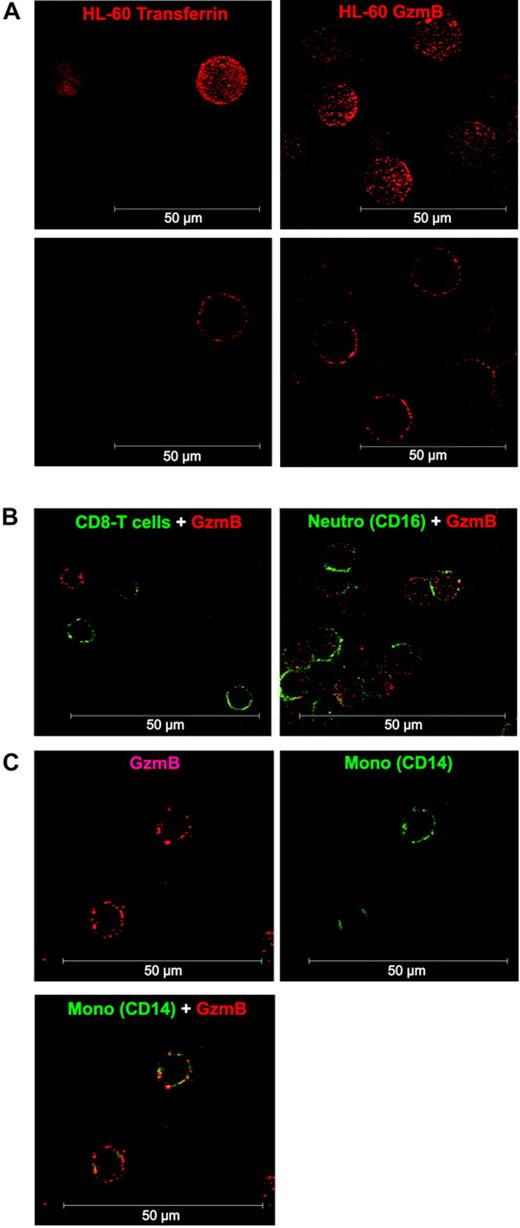
We also analyzed the binding pattern of GzmB-Bio to the human monocytic cell line HL-60 and to CD8+ T cells, granulocytes, and monocytes by confocal laser scanning microscopy (Figure 2). Experimental conditions for GzmB binding and detection were the same as in our FACS analyses (nonfixed cells, 4°C, NaN3 in the buffer, detection with labeled streptavidin). Thus, GzmB-Bio binding took place exclusively on the cell surface (Figure 2A). The binding occurred in a peculiarly punctual fashion. In agreement with our flow cytometry findings, no binding could be observed on CD8+ T cells (Figure 2B). As already seen by FACS, neutrophils showed homogeneous, but low, binding of GzmB-Bio (Figure 2B). As expected, we found strong surface binding to monocytes (Figure 2C).
Confocal analysis of GzmB binding to HL-60 cells and human leukocytes. (A) Comparison of TF-Bio (left column) and GzmB-Bio (right column) binding to HL-60 cells. Binding at 4°C occurred exclusively on the cell surface, as demonstrated by the 3-dimensional reconstruction (top row) or the section view (bottom row). (B) Costaining of lymphocytes with anti-CD8 FITC (left; section view) or granulocytes with anti-CD16 FITC (right; 3-dimensional reconstruction) and GzmB-Bio/SA-Cy3. (C) Double staining of monocytes with anti-CD14 FITC and GzmB-Bio/SA-Cy3. Shown is the section view of GzmB-Bio binding (top left) or CD14-FITC (top right) and the overlay of both (bottom). Neutro indicates neutrophil granulocytes. White scale bar = 50 μm.
Confocal analysis of GzmB binding to HL-60 cells and human leukocytes. (A) Comparison of TF-Bio (left column) and GzmB-Bio (right column) binding to HL-60 cells. Binding at 4°C occurred exclusively on the cell surface, as demonstrated by the 3-dimensional reconstruction (top row) or the section view (bottom row). (B) Costaining of lymphocytes with anti-CD8 FITC (left; section view) or granulocytes with anti-CD16 FITC (right; 3-dimensional reconstruction) and GzmB-Bio/SA-Cy3. (C) Double staining of monocytes with anti-CD14 FITC and GzmB-Bio/SA-Cy3. Shown is the section view of GzmB-Bio binding (top left) or CD14-FITC (top right) and the overlay of both (bottom). Neutro indicates neutrophil granulocytes. White scale bar = 50 μm.
