Abstract
Rearrangements involving the MLL gene on chromosome band 11q23 are a hallmark of therapy-related acute myeloid leukemias following treatment with topoisomerase II poisons including etoposide. Therapy-related and de novo genomic translocation breakpoints cluster within a well-characterized 8.3-kb fragment of MLL. Repair of etoposide-stabilized DNA topoisomerase II covalent complexes may initiate MLL rearrangements observed in patients. We used a culture system of primary human hematopoietic CD34+ cells and inverse polymerase chain reaction to characterize the spectrum of stable genomic rearrangements promoted by etoposide exposure originating within an MLL translocation hotspot in therapy-related leukemia. Alterations to the region were observed at a readily detectable frequency in etoposide-treated cells. Illegitimate repair events after minimal repair included MLL tandem duplications and translocations, with minor populations of deletions or insertions. In stably repaired cells that proliferated for 10 to 14 days, the significant majority of illegitimate events were MLL tandem duplications, and several deletions, inversions, insertions, and translocations. Thus, etoposide promotes specific rearrangements of MLL consistent with the full spectrum of oncogenic events identified in leukemic samples. Although etoposide-initiated rearrangements are frequent, only a small subset of translocations occurs in cells that proliferate significantly.
Introduction
Genome integrity is controlled by multiple damage surveillance pathways, cell-cycle checkpoints, and repair mechanisms.1-3 Despite these safeguards, illegitimate repair of chromosomal double-strand breaks (DSBs), such as those produced by chemotherapy agents that target topoisomerase II (topo II), can promote chromosomal rearrangements and, ultimately, tumorigenesis. Topo II is an essential cellular enzyme that catalyzes changes in DNA topology via its cleavage-religation equilibrium. Topo II–targeted drugs that are poisons of this enzyme stabilize topo II–DNA covalent complexes, most often by decreasing the rate of religation in a dose-dependent manner. Disruption of the cleavage-religation reaction results in accumulation of DSBs, p53 activation, and induction of apoptosis or repair.4,5 Chromosomal DSBs are potent inducers of recombination, stimulating the exchange of homologous sequences between 2 DNA duplexes 1000-fold6-8 not only between sister chromatids, but also between homologs, and sequence repeats on heterologous chromosomes.9-12 DSBs may be repaired by homologous recombination or nonhomologous end joining (NHEJ), and both repair mechanisms have been associated with chromosomal translocations, a hallmark of leukemias, lymphomas, and soft-tissue sarcomas.13-15
There is clear evidence that exposure to chemotherapy or irradiation can result in subsequent development of therapy-related leukemias, which occur in 1% to 15% of patients exposed to DNA-damaging agents in anticancer regimens.16,17 Exposure to topo II poisons such as etoposide is predominantly associated with therapy-related leukemias characterized by rearrangements of the MLL gene on chromosome band 11q23.16 MLL spans 100 kb, encodes a 430-kDa protein homologous to the Drosophila trithorax gene, and has important functions in embryogenesis and hematopoiesis.18,19 Mixed-lineage leukemia (MLL) is a critical transcriptional regulator and numerous translocations involving MLL suggest that gain of function contributes to the critical leukemogenic lesion.20
The MLL gene is involved in rearrangements and translocations with multiple partner genes, many of which are uncharacterized. Most MLL aberrations initiate within a particular 8.3-kb BamH1 fragment of the gene located between exons 5 and 11 known as the breakpoint cluster region (bcr).21-23 The European Union's Concerted Action Workshop examined the spectrum of MLL rearrangements in patient samples. Of the samples, 16% were internal duplications, deletions, and inversions, while the remaining 84% were reciprocal translocations with as many as 40 different partner genes.24 This particularly broad spectrum of rearrangements is a hallmark of MLL rearrangements and may reflect an inherent recombinogenic nature of the locus, or the central role of the MLL protein in proper hematopoietic development and differentiation such that rearrangements often possess leukemogenic potential. Individual genetic factors have also been identified to predispose some patients to therapy-related leukemia with MLL alterations.25
The distribution of etoposide-induced aberrations observed in therapy-related leukemia is nonrandom.26,27 However, the mechanisms that lead to specific rearrangements remain unclear. Etoposide-associated genomic alterations may coincide with topo II cleavage activity particularly within MLL.28-31 Etoposide addition to in vitro topo II cleavage assays enhances DNA topo II cleavage complexes within MLL and partner genes near translocation breakpoint sites identified in treatment-related leukemias.32-34 Topo II cleavage complexes with the MLL bcr in hematopoietic progenitor cells are detectable after etoposide exposure.35 Rearrangement of this region can be detected by Southern blotting after 16 to 24 hours of continuous etoposide exposure in many hematopoietic cell lines.30,36,37 MLL bcr cleavage and rearrangements also are detectable following exposure of cells to multiple nongenotoxic agents, possibly due to apoptotic DNA fragmentation.38-41
The observation that leukemias with MLL rearrangements are of both myeloid and lymphoid lineages and analysis of their gene expression profiles suggest that the MLL rearrangement and disease itself initiate within an undifferentiated hematopoietic stem cell.42 However, there are minimal data on the clastogenic effects of etoposide on the hematopoietic CD34+ stem cell–enriched population, the emergence of stable MLL rearrangements, or the proliferative potential of cells that contain illegitimate repaired rearrangement products. To provide direct evidence for stimulation of MLL rearrangements by etoposide in primary human CD34+ cells, we established an in vitro cell culture system of etoposide exposure and recovery, and used inverse polymerase chain reaction (IPCR) to directly determine the potential of etoposide exposure to lead to stable genome rearrangements originating within an MLL translocation breakpoint hotspot in therapy-related leukemia.32,43-45 Alterations to this region in etoposide-treated cells were readily detected. The spectrum of illegitimate repair products detectable after minimal 2 to 3 hours recovery included frequent MLL partial tandem duplications (PTDs, 44%) and translocations (44%), with minor populations that contained deletions or insertions. However, in stably repaired cells capable of continued proliferation for 10 to 14 days, most illegitimate repair events were MLL PTDs (79%), and also included deletions, inversions, insertions, and translocations. Several clones had breakpoints that localize to MLL bcr sequences identified at rearrangement junctions in therapy-related leukemias.32,46-49 These data indicate that although etoposide-initiated rearrangements of MLL are frequent, only a small subset of chromosomal translocations occurs in cells that are capable of continued proliferation and, ultimately, leukemogenesis.
Materials and methods
Cell culture
CD34+ cells were isolated from human umbilical cord blood (CB) specimens on ficoll gradient and positive selection through anti-CD34+ columns (Miltenyi, Auburn, CA). The purity of isolated CD34+ cells was determined by phycoerythrin (PE)–conjugated anti-CD34+ antibody (Becton Dickinson, San Jose, CA) and flow cytometry. Alternatively, frozen adult peripheral blood CD34+ cells were obtained from National Cancer Institute (NCI) core facility, Seattle, WA. Other cell lines obtained from American Type Culture Collection (Manassas, VA) included TF-1 (RPMI supplemented with glutamine [GIBCO, Carlsbad, CA], 10% fetal bovine serum [FBS]), WS1 fibroblasts (Dulbecco modified Eagle medium [DMEM], 10% FBS), M059K, and M059J (DMEM/HAM-F12, 10% FBS). Use of human cells was approved by Columbia University (New York) institutional review board (IRB no. 15183). Informed consent was provided according to the Declaration of Helsinki. Cells were cultured in Iscoves modified Dulbecco medium (IMDM) supplemented with 25% BIT9500 (Stem Cell Technology, Vancouver, BC, Canada), Fms-like tyrosine kinase 3 ligand (Flt-3L), thrombopoietin (TPO), and 100 ng/mL stem cell factor (SCF; Peprotech, Rocky Hill, NJ) for 2 to 4 days. CD34+ cells were exposed to 20 to 50 μM etoposide (Sigma-Aldrich, St Louis, MO; 20 mM stock solution prepared in dimethylsulfoxide [DMSO]) for 1 hour and recovered in myeloid differentiating media (IMDM, BIT9500, 10 ng/mL interleukin-3 [IL-3], 10 ng/mL granulocyte colony-stimulating factor [G-CSF], 10 ng/mL granulocyte-macrophage colony-stimulating factor [GM-CSF] [Peprotech]). Myeloid differentiation was confirmed after 7 to 14 days by PE-conjugated anti-CD11b antibody (Becton Dickinson) or anti–PE-conjugated anti-CD34+ antibody.
Analysis of DNA damage, apoptosis, and recovery
Pulsed-field gel electrophoresis was performed to detect high-molecular-weight DNA fragmentation. Cells (200 000) were immobilized in 1.5% low-melting-point agarose. Plugs were treated with extraction buffer (10 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 9.5; 10 mM NaCl; 25 mM EDTA [ethylenediaminetetraacetic acid]; 1 mM ethylene glycol–tetraacetic acid [EGTA]; 1.5% sodium dodecyl sulfate [SDS]; 0.1% β-mercaptoethanol) and washed 3 times in mM EDTA and 1 mM Tris-HCl (pH 8); high-molecular-weight DNA was resolved on a 1% agarose gel using the field inversion gel electrophoresis mode of the clamped homogeneous electric field Mapper (Bio-Rad, Hercules, CA). DNA entry into the gel was facilitated by continuous electrophoresis for 20 minutes at 6 V/cm. Field inversion followed at 5 V/cm, a 3:1 ratio of forward to reverse switch times, linearly ramping 16 hours at 14°C. Fragmentation was viewed after ethidium bromide staining. Apoptosis was evaluated using an annexin V–fluorescein isothiocyanate (FITC) apoptosis detection kit (PharMingen, San Diego, CA). Cell survival and proliferative recovery were assessed using Alamar Blue (Biosource, Camarillo, CA) or counting by hemacytometer.
IPCR
Genomic DNA was digested with XbaI, extracted with phenol-chloroform, and ethanol-precipitated. DNA fragments (5-μg each) were circularized with 2000 units T4 DNA ligase in 300 μL at room temperature. DNA was purified by phenol-chloroform extraction using phase-lock gels (Eppendorf) followed by ethanol-precipitation. DNA (200 ng) was used for the first PCR. For nested PCR, 1 μL of the first PCR was used. Final PCR products were separated on a 1% agarose gel. There were 2 sets of nested primers used to amplify MLL and putative fusion partners: MF1-5′TCTACAAGTGCCAGGGGTCT3′; MF2-5′AATAGCATGCTGCCTGCACTGCACTCCTAA3′; MR1-5′CCCGACGTGGATTTTCTTTA3′; MR2-5′GATCGTAGGATATGTCCCTTATAAATGACAAACTACTGCTTCC3′. For the first PCR, the reaction was as follows: 35 cycles of 94°C for 30 seconds, 54°C for 45 seconds, or 68°C for 3 minutes + 20 seconds each cycle after the 10th cycle. For the second PCR, the reaction was as follows: 35 cycles of 94°C for 30 seconds, 56°C for 45 seconds, or 68°C for 3 minutes + 20 seconds each cycle after the 10th cycle. In some cases, PvuII digestion of DNA prior to PCR eliminated amplification of competing germ-line product. The entire PCR reaction, or individual IPCR products excised and purified from gels, was cloned into pCR2.1 TOPO (Invitrogen, Frederick, MD). Resultant cloned PCR products were sequenced with M13 forward and reverse primers, and, for larger products, with internal MLL-specific primers. Nucleotide sequences were compared with MLL and analyzed using the National Center for Biotechnology Information Basic Local Alignment Search Tool (BLAST NCBI, Bethesda, MD).