Inhibition of binding by other basic serine proteases
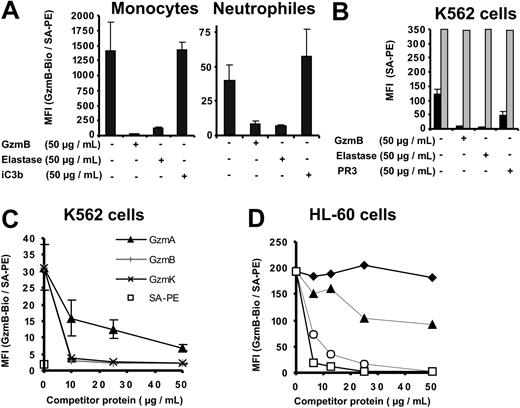
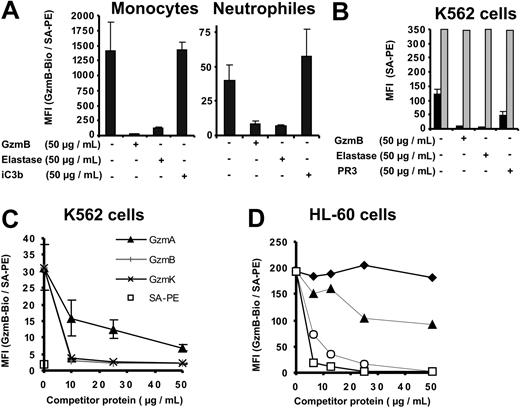
To learn more about the specificity and characteristics of GzmB binding sites on cellular surfaces, we performed competition experiments with structurally related serine proteases from lymphocytes and neutrophils. Preincubation of K562 cells, human PBMCs, and granulocytes with a 5-fold molar excess of unlabeled recombinant GzmB or neutrophil-derived human leukocyte elastase (HLE) completely abolished GzmB-Bio binding to monocytes, neutrophils, and K562 cells (Figure 3A-B). During these incubations with labeled GzmB, free, unlabeled proteases were continuously present. In binding experiments with K562 cells, we compared the inhibitory potential of HLE with that of proteinase 3 (PR3). These 2 neutrophil serine proteases differ by their isoelectric points (9.1 and 11.3 for mature PR3 and HLE, respectively).24 Consistent with a lower IEP, PR3 only partially inhibited the binding of GzmB-Bio, suggesting that ionic interactions were involved in surface binding. To exclude the degradation of GzmB and GzmB binding sites by HLE, we also assessed inactive HLE (preincubated with methoxysuccinyl-Ala-Ala-Pro-Val-chloromethyl ketone, an irreversible inhibitor) and found the same inhibition of GzmB-binding to K562 cells (data not shown). Competitive inhibition with the GzmB-related enzymes granzyme A (GzmA) and granzyme K (GzmK) was also performed at different concentrations (Figure 3C). We found that GzmK inhibited GzmB-Bio binding as efficiently as nonlabeled GzmB, whereas GzmA was less competitive. Even the highest GzmA concentrations of 50 μg/mL did not inhibit GzmB-Bio binding completely. The membrane affinities of HLE, GzmB, GzmK, GzmA, and PR3 appeared to be related to their calculated IEPs and positive surface charges (11.3, 10.4, 10.2, 9.2, 9.1). Assuming that these interactions were due to the overall positive surface potential of granule-associated serine proteases, we investigated the binding of GzmB-Bio to HL-60 cells in the presence of other basic proteins, bovine cationic trypsin, enzymatically inactivated by the irreversible inhibitor TLCK, and lysozyme (Figure 3D). Although its IEP was approximately 10.5, trypsin-TLCK did not compete for GzmB binding sites. Moreover, lysozyme, which is known to interact with negatively charged carbohydrates,25 inhibited the binding of GzmB slightly at low concentrations and only partially at high molar concentrations. These findings suggest that the complementary binding motif recognized by GzmB was shared by closely related granule proteases but was not a general binding site for cationic proteins. The proteolytically inactive GzmBS195A mutant and the unlabeled wild-type GzmB competed equally well for the binding of GzmB-Bio (Figure 3D), indicating that proteolytically inactive and native glycosylated GzmB showed similar binding properties (Figure 3D). Interactions of GzmB-Bio with membranes through biotin moieties were also ruled out because the binding of TF-Bio, another biotinylated ligand, on K562 cells was not reduced by unlabeled competitor proteases (Figure 3B). The integrin Mac-1 (CD11b/CD18, αMβ2), also called complement receptor 3 (CR3), is a receptor for iC3b but is also regarded as a receptor for HLE and PR3 on neutrophils.26 Given that PR3 and HLE compete with iC3B for binding to Mac-1, we included iC3b in our inhibition experiments (Figure 3A), but the Mac-1 ligand did not suppress the binding of GzmB on monocytes or neutrophils. All binding and competition experiments were performed in the presence of 3% fetal calf serum (FCS), which prevents unspecific interactions of competitor proteins with GzmB and cellular membranes. Binding of GzmB-Bio to K562 cells was also not changed in the presence of 5% BSA (data not shown).
Specific competition for cell surface–binding sites. (A-D) The different cell preparations were preincubated with the indicated competitor proteins at the indicated concentrations. GzmB-Bio or TF-Bio was then added to a final concentration of 10 μg/mL. (A) Inhibition of GzmB-Bio binding to monocytes and neutrophil granulocytes by nonlabeled GzmB and elastase but not by iC3b. (B) GzmB-Bio (▪) binding to K562 cells is inhibited by free nonlabeled GzmB, elastase, and proteinase 3 (PR3). In contrast, TF-Bio (▦) is not inhibited by either of these proteases. (C) Binding of GzmB to K562 cells is efficiently competed by GzmK (×) or nonlabeled GzmB (+) but not by GzmA (▴). SA-PE alone is indicated by □. Panels A-C depict the means of triplicate measurements with their standard deviations (error bars). (D) Native glycosylated GzmB (○) inhibits GzmB-Bio binding to HL-60 cells in a fashion similar to that for inactive GzmBS195A (□), whereas other basic proteins, such as trypsin-TLCK ( ) and lysozyme (▴), display no or very low inhibition.
) and lysozyme (▴), display no or very low inhibition.