Chromosomal analysis
Cells were incubated with ethidium bromide (10 μg/mL) for 15 minutes and then Colcemid (GIBCO) for 2 hours. Cells were swelled in hypotonic 0.54% KCl solution, and fixed in methanol-acetic acid (3:1) 3 times. Metaphase spreads were analyzed by fluorescence in situ hybridization (FISH) using chromosome (chr) 11-cyanin3 (Cy3) and chr 4-FITC whole chromosome probes according to the manufacturer's protocol (Vysis, Downers Grove, IL). Aberrations were scored in approximately 100 metaphases. In parallel, metaphase and interphase spreads were analyzed by FISH using an MLL locus-specific identifier spectrum green and spectrum orange split-probe (LSI; Vysis) according to the manufacturer's protocol. Approximately 250 metaphases or interphases were scored per sample. Images were captured with a Nikon Microphot fluorescent microscope equipped with CCD camera and filters (Nikon, Tokyo, Japan), and obtained with CytoVision software (Applied imaging Inc., San Jose, CA).
Results
Human CD34+ cell etoposide exposure
We established an in vitro culture and IPCR system to directly determine the potential of etoposide exposure to lead to stable genome rearrangements originating within the MLL locus on chromosome band 11q23 in primary stem cell–enriched CD34+ cells. Fresh CB CD34+ cells were isolated by ficoll gradient and positive selection through anti-CD34+ columns. Peripheral blood CD34+ cells from adult individuals were obtained from NCI. Staining of cells immediately following isolation with PE-conjugated anti-CD34+ antibody and analysis by flow cytometry confirmed more than 90% purity in all samples (data not shown). Although CB cells represent an immature population and possibly contain cells with higher replication and repair potential, both cell sources represent similar mobilized peripheral blood CD34+ cells induced by stress conditions and cytokines, and both gave similar results. Depending on yield, between 5 and 8 individual samples were pooled to obtain 3 to 5 × 106 cells per experiment, and 4 independent experiments were performed. CD34+ cells were cultured for 2 to 4 days in IMDM supplemented with SCF, TPO, and Flt-3L to optimally maintain and expand stem cells and progenitors.50 To determine the clastogenic effects of etoposide, CD34+ cells were exposed for 1 hour with a physiologically relevant dose of 20 to 50 μM etoposide, in agreement with pharmacokinetic studies demonstrating peak plasma levels of 25 to 75 μM in patients.51 Etoposide was subsequently washed away, and cells were allowed to repair DNA damage and proliferate. Following 7 to 14 days of expansion in culture, flow cytometry confirmed that the majority of cells were CD34- and, on average, 40% were CD11b+, indicating substantial myeloid differentiation during this period (data not shown). We did not examine directly the percentage of lymphoid cells in expanded populations. However, culture conditions used were expected to greatly favor myeloid differentiation.
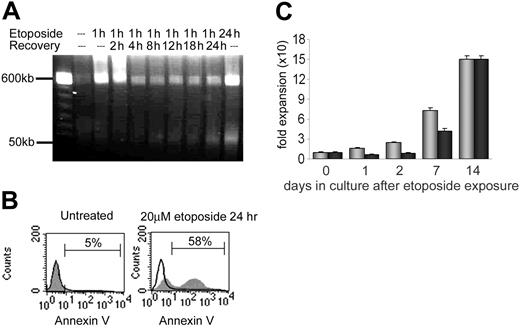
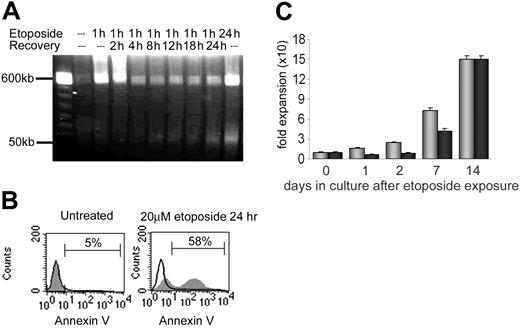
The overall level of DNA damage, chromatin fragmentation, and cell viability following etoposide was assessed by pulse field gel electrophoresis (Figure 1A) and annexin V staining (Figure 1B). Fragments of more than 400 kb were immediately visible after 1 hour of etoposide exposure. This population disappeared within 2 to 4 hours after removal of etoposide, either due to repair of DNA damage or due to apoptosis and clearance of dead cells from the population. By 8 hours after etoposide removal, 50-kb fragments were detected, corresponding to DNA fragmentation by caspase-activated deoxynucleases and early apoptosis in a portion of treated cells. Of cells, 20% to 60% were viable 24 to 48 hours after treatment (Figure 1B), depending on the individual sample (data not shown). Incubation of cells for 10 to 14 days demonstrated recovery of proliferative capacity of the remaining viable cells. By 14 days, numbers of control and etoposide-treated cells increased 155-fold and 150-fold, respectively (Figure 1C). As expected, continuous exposure of CD34+ cells to 20 μM etoposide for 24 hours resulted in decreased cell survival (0%-5%) and significantly more prominent band formation of more than 400 kb and 50 kb as well as decreased cell viability (Figure 1A). Although there is likely an etoposide dose-response effect in CD34+ cells, these cells were not further analyzed.