Specific competition for cell surface–binding sites. (A-D) The different cell preparations were preincubated with the indicated competitor proteins at the indicated concentrations. GzmB-Bio or TF-Bio was then added to a final concentration of 10 μg/mL. (A) Inhibition of GzmB-Bio binding to monocytes and neutrophil granulocytes by nonlabeled GzmB and elastase but not by iC3b. (B) GzmB-Bio (▪) binding to K562 cells is inhibited by free nonlabeled GzmB, elastase, and proteinase 3 (PR3). In contrast, TF-Bio (▦) is not inhibited by either of these proteases. (C) Binding of GzmB to K562 cells is efficiently competed by GzmK (×) or nonlabeled GzmB (+) but not by GzmA (▴). SA-PE alone is indicated by □. Panels A-C depict the means of triplicate measurements with their standard deviations (error bars). (D) Native glycosylated GzmB (○) inhibits GzmB-Bio binding to HL-60 cells in a fashion similar to that for inactive GzmBS195A (□), whereas other basic proteins, such as trypsin-TLCK ( ) and lysozyme (▴), display no or very low inhibition.
) and lysozyme (▴), display no or very low inhibition.
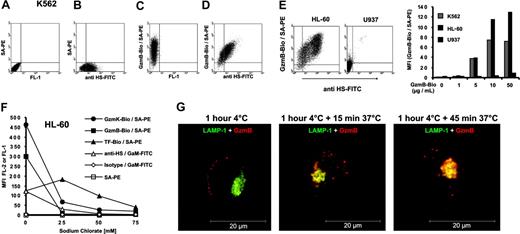
Correlation between GzmB binding and heparan sulfate expression
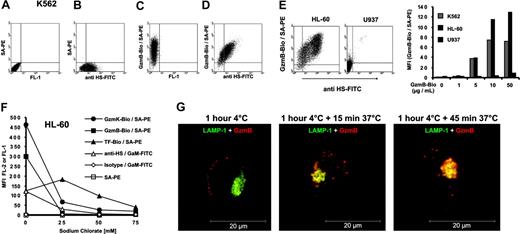
Because the more positively charged granzymes competed better for GzmB-binding sites, we assumed that ionic interactions played a critical role in the interaction with cell surface membranes. HS chains, which are attached to various proteoglycan core proteins on the cell membrane, are a major source of strong negative charges on the cell surface. We used an HS-specific monoclonal antibody that recognizes certain domains in a subset of HS chains with N-sulfated glucosamine residues (10E4-epitope). In fact, we found a close correlation between HS expression (Figure 4B) and the extent of GzmB binding (Figure 4C) on the surface of individual K562 cells, as revealed by double-staining experiments (Figure 4D). We also analyzed the monocytic cell lines HL-60 and U937 for HS expression and GzmB binding. HL-60 cells showed strong HS expression and bound the greatest amounts of GzmB among all cell lines tested (Figure 4E). In contrast to HL-60 cells, U937 cells did not express the peculiar HS-associated 10E4 epitope and also did not bind GzmB. Because the motif of the 10E4 binding site in HS is still unclear, we reduced the sulfate density of HS chains by chlorate, which competitively inhibits the formation of the high-energy sulfate donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS).27 With chlorate treatment of HL-60 cells, GzmB-Bio– and 10E4-specific binding sites were lowered in a concentration-dependent manner (Figure 4F). Chlorate concentrations of 25 mM in the cell culture medium sufficed to reduce GzmB binding to near background levels. Chlorate concentrations of 75 mM, however, suppressed the growth of cells and resulted in reduced TF-Bio binding, suggesting that this concentration elicited multiple unfavorable effects. Because HS is known as an internalizing receptor of many proteins, we investigated whether HS-bound GzmB is taken up by HL-60 cells (Figure 4G). To analyze exclusively the uptake of HS-bound GzmB, we preincubated HL-60 cells with GzmBAlexa633 at 4°C and removed nonbound GzmB from the medium by washing. Thereafter, the cells were incubated at 37°C for 15 and 45 minutes. We found that surface-bound GzmBAlexa633 was rapidly internalized into vesicular and lysosomal compartments. Already after 15 minutes of incubation at 37°C, colocalization with lysosomal compartments started to occur and was nearly completed after 45 minutes of incubation (Figure 4G).
GzmB binding and uptake by cell surface heparan sulfate. (A-D) K562 cells were stained with an FITC-labeled monoclonal antibody against heparan sulfate and GzmB-Bio/SA-PE. (A) Staining without the GzmB-Bio and anti HS-FITC antibody. (B) Staining with anti HS-FITC and SA-PE, omitting GzmB-Bio. (C) Single staining with GzmB-Bio and SA-PE, omitting anti HS-FITC. (D) Double staining with GzmB-Bio/SA-PE and anti HS-FITC. Note the clear correlation between GzmB-Bio binding and heparan-sulfate expression. (E) Binding of GzmB and HS expression of K562, HL-60, and U937 cells. (Left) FACS representation of anti–HS-FITC versus GzmB-Bio/SA-PE staining of HL-60 and U937 cells. (Right) The diagram shows the MFI of GzmB-Bio binding at the indicated GzmB-Bio concentrations. (F) Sodium chlorate treatment diminishes granzyme binding to cell surfaces. HL-60 cells were treated with the indicated sodium chlorate concentrations, and binding of the indicated proteins was investigated. (G) HS-bound GzmB is rapidly internalized. GzmBAlexa633 (40 μg/mL) was bound to HL-60 cells for 1 hour at 4°C (left). Thereafter, cells were washed twice and incubated at 37°C for 15 (middle) or 45 minutes (right). Shown is an analysis by confocal fluorescence microscopy of permeabilized HL-60 cells. Merged images of 2 cytoplasmic planes of z series are shown. GzmBAlexa633 is shown in red, and the lysosomal antigen LAMP-1 (CD107a) is shown in green. Image processing was identical for all images, and control primary antibody staining for this marker appeared black under these conditions. Already after 15 minutes, a clear colocalization (in yellow) between GzmB and lysosomes can be observed. Scale bar = 20 μm.