Cytotoxicity of etoposide on primary human CD34+ cells. (A) Pulse field electrophoresis of genomic DNA to detect DNA fragmentation following 20 μM etoposide exposure and 0 hour to 24 hours recovery times. Fragment populations of more than 400 kb and 50 kb are indicated on the left. (B) Annexin V detection of apoptotic cells. Black line histogram indicates unstained control; grey histogram, annexin V–positive population. Left panel: untreated CD34+ cells. Right panel: CD34+ cells following 20 μM etoposide exposure for 24 hours. (C) Recovery and proliferation of CD34+ cells in culture after exposure to etoposide. There were 3 independent experiments performed, and standard deviations indicated by error bars. The difference in expansion between the untreated and treated samples is statistically significant on days 1, 2, and 7. Grey bars indicate untreated CD34+ cells; black bars, CD34+ cells following 20 μM etoposide exposure.
Cytotoxicity of etoposide on primary human CD34+ cells. (A) Pulse field electrophoresis of genomic DNA to detect DNA fragmentation following 20 μM etoposide exposure and 0 hour to 24 hours recovery times. Fragment populations of more than 400 kb and 50 kb are indicated on the left. (B) Annexin V detection of apoptotic cells. Black line histogram indicates unstained control; grey histogram, annexin V–positive population. Left panel: untreated CD34+ cells. Right panel: CD34+ cells following 20 μM etoposide exposure for 24 hours. (C) Recovery and proliferation of CD34+ cells in culture after exposure to etoposide. There were 3 independent experiments performed, and standard deviations indicated by error bars. The difference in expansion between the untreated and treated samples is statistically significant on days 1, 2, and 7. Grey bars indicate untreated CD34+ cells; black bars, CD34+ cells following 20 μM etoposide exposure.
IPCR analysis of etoposide-induced MLL repair products
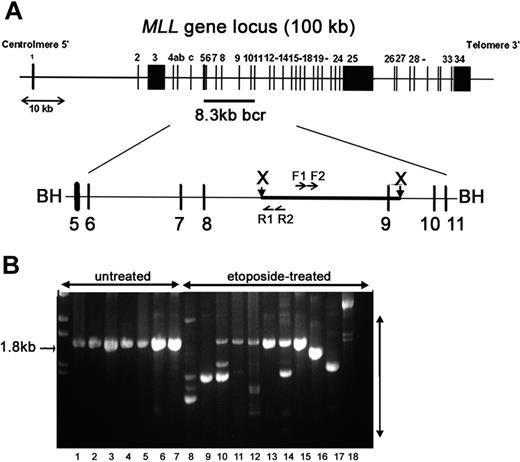
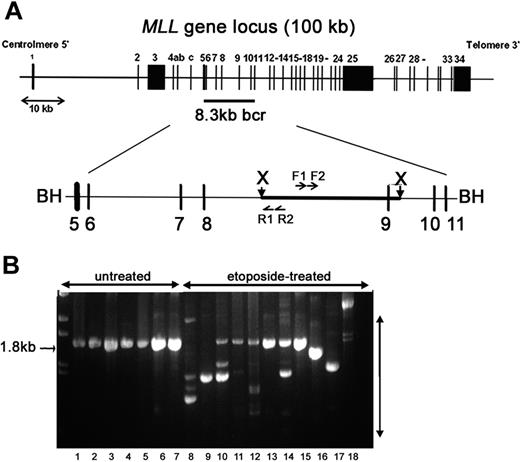
To determine the direct potential for etoposide to lead to MLL rearrangements and the stability of rearrangements in viable cells, cells were harvested either 2 to 3 hours (short recovery period) or 10 to 14 days (long recovery period) after removal of etoposide. We used IPCR to determine the fidelity of repair in a 1.8-kb region in the 3′ portion of MLL intron 8 that contains a translocation breakpoint hotspot in treatment-related leukemia and exon 9 that contains sequence homology to a putative topo II recognition sequence and is sensitive to DNaseI and multiple cytotoxic agents (Figure 2A).23,43,44 IPCR of untreated samples gave almost exclusively a germ-line 1.8-kb product that was confirmed by sequencing (Figure 2B). In contrast, IPCR of etoposide-treated cells also gave readily detectable variable-sized bands representing alterations to the region (Figure 2B). As visualized by gel electrophoresis (Figure 2B), IPCR favors amplification of smaller rearranged products (300-700 bp) with some larger (1.5-2 kb) products; thus, the total number of rearrangements in each etoposide-treated population is likely higher. More clones were observed from IPCR reactions harvested after a short recovery compared with samples harvested after a long recovery. Any decrease in detectable illegitimate repair products between short and long recovery times is consistent with chromatin fragmentation and apoptosis of cells with unrepaired damage 12 to 24 hours after etoposide exposure, but could also reflect competitive expansion of specific clones over time. Parallel experiments in multiple hematologic and other adherent cell lines gave similar IPCR results (Figure 2B), but individual clones were not characterized.
IPCR analysis to detect etoposide-induced rearrangements initiating within the MLL bcr. (A) Schematic representation of the MLL gene locus on chromosome band 11q23. The 8.3-kb MLL bcr, flanked by BamHI sites, includes exons 5 to 11 and intervening introns. XbaI sites are 2.6 kb apart within the MLL bcr. Genomic DNA was digested with XbaI and circularized. Nested PCR reactions were carried out with F1-R1 primer pair, followed by F2-R2 primer pair. BH (BamH1); X (XbaI); F1, F2, R1, R2 are primers used for PCR (see “Materials and methods”). (B) Representative IPCR products. Lanes 1-7: untreated controls that give the expected 1.8-kb germ-line product. Lanes 8-18: etoposide-treated samples that all give alternative products representing possible rearrangements of MLL. Pretreatment of samples shown in lanes 8 to 18 with PvuII eliminated some or all detectable germ-line product to facilitate isolation of individual alternative sized products. Lanes 8-12: multiple independent CD34+ cell samples (long recovery) with all germ-line products eliminated by PvuII in lanes 8-9; lane 13: TF-1 (short recovery); lane 14: WS1 (short recovery); lanes 15-16: M059K (short recovery); lane 17: M059J (short recovery); and lane 18: cord blood mononuclear cells (short recovery). Parallel experiments in multiple hematologic and other adherent cell lines gave similar IPCR results (B), but individual repair clones have not been fully characterized.