GzmB binding and uptake by cell surface heparan sulfate. (A-D) K562 cells were stained with an FITC-labeled monoclonal antibody against heparan sulfate and GzmB-Bio/SA-PE. (A) Staining without the GzmB-Bio and anti HS-FITC antibody. (B) Staining with anti HS-FITC and SA-PE, omitting GzmB-Bio. (C) Single staining with GzmB-Bio and SA-PE, omitting anti HS-FITC. (D) Double staining with GzmB-Bio/SA-PE and anti HS-FITC. Note the clear correlation between GzmB-Bio binding and heparan-sulfate expression. (E) Binding of GzmB and HS expression of K562, HL-60, and U937 cells. (Left) FACS representation of anti–HS-FITC versus GzmB-Bio/SA-PE staining of HL-60 and U937 cells. (Right) The diagram shows the MFI of GzmB-Bio binding at the indicated GzmB-Bio concentrations. (F) Sodium chlorate treatment diminishes granzyme binding to cell surfaces. HL-60 cells were treated with the indicated sodium chlorate concentrations, and binding of the indicated proteins was investigated. (G) HS-bound GzmB is rapidly internalized. GzmBAlexa633 (40 μg/mL) was bound to HL-60 cells for 1 hour at 4°C (left). Thereafter, cells were washed twice and incubated at 37°C for 15 (middle) or 45 minutes (right). Shown is an analysis by confocal fluorescence microscopy of permeabilized HL-60 cells. Merged images of 2 cytoplasmic planes of z series are shown. GzmBAlexa633 is shown in red, and the lysosomal antigen LAMP-1 (CD107a) is shown in green. Image processing was identical for all images, and control primary antibody staining for this marker appeared black under these conditions. Already after 15 minutes, a clear colocalization (in yellow) between GzmB and lysosomes can be observed. Scale bar = 20 μm.
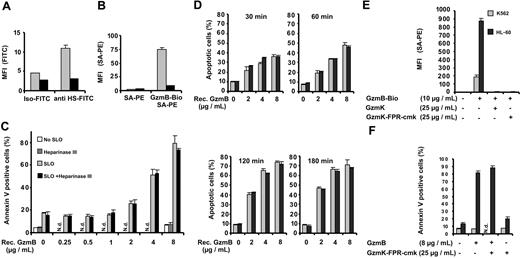
Interactions between GzmB and heparan sulfate chains
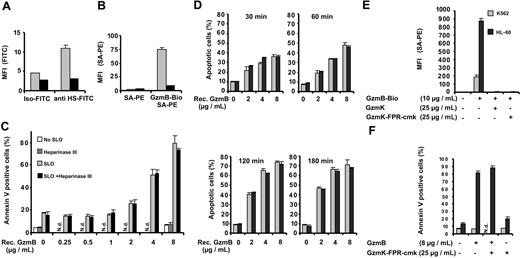
The correlation between GzmB binding and HS expression suggested to us a direct interaction between these 2 oppositely charged molecules. Therefore, we tested GzmB-Bio binding to K562 cells after heparinase III treatment (Figure 5A-B). Figure 5A shows the treatment of K562 cells with heparinase III (heparin lyase, heparitinase from Flavobacterium heparinum, E.C.4.2.2.8) and the concomitant drastic reduction of HS epitopes on the cell surface. GzmB-Bio binding was almost totally abolished after heparinase III treatment (Figure 5B). Similarly, heparinase III treatment drastically reduced binding of GzmB to HL-60 cells (data not shown). This finding clearly proves our suggestion that most binding sites on cellular surfaces are HS chains or a subset of these highly heterogeneous glycosaminoglycan chains associated with several carrier proteins. Given that HS chains are internalized and degraded together with their cargo in acidic endosomes, we wondered whether binding to HS chains had any influence on apoptosis induction by GzmB. Therefore, we compared the susceptibility of heparinase III–treated and mock-treated K562 cells toward GzmB-induced apoptosis (Figure 5C-D). Heparinase III–treated cells, however, showed the same degree of susceptibility to GzmB and sublytic amounts of SLO as untreated cells over the entire range of GzmB concentrations (Figure 5C) and over different treatment times (Figure 5D). As a second means to exclude the potential action of HS chains, we blocked GzmB-binding sites with an excess of enzymatically inactive GzmK (GzmK-FPR-cmk) (Figure 5E). Again, apoptosis induction by GzmB and sublytic SLO amounts was not functionally altered after saturation of HS chains with GzmK (Figure 5F).
Influence of surface heparan sulfate on GzmB binding and apoptosis induction. (A-C) K562 cells were treated with heparinase III and analyzed for cell surface HS, GzmB-Bio binding, and susceptibility to apoptosis induction. (A) Binding of anti–HS-FITC is reduced to background levels after heparinase III treatment. Buffer-treated (▦) or heparinase-treated (▪) cells were stained in triplicate with the FITC-labeled anti-HS monoclonal antibody (anti HS-FITC) or with an FITC-labeled IgM (immunoglobulin M)–isotype control antibody (Iso-FITC). (B) Heparinase III treatment drastically reduces GzmB binding to K562 cells. Bar shading indicates same treatments as in panel A. (C) In vitro apoptosis induction is not impaired by heparinase III treatment. K562 cells, treated as described for panel A, were subjected to GzmB-SLO–induced apoptosis. The cells were treated as indicated either with buffer or with heparinase III and thereafter were incubated with the indicated concentrations of recombinant GzmB without SLO or with sublytic concentrations of SLO. After 5-hour treatment, apoptosis was determined by FACS after staining with annexin V–FITC and PI. Shown is the mean ± SD of triplicates of the percentage of annexin V–positive cells encompassing early apoptotic cells (PI-negative) and late-stage apoptotic cells (PI-positive). (D) K562 cells were treated as described in panel C, but cells were analyzed after consecutive time points. Shown is the percentage of apoptotic cells (annexin V–positive and PI-negative). Similarly when necrotic cells were included, no significant differences between buffer and heparinase-treated cells were observed (data not shown). ▦ indicates SLO alone; ▪, SLO with heparinase. (E) Inhibition of GzmB-Bio binding to K562 (▦) or HL-60 (▪) cells by enzymatically inactive GzmK (GzmK-FPR-cmk). Active GzmK and chemical irreversibly inactivated GzmK-FPR-cmk (both 25 μg/mL) totally abolished GzmB-Bio (10 μg/mL) binding. (F) Apoptosis induced by GzmB delivered by SLO is not inhibited when GzmB-binding is blocked. The experiment was performed as described for panel C, with K562 cells preincubated with GzmK-FPR-cmk. ▪ indicates presence, ▦ absence of SLO. N.d. indicates not determined.
Influence of surface heparan sulfate on GzmB binding and apoptosis induction. (A-C) K562 cells were treated with heparinase III and analyzed for cell surface HS, GzmB-Bio binding, and susceptibility to apoptosis induction. (A) Binding of anti–HS-FITC is reduced to background levels after heparinase III treatment. Buffer-treated (▦) or heparinase-treated (▪) cells were stained in triplicate with the FITC-labeled anti-HS monoclonal antibody (anti HS-FITC) or with an FITC-labeled IgM (immunoglobulin M)–isotype control antibody (Iso-FITC). (B) Heparinase III treatment drastically reduces GzmB binding to K562 cells. Bar shading indicates same treatments as in panel A. (C) In vitro apoptosis induction is not impaired by heparinase III treatment. K562 cells, treated as described for panel A, were subjected to GzmB-SLO–induced apoptosis. The cells were treated as indicated either with buffer or with heparinase III and thereafter were incubated with the indicated concentrations of recombinant GzmB without SLO or with sublytic concentrations of SLO. After 5-hour treatment, apoptosis was determined by FACS after staining with annexin V–FITC and PI. Shown is the mean ± SD of triplicates of the percentage of annexin V–positive cells encompassing early apoptotic cells (PI-negative) and late-stage apoptotic cells (PI-positive). (D) K562 cells were treated as described in panel C, but cells were analyzed after consecutive time points. Shown is the percentage of apoptotic cells (annexin V–positive and PI-negative). Similarly when necrotic cells were included, no significant differences between buffer and heparinase-treated cells were observed (data not shown). ▦ indicates SLO alone; ▪, SLO with heparinase. (E) Inhibition of GzmB-Bio binding to K562 (▦) or HL-60 (▪) cells by enzymatically inactive GzmK (GzmK-FPR-cmk). Active GzmK and chemical irreversibly inactivated GzmK-FPR-cmk (both 25 μg/mL) totally abolished GzmB-Bio (10 μg/mL) binding. (F) Apoptosis induced by GzmB delivered by SLO is not inhibited when GzmB-binding is blocked. The experiment was performed as described for panel C, with K562 cells preincubated with GzmK-FPR-cmk. ▪ indicates presence, ▦ absence of SLO. N.d. indicates not determined.