IPCR analysis to detect etoposide-induced rearrangements initiating within the MLL bcr. (A) Schematic representation of the MLL gene locus on chromosome band 11q23. The 8.3-kb MLL bcr, flanked by BamHI sites, includes exons 5 to 11 and intervening introns. XbaI sites are 2.6 kb apart within the MLL bcr. Genomic DNA was digested with XbaI and circularized. Nested PCR reactions were carried out with F1-R1 primer pair, followed by F2-R2 primer pair. BH (BamH1); X (XbaI); F1, F2, R1, R2 are primers used for PCR (see “Materials and methods”). (B) Representative IPCR products. Lanes 1-7: untreated controls that give the expected 1.8-kb germ-line product. Lanes 8-18: etoposide-treated samples that all give alternative products representing possible rearrangements of MLL. Pretreatment of samples shown in lanes 8 to 18 with PvuII eliminated some or all detectable germ-line product to facilitate isolation of individual alternative sized products. Lanes 8-12: multiple independent CD34+ cell samples (long recovery) with all germ-line products eliminated by PvuII in lanes 8-9; lane 13: TF-1 (short recovery); lane 14: WS1 (short recovery); lanes 15-16: M059K (short recovery); lane 17: M059J (short recovery); and lane 18: cord blood mononuclear cells (short recovery). Parallel experiments in multiple hematologic and other adherent cell lines gave similar IPCR results (B), but individual repair clones have not been fully characterized.
We isolated, cloned, and sequenced 67 novel IPCR products from primary CD34+ cells to characterize etoposide-induced illegitimate repair events. Of the products, 34 were from samples harvested after the short recovery period, and 33 were from samples harvested after the long recovery period (Table 1). After short recovery, half of the analyzed repair events (16 [47%] of 34) were intragenic MLL rearrangements. Almost all of these events were PTDs. The most extensive PTD contained a fusion between intron 8 and intron 4, 5′ of the MLL bcr. One product contained a deletion of more than 600 bp. The other half of repair products (18 [53%] of 34) contained fusions of MLL to new sequence. The majority of these (15 of 18) were apparent chromosomal translocations. These events are in agreement with other reports that DNA-damaging agents induce MLL translocations immediately after exposure and a short recovery period.38,52 The remaining 3 events contained insertions of novel sequences ranging from 39 bp to 180 bp.
By contrast, after a long recovery almost all analyzed repair events (31 [94%] of 33) were intragenic MLL rearrangements. Similar to products analyzed after short recovery, most of these were PTDs (26 [79%] of 33). In addition, one product contained a deletion of approximately 100 bp, and 4 were inversions of sequence within the MLL bcr. Only a small minority of products (2 [6%] of 33) contained fusions of MLL to new sequence; one contained an insertion of 180 bp, and one was an apparent translocation. Thus, products derived from etoposide-treated cells and repaired within 2 to 3 hours showed a significantly higher proportion of translocations compared with the spectrum of repair products isolated from cells that proliferated for 14 days (53% versus 6%, P = .000 02). As an additional control for PCR contamination or artifact within samples, multiple aliquots of isolated DNA from the same sample were used for independent IPCR amplifications. In several cases, we cloned and sequenced the same repair products from 2 different aliquots of the same sample, and in one case, the same repair product was present in 3 different aliquots of the DNA sample. Identification of these events in multiple aliquots after a long recovery is further evidence that they are contained within viable and proliferating cells.
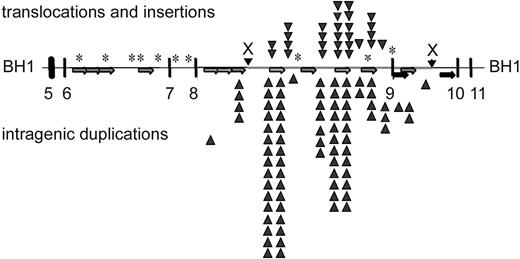
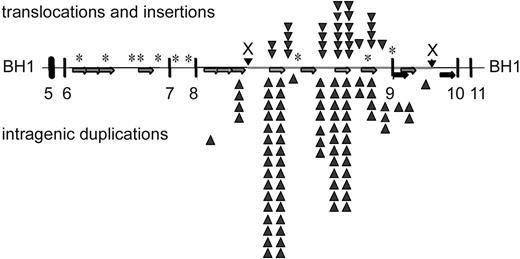
Overall, the breakpoint junctions were similar to breakpoints sequenced from leukemic cells. Several cloned rearrangement junctions localized to MLL bcr sequences that have been identified at breakpoint junctions in therapy-related leukemias.32 Microhomologies of 1 to 20 bp were present at approximately 70% of the breakpoint junctions (Figure 3). In 3 cases, a longer patch of overlapping homologous sequence of 21 bp, 277 bp, and 350 bp was observed at the breakpoint junction, suggesting single-strand annealing as the DNA repair mechanism.53 The distribution of etoposide-induced breakpoints in the repair products is displayed against the localization of repetitive elements within the MLL bcr in Figure 4. PTD breakpoints (86%) and translocation breakpoints (60%) localized within repetitive elements. Of the 35 PTD breakpoints, 31 contained Alu-Alu or LINE-LINE fusions, supporting previous reports of repetitive element-mediated recombination to generate MLL PTDs in acute myeloid leukemia (AML).54,55 Of the 20 novel sequences fused to MLL, 16 were of sufficient length to allow identification by BLAST. Apparent translocations fused MLL to sequence located on chromosome bands 3q26, 3p25, 4q31-32, 4p15, 6q21, 7q31-32, 9q21-22, 12q21 (2 clones), 14q21, 15q12-13, 17q24 (3 clones), and Xq22-23. Although several of these chromosomal bands contain known partner genes of MLL (3q25-26–GMPS, 6q21-AF6q21, 17q24-LASP1, and Xq22-23–Septin656-59 ), the partner sequences in the induced clones did not match known MLL partners.