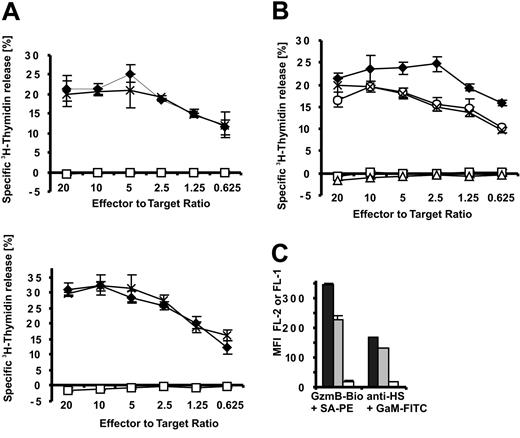
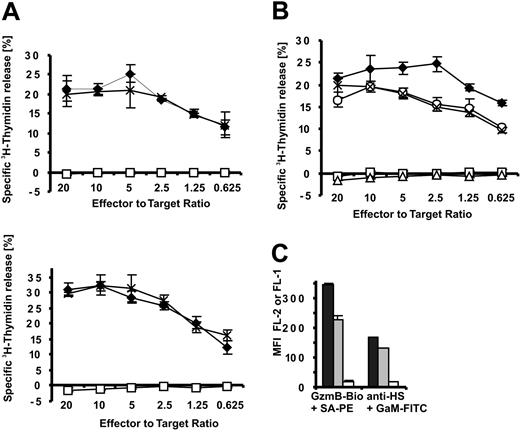
Because these experiments with soluble GzmB and SLO do not fully address the potential function of GzmB-HS interactions during killer cell attack, it was essential to assess the importance of HS chains in killer cell–mediated apoptosis induction. This intricate problem was thus approached in the following ways. The extent of DNA fragmentation within the first 3.5 hours is accepted as a reliable indicator of GzmB delivery into the cytosol17,28-30 of target cells and was, therefore, determined by the 3H-thymidine release assay in our experiments. We used human target (HL-60, K562) and effector (NK92) cells at various effector-target cell ratios (Figure 6A). As shown in Figure 3D, high concentrations of recombinant inactive GzmBS195A (25 μg/mL) are able to saturate the GzmB-binding sites on HS and, moreover, should block additional hypothetical GzmB receptors. In our large series of killing experiments, blocking of heparan sulfates by GzmBS195A had no (negative) impact on DNA fragmentation over the entire range of effector-target cell ratios. EGTA (ethyleneglycotetraacetic acid), which blocks granule secretion and perforin activity, or the pan-caspase inhibitor zVAD-fmk completely abolished 3H-thymidine release in the experiments presented (Figure 6A-B and data not shown), indicating that DNA fragmentation was initiated by granule exocytosis and caspase activation.
GzmB binding to heparan sulfate is dispensable for NK cell effector function. Apoptosis induction of HL-60 and K562 cells by the human NK92 cell line was assessed by 3H-thymidine release. (A) To block binding sites on target cells, HL-60 (top) and K562 cells (bottom) were preincubated with GzmBS195A, which was then kept constant at 25 μg/mL during the entire killing experiment (3.5 hours).  indicates medium; ×, GzmBS195A; □, EGTA/MgCl2. (A-B) EGTA/MgCl2 (6 mM/3 mM) and the pan-caspase inhibitor z-Val-Ala-Asp-Fluoromethylketone (z-VAD-fmk) (50 μM) were added in control experiments to verify perforin and caspase involvement. (B) Apoptosis induced by NK cell–delivered GzmB is independent of GzmB cell surface binding. To abrogate GzmB binding, we incubated HL-60 cells for 36 hours in 50 mM sodium chlorate. Thereafter, sodium chlorate concentrations were maintained at 10 mM (3.5 hours) (indicated by ×). As a control, nontreated HL-60 cells were coincubated with NK92 cells in the presence of 10 mM sodium chlorate (○).
indicates medium; ×, GzmBS195A; □, EGTA/MgCl2. (A-B) EGTA/MgCl2 (6 mM/3 mM) and the pan-caspase inhibitor z-Val-Ala-Asp-Fluoromethylketone (z-VAD-fmk) (50 μM) were added in control experiments to verify perforin and caspase involvement. (B) Apoptosis induced by NK cell–delivered GzmB is independent of GzmB cell surface binding. To abrogate GzmB binding, we incubated HL-60 cells for 36 hours in 50 mM sodium chlorate. Thereafter, sodium chlorate concentrations were maintained at 10 mM (3.5 hours) (indicated by ×). As a control, nontreated HL-60 cells were coincubated with NK92 cells in the presence of 10 mM sodium chlorate (○).  indicates medium; □, EGTA/MgCl2; and ▵, z-VAD-fmk. (C) Disappearance of GzmB-binding sites on sodium chlorate treatment. HL-60 cells were treated with sodium chlorate at 50 mM for 36 hours and then at 10 mM for 3.5 hours (□) and were compared with cells treated only for 3.5 hours at 10 mM (▦). ▪ indicates medium. Binding of GzmB-Bio and the 10E4 HS epitope was detected with streptavidin-PE (SA-PE) and FITC-labeled goat antimouse antibodies (GaM-FITC) by FACS analysis.
indicates medium; □, EGTA/MgCl2; and ▵, z-VAD-fmk. (C) Disappearance of GzmB-binding sites on sodium chlorate treatment. HL-60 cells were treated with sodium chlorate at 50 mM for 36 hours and then at 10 mM for 3.5 hours (□) and were compared with cells treated only for 3.5 hours at 10 mM (▦). ▪ indicates medium. Binding of GzmB-Bio and the 10E4 HS epitope was detected with streptavidin-PE (SA-PE) and FITC-labeled goat antimouse antibodies (GaM-FITC) by FACS analysis.
GzmB binding to heparan sulfate is dispensable for NK cell effector function. Apoptosis induction of HL-60 and K562 cells by the human NK92 cell line was assessed by 3H-thymidine release. (A) To block binding sites on target cells, HL-60 (top) and K562 cells (bottom) were preincubated with GzmBS195A, which was then kept constant at 25 μg/mL during the entire killing experiment (3.5 hours).  indicates medium; ×, GzmBS195A; □, EGTA/MgCl2. (A-B) EGTA/MgCl2 (6 mM/3 mM) and the pan-caspase inhibitor z-Val-Ala-Asp-Fluoromethylketone (z-VAD-fmk) (50 μM) were added in control experiments to verify perforin and caspase involvement. (B) Apoptosis induced by NK cell–delivered GzmB is independent of GzmB cell surface binding. To abrogate GzmB binding, we incubated HL-60 cells for 36 hours in 50 mM sodium chlorate. Thereafter, sodium chlorate concentrations were maintained at 10 mM (3.5 hours) (indicated by ×). As a control, nontreated HL-60 cells were coincubated with NK92 cells in the presence of 10 mM sodium chlorate (○).
indicates medium; ×, GzmBS195A; □, EGTA/MgCl2. (A-B) EGTA/MgCl2 (6 mM/3 mM) and the pan-caspase inhibitor z-Val-Ala-Asp-Fluoromethylketone (z-VAD-fmk) (50 μM) were added in control experiments to verify perforin and caspase involvement. (B) Apoptosis induced by NK cell–delivered GzmB is independent of GzmB cell surface binding. To abrogate GzmB binding, we incubated HL-60 cells for 36 hours in 50 mM sodium chlorate. Thereafter, sodium chlorate concentrations were maintained at 10 mM (3.5 hours) (indicated by ×). As a control, nontreated HL-60 cells were coincubated with NK92 cells in the presence of 10 mM sodium chlorate (○).  indicates medium; □, EGTA/MgCl2; and ▵, z-VAD-fmk. (C) Disappearance of GzmB-binding sites on sodium chlorate treatment. HL-60 cells were treated with sodium chlorate at 50 mM for 36 hours and then at 10 mM for 3.5 hours (□) and were compared with cells treated only for 3.5 hours at 10 mM (▦). ▪ indicates medium. Binding of GzmB-Bio and the 10E4 HS epitope was detected with streptavidin-PE (SA-PE) and FITC-labeled goat antimouse antibodies (GaM-FITC) by FACS analysis.
indicates medium; □, EGTA/MgCl2; and ▵, z-VAD-fmk. (C) Disappearance of GzmB-binding sites on sodium chlorate treatment. HL-60 cells were treated with sodium chlorate at 50 mM for 36 hours and then at 10 mM for 3.5 hours (□) and were compared with cells treated only for 3.5 hours at 10 mM (▦). ▪ indicates medium. Binding of GzmB-Bio and the 10E4 HS epitope was detected with streptavidin-PE (SA-PE) and FITC-labeled goat antimouse antibodies (GaM-FITC) by FACS analysis.
As an independent test for the role of HS in GzmB delivery, we compared the NK susceptibility of highly HS-positive target cells (HL-60) before and after inhibition of HS biosynthesis. HL-60 cells exposed to 50 mM sodium chlorate over 36 hours strongly diminished the sulfation density of HS chains and reduced GzmB binding by more than 95%. We found a concentration-dependent inhibition of killer cell activity against untreated HL-60 cells by high sodium chlorate concentrations (data not shown); hence, we reduced the sodium chlorate concentration to 10 mM during coincubations with NK cells. This procedure retained the loss of GzmB-binding sites for at least 3.5 hours (Figure 6C). On the other hand, the short treatment of untreated HL-60 cells with 10 mM sodium chlorate for 3.5 hours led to a reduction of GzmB binding only by approximately one third (Figure 6C). Killer cell activity and 3H-thymidine release were only slightly suppressed when nontreated HL-60 and NK92 killer cells were coincubated in 10 mM sodium chlorate for 3.5 hours (Figure 6B). HL-60 cells preexposed to 50 mM sodium chlorate for 36 hours displayed the same degree of DNA fragmentation as cells exposed to 10 mM sodium chlorate during the coincubation only. These findings clearly indicate that GzmB import through membrane receptors is not a critical factor during killer cell attack.
Discussion
In this study, we explored the possibility that nonglycosylated GzmB binds to cell surfaces and is imported into target cells independently of CI-MPR.14 To this end, we used recombinant GzmB, which was produced in E coli and was refolded, to obtain catalytically active, nonglycosylated enzyme in large homogenous quantities with the same substrate specificity and activity as the natural enzyme.18 Absence of carbohydrates on GzmB facilitated the identification and analyses of GzmB interactions, with cellular surfaces ignored thus far.