Representative etoposide-induced breakpoint sequence junctions. Left panel: representative sequence junctions of translocations of MLL gene to heterologous chromosomes. Red indicates sequence 3′ of the breakpoint junction, and green indicates sequence 5′ of the breakpoint junction. Right panel: representative sequence junctions of MLL PTDs. Underlined red sequences indicate overlapping homology between fusion partners. Bold black numbers indicate insertions. Numbers in brackets correspond to nucleotide lengths of longer insertions or homologies. For instance, in the top clone, a 2.1-kb PTD was produced following an Alu-Alu fusion with 23 bp overlapping homology.
Representative etoposide-induced breakpoint sequence junctions. Left panel: representative sequence junctions of translocations of MLL gene to heterologous chromosomes. Red indicates sequence 3′ of the breakpoint junction, and green indicates sequence 5′ of the breakpoint junction. Right panel: representative sequence junctions of MLL PTDs. Underlined red sequences indicate overlapping homology between fusion partners. Bold black numbers indicate insertions. Numbers in brackets correspond to nucleotide lengths of longer insertions or homologies. For instance, in the top clone, a 2.1-kb PTD was produced following an Alu-Alu fusion with 23 bp overlapping homology.
Summary of etoposide-induced breakpoints within the MLL bcr. Localization of repetitive elements in 8.3-kb MLL bcr, and distribution of breakpoints associated with translocations and insertions (above the line) or intragenic duplications (below the line). Alu elements in same orientation (light gray arrows; 799-1109, 1119-1420, 1423-1716, 1927-2215, 3973-4268, 4765-5094, 6072-6372, 7163-7427), LINE elements (black arrows; 3490-3589, 3691-3972, 4269-4612, 5610-5998), low copy MER repeat (dark gray arrows; 6959-7162, 7428-7558). *Sequence homology to a putative topo II recognition sequence (according to Gu et al22 ).
Summary of etoposide-induced breakpoints within the MLL bcr. Localization of repetitive elements in 8.3-kb MLL bcr, and distribution of breakpoints associated with translocations and insertions (above the line) or intragenic duplications (below the line). Alu elements in same orientation (light gray arrows; 799-1109, 1119-1420, 1423-1716, 1927-2215, 3973-4268, 4765-5094, 6072-6372, 7163-7427), LINE elements (black arrows; 3490-3589, 3691-3972, 4269-4612, 5610-5998), low copy MER repeat (dark gray arrows; 6959-7162, 7428-7558). *Sequence homology to a putative topo II recognition sequence (according to Gu et al22 ).
Karyotypic analysis of etoposide-induced chr 11 aberrations
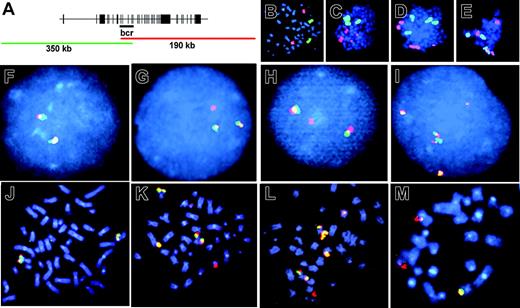
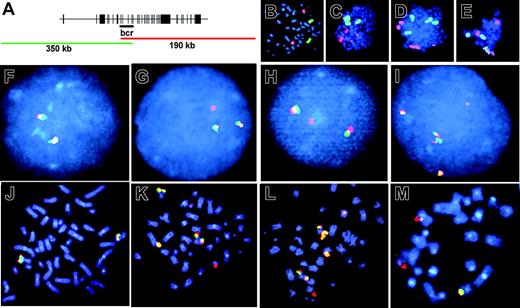
We visualized aberrations of the MLL locus at the chromosomal level by FISH using a dual-color fluorescent split signal MLL probe (Figure 5A). This technique captures all structural aberrations of the MLL bcr corresponding to fragmented chromosome material including translocations, dicentrics, amplification, and loss of MLL sequence. There were 3 independent primary CD34+ samples divided into parallel untreated or etoposide-treated aliquots, cultured, and proliferated for 7 to 14 days, and harvested on the same day. At least 190 metaphase spreads from each sample were scored. In untreated samples, 4 of 640 cells separated the overlapping fluorescent probes, and no cells contained multiple signal aberrations (0 of 640). By contrast, following etoposide exposure and proliferation for 7 to 14 days, 59 of 850 cells separated the overlapping fluorescent probes, indicating that they contained MLL translocations (P < 10-11; Figure 5, compare F with G, H). In some cases the second fluorescent signal was not visible. These events could be either translocations involving loss of a significant amount of chromosome band 11q23 material during the translocation event or deletions of the whole chr 11 end. Interestingly, 46 of 850 cells contained multiple signal aberrations (P < 10-13; Figure 5, compare F with I, compare J with K-M) indicating that etoposide treatment can promote complex MLL locus rearrangements within a single cell, which is consistent with complex MLL rearrangements that occasionally occur in patients.60-62 The percentage of gross chromosomal translocations that included chr 11 was confirmed by FISH using a chr 11–Cy3 probe (Figure 5B-E). Because a predominant MLL translocation fusion partner is AF-4 located on chromosome band 4q21, we compared the frequency of translocations that included chr 4 using a chr 4–FITC probe (Figure 5B-E). Following etoposide exposure and long recovery, translocations involving chr 11 were observed in 10% of metaphases, and translocations involving chr 4 were observed in 5% of metaphases. In no case, did we observe a t(4;11). However, the cell culture conditions favored myeloid expansion, and the t(4;11)(q21;q23) fusing MLL and AF-4 occurs mainly in acute lymphoblastic leukemia (ALL) and less often in monoblastic AML. Aberrations involving either chromosome were observed in 1% of untreated controls, in agreement with the expected frequency of spontaneously detectable aberrations of cells in culture.63,64
Fluorescence in situ hybridization to visualize etoposide-induced chr11 rearrangements in cells after long recovery period. (A) Schematic overlap between fluorescent probes of 11q23 locus–specific identifier (LSI). Intact 11q23 is seen as yellow signal (overlap of green and red). Aberration of 11q23 is seen as separation of probes into independent green and red signals. (B-E) Whole chromosome painting with chr 11–Cy3 (red) and chr 4–FITC (green). (B) Untreated CD34+ control. (C-E) Representative metaphase spreads that contain chr 11 aberrations. (F-I) LSI of chromosome band 11q23 in interphase and metaphase cells. (F,J) Untreated CD34+ control. (G-H) Representative split signals observed in 59 of 850 etoposide-treated and analyzed cells. (I, K-M) Representative complex rearrangements observed in 46 of 850 etoposide-treated and analyzed cells. Images were viewed at 600×, 1.4 aperture, with Immersol oil immersion (Zeiss, Oberkochen, Germany).
Fluorescence in situ hybridization to visualize etoposide-induced chr11 rearrangements in cells after long recovery period. (A) Schematic overlap between fluorescent probes of 11q23 locus–specific identifier (LSI). Intact 11q23 is seen as yellow signal (overlap of green and red). Aberration of 11q23 is seen as separation of probes into independent green and red signals. (B-E) Whole chromosome painting with chr 11–Cy3 (red) and chr 4–FITC (green). (B) Untreated CD34+ control. (C-E) Representative metaphase spreads that contain chr 11 aberrations. (F-I) LSI of chromosome band 11q23 in interphase and metaphase cells. (F,J) Untreated CD34+ control. (G-H) Representative split signals observed in 59 of 850 etoposide-treated and analyzed cells. (I, K-M) Representative complex rearrangements observed in 46 of 850 etoposide-treated and analyzed cells. Images were viewed at 600×, 1.4 aperture, with Immersol oil immersion (Zeiss, Oberkochen, Germany).
Discussion
We established an in vitro culture system and IPCR to characterize the spectrum of stable, and potentially leukemogenic, genomic rearrangements initiating within a therapy-related translocation hotspot of the MLL bcr induced by etoposide exposure. IPCR of etoposide-treated CD34+ cell samples gave readily detectable, variable-sized bands representing alterations to the region. Importantly, in several instances, the same aberrations were obtained in different aliquots of a single DNA sample used for independent amplifications. In samples that repaired DNA damage and proliferated 10 to 14 days, this result is likely due to expansion of aberrant clones. However, in samples harvested 2 to 3 hours after etoposide exposure when no proliferation of cells is expected, this result suggests that some sequences are preferential substrates for etoposide-induced cleavage and illegitimate recombination repair events. We estimate that the relative frequency of the stable MLL illegitimate repair products is 2 to 4 per 20 000 etoposide-treated cells (1.5 × 10-4). This calculation is based on the relative sensitivity of PCR to amplify 1 copy per 10 000 cells, and 200 ng DNA used per PCR reaction corresponding to 40 000 cells, and 50% cell death induced by etoposide at this dose range.
Our results imply that the initial formation of MLL locus aberrations including translocations are a general phenomenon, and can develop independently of individual genetic factors; however, any dysfunction or variation of DNA damage sensor and repair proteins might be expected to influence both the frequency and spectrum of repair products. After etoposide removal, DSBs rapidly disappear in 2 to 4 hours, and, within this short period, we detected multiple rearrangements with a high ratio of MLL translocations. The major decrease in cell number occurred 12 to 24 hours after treatment, presumably as cells with unrepaired DNA breaks led to apoptosis and cell death. Cells that appropriately repaired damage or acquired nonlethal DNA rearrangements resumed proliferation after 2 to 4 days in culture. However, the development and final emergence of a malignant clone responsible for therapy-related leukemia depends on the survival and proliferative potential and possibly secondary alterations leading to the selection and clonal expansion of the cell in which repair has occurred. The early clearance and delayed selection of rearranged clones is consistent with the development of MLL-rearranged therapy-related leukemias in a small fraction of patients receiving chemotherapy regimens that include topo II poisons.
The specific etoposide-induced rearrangements initiating within MLL reported here are consistent with the full spectrum of tandem duplications, translocations, and complex rearrangements identified in cells from patients with therapy-related leukemias.55,65,66 The spectrum of illegitimate repair products detectable after minimal 2 to 3 hours recovery included frequent MLL PTDs and translocations. However, in cells that repaired damage and continued to proliferate for 10 to 14 days, the significant majority of illegitimate repair events were MLL PTDs, and a single translocation. This shift in the spectrum of detectable rearrangements may result from clearance of unstable clones through apoptosis or competitive expansion of certain clones due to proliferative advantage. Although our data indicate that stable MLL PTDs are a frequent outcome of etoposide-induced DNA damage, it is likely that many are not fully leukemogenic and that additional alterations are needed. Smaller duplications would not be expected to alter MLL protein function, but produced mechanistically similar to those in leukemias in patients.54 Since our culture conditions favor expansion of myeloid precursors, PTDs also seem consistent with the clinical association of MLL PTDs with AML but not ALL. Conditions generating lymphoid precursors might result in expansion of clones with different spectrum of products.