Depending on the cell type, we observed binding of GzmB to cell surfaces already at concentrations of 1 μg/mL. Freshly prepared monocytes and B cells had the strongest binding capacity, whereas T and NK cells remained completely negative after exposure to GzmB. The strongly diminished binding and reuptake of GzmB by T and NK cells most likely facilitates the delivery of GzmB into the postsynaptic target cell. Furthermore, strongly reduced binding of granzymes by NK and T cells may protect these cells against the potentially harmful reuptake of secreted granzymes at the presynaptic membrane. During biosynthesis, granzymes are packaged together with chondroitin sulfate A into lysosome-like granules and are stored at pH 5.2. At this pH, even fully processed serine proteases are basically inactive and tightly bound to the carbohydrates of the serine-glycine core peptide.31 Sequestration and complexation of granzymes into acidic storage granules is believed to result in low diffusible concentrations of granzymes during biosynthesis. Absence of HS chains on killer cells is supportive of this sequestration process and minimizes the shuffling of granzymes onto the cell surface of killer cells. The synaptic cavity between target and killer cells, however, is most likely neutral or only slightly acidic. At this pH, the granzyme-proteoglycan interactions are weaker, and the freely diffusible fraction of granzymes increases.31 Low affinity of effector cell membranes for GzmB would prevent reuptake and accumulation of high GzmB amounts in endocytic vesicles of effector cells. This and additional independent mechanisms, previously described, most likely protect effector cells against self-lysis. Cathepsin B appears to be translocated to the synaptic cleft during target cell attack and has been shown to degrade perforin,32 whereas cytosolic expression of the serpin proteinase inhibitor 9 (PI-9) by effector cells can neutralize active GzmB leaking into the cytosol.33,34
Monocytes displayed strong and homogeneous binding of GzmB, whereas the B-cell population was highly heterogeneous. PR3 and HLE, which were present in azurophil granules of neutrophils, were able to block these GzmB-binding sites, indicating that all basic granule-associated serine proteases tested competed for highly similar surface structures on leukocyte populations. In vivo, other basic ligands may displace these proteases from acidic glycosaminoglycans, depending on the local amounts of individual proteases and ligands at the site of inflammation. Uptake and endosomal transport of proteases into monocytes and B cells may well take place at sites of neutrophil activation and degranulation. Indeed, it was recently shown that B lymphocytes, which do not synthesize cathepsin G, contain the active enzyme in their endosomes and use it in antigen processing.35
Because GzmB competes with GzmK, HLE, and PR3 for the same binding sites on K562 cells and on monocytes with equal efficacy, we suggest that binding to all other cell types occurs through the same surface structure, most likely HS groups. Soluble heparin, however, does not prevent surface binding (data not shown), indicating that specific binding motifs are involved in this process. When HS were enzymatically removed from the surfaces of K562 cells, we found an almost complete loss of binding sites (Figure 5). We conclude that HS chains, attached to a limited number of membrane-anchored proteins, are the most relevant receptors for GzmB on the plasma membrane. HS-binding ligands are known to show unique specificity and affinity for certain oligosaccharide structures, which are made by the concerted action of a subset of glycosyltransferases and sulfotransferases in a cell type–specific manner. Differential binding of GzmB to certain leukocyte populations most likely reflects the variable expression of these biosynthetic enzymes rather than the expression profile of the various proteoglycan carrier proteins.
Given that cell surface HS functions as a widely exploited coreceptor, especially for the cytosolic uptake of basic fibroblast growth factor 2 (FGF2)36 and for membrane-traversing viruses,37-39 we tested the intriguing hypothesis that GzmB delivery to the cytosol and concomitant apoptosis induction is improved by the binding to HS receptors. First, we investigated whether HS-bound GzmB is taken up into intracellular compartments, and indeed we found a rapid internalization of GzmB accumulating in lysosomal structures through HS. We then compared the susceptibility of HS-positive and -negative cells to increasing extracellular concentrations of GzmB directly, but no differences in the number of apoptotic cells between heparinase III– and buffer-treated cells were observed.
To evaluate the importance of HS receptors for NK-mediated killing, we used different target cell types with high and intermediate HS expression and saturated the GzmB-binding sites with catalytically inactive GzmB. Incubation of effector and target cells in the presence of EGTA largely reduced the release of 3H-thymidine, indicating that NK-mediated apoptotic cell death was dependent on granule release within the initial 3.5-hour time period. In these experiments, we could not discern any correlation between NK susceptibility and HS density on target cells and could not prevent target cell killing by coincubation with high concentrations of inactive GzmB. As an alternative way to inhibit binding to HS-structures, we used HL-60 cells treated with high concentrations of sodium chlorate as target cells. Again, no impact of HS binding on apoptosis induction by killer cells was observed.
Although our observations do not prove a direct link between GzmB delivery and rapid onset of DNA fragmentation for our system, previous evidence obtained by similar killing experiments suggests a unique and universal role of GzmB in granule-dependent DNA fragmentation.17,28-30 The only freely available GzmB-specific inhibitor is Z-Ala-Ala-Asp-CH2Cl, which, however, shows poor efficacy in whole cell models of apoptosis induction. To overcome this problem, researchers at Merck Research Laboratories developed and optimized selective small molecule inhibitors against GzmB and showed that 2 of these new compounds blocked killer–cell mediated DNA fragmentation in the same cell culture system, which we used.40 Their assays made use of human NK-92 natural killer cells and K562 target cells whose DNA had been labeled with sodium iodide I 125 and convincingly demonstrated the role of GzmB in DNA fragmentation.
It is nevertheless difficult to tell where the fatal number of GzmB molecules crosses the perforin-primed plasma membrane during killer cell attack. Given that only a small fraction of cytolytic granules is locally released from the effector cell, a single local pulse of coreleased perforin and GzmB is expected to be most effective at the time and site of its release and will subsequently lose its apoptotic potential by the dispersion of membranes and solutes into various subcellular compartments. The membrane-inserting potential of perforin is rapidly lost at neutral pH in the presence of calcium and thus cannot traverse large distances as a soluble protein. Direct binding and insertion of perforin into the target membrane at the conjugation site most likely represents the crucial hit for apoptosis induction. The subsequent responses of the target cell may rather reflect the desperate situation and futile attempts to repair the locally damaged membrane.41 The data presented in this study and by others17 challenge the model of receptor-mediated endocytosis as a major event in target cell killing.
Prepublished online as Blood First Edition Paper, November 4, 2004; DOI 10.1182/blood-2004-06-2180.
Supported by the German Research Council (SFB 571 and grant JE194-2) and the German Cancer Aid Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank E. Stegmann, B. Mosetter, W. Lasinger, and H. Reimann for excellent technical assistance, Chris Bleackley for native GzmB, V. Vargas for help in the preparation of PBMCs, and H. Wekerle for his continuous interest in the project.


 representes monocytes;
representes monocytes;  , B cells; and ▦, CD8+ T cells, (F) Jurkat (
, B cells; and ▦, CD8+ T cells, (F) Jurkat (