The low ratio of deletions is likely due to the predominant isolation and analysis of products that differed significantly in size relative to the genomic 1.8-kb MLL fragment. The smallest deletion was 100 bp, similar to another study in which small deletions of 100 to 200 bp of the TEL gene locus were detected in 12 of 17 clones from ALL cell lines after serum starvation.67 Larger deletions that include the primers used for IPCR would also be excluded from this analysis. Alternatively, the lack of deletions could reflect locus- and chromosome-specific differences or differences in the preferred type of DNA damage induced by etoposide. This study also revealed 3 complex repair events with insertion of foreign sequences followed by a PTD of MLL sequence. A similar sequence abnormality was described in a case of B-lineage ALL in which a duplicated portion of MLL was interrupted by a 72-bp insertion of AF9 genomic sequence from 9p22.68 Together with the complex events detected by cytogenetic analysis, these data indicate that the spectrum of potentially leukemogenic MLL repair events detected in this experimental system is similar to events identified in leukemia in patients. Additional studies in patients undergoing treatment would determine whether there is also a shift in repair products from early times following etoposide exposure to when stable leukemia-associated clones have been established.
FISH analysis correlated with the molecular analysis. FISH analysis using the MLL-specific probe identified aberrations within this locus in 12% of cells (105 of 850) Similarly, using FISH and whole chromosome fluorescent probes, we detected chr 11 aberrations twice as frequently as chr 4 aberrations. This is in accordance with results of cytogenetic analysis of peripheral blood mononuclear cells after 48 hours' continuous exposure to etoposide in which chr 11 was altered 2 times more frequently than expected based on relative length, and chr 4 was altered at a lower frequency than expected.26 This result also is consistent with the predilection for translocations involving chr 4 to occur in cells of lymphoid lineage and the culture conditions used here that would favor myeloid outgrowth.
Among the observed intragenic rearrangements, most were PTDs with breakpoints localized within repetitive elements, which were detected more frequently in this model system than seen in patient samples. Alterations of the MLL locus in leukemic cells have been found to involve PTDs at or near Alu repeats.54,69 It has been proposed that Alu elements promote homology-directed replication slippage or homology-mediated illegitimate DSB repair between sister chromatids or homologs. A similar repair mechanism could account for both leukemia-associated MLL PTDs and those induced in this experimental system. PTDs have not been detected in other reports of human cells treated with etoposide or after apoptotic stimuli, although others have not analyzed cells after extensive proliferation or events at the sequence level. A recent report on rearrangements of the MLL locus in mouse embryonic stem (ES) cells does not provide evidence of etoposide-induced PTDs in MLL after 48 hours of recovery.52 This difference might be due to the sequence divergences between the MLL locus of the 2 species, the general lack of Alu elements in the rodent genome, or different repair pathways used for DSB repair in ES cells compared with hematopoietic CD34+ progenitors. Homologous recombination is a predominant DSB repair pathway in ES cells, but the relative reliance of repair pathways in the stem cell–enriched CD34+ cell population is not known. However, mouse ES cells can repair I-SceI endonuclease-induced DSBs to form stable intragenic duplications and chromosomal translocations between introduced neomycin gene sequence repeats (frequency, 1 × 10-4), indicating that ES cells and hematopoietic progenitors can use the same repair pathways in at least some instances.11,70
It is notable that a significant majority of both the PTD breakpoints (86%) and translocation breakpoints (60%) localized within repetitive elements (Figure 4). The MLL bcr contains 8 Alu, 4 LINE, and 2 low-copy MER repetitive elements that span 3824 bp (46% of the bcr).22 Thus, the percentage of both internal rearrangements (86% vs 46%) and translocations (60% vs 46%) located within repetitive elements is higher than expected by chance. The presence of Alu elements previously identified at or near breakpoint junctions in leukemic cells suggested that such sequences are capable of illegitimate invasion into a homologous Alu sequence during DSB repair or restart of a collapsed replication fork.54 Of the 35 recovered intragenic duplications, 31 contain Alu-Alu or LINE-LINE element breakpoint junctions, demonstrating experimentally that homology-mediated invasion is a predominant mechanism to repair etoposide-stimulated DNA damage. Further, the spectrum of repair products identified immediately after etoposide exposure demonstrates that illegitimate homology-mediated repair can occur with similar frequency between homologs or sister chromatids as between heterologs. Repair mechanism choice may be locus specific. In support of this, the MLL bcr fragment may act in cis to promote spontaneous recombination 3- to 4-fold between simian virus 40 molecules that can be elevated by etoposide treatment.71
Homology-mediated DSB repair in mammalian cells requires members of the RAD52 epistasis group.72 Rad51 is central to most homologous recombination events; Rad51 forms nucleoprotein filaments on ssDNA, and mediates homologous pairing and strand exchange between DNA duplexes,73,74 although single-strand annealing can be promoted by Rad52 in the absence of Rad51.75 Whether the homology-mediated events observed here are Rad51 dependent or are sufficiently promoted by the annealing activity of Rad52 is not clear. Aberrant overexpression of Rad51 is tolerated for cell viability, but leads to multiple cell defects including impaired cell-cycle progression, promiscuous recombination, and aneuploidy.76-79 Elevated levels of Rad51 detected in multiple tumor cell types may contribute to genomic instability by stimulating recombination between short repetitive elements and homologous sequences.76,78,80 Expression of Rad51 is predictive of B-cell chronic lymphocytic leukemia patient response to nitrogen mustards,81,82 as well as small-cell lung cancer cell sensitivity to VP-16.83 Thus, the ability to modulate damage sensor and repair proteins will impact the ability of the CD34+ subpopulation to protect against illegitimate repair of etoposide-induced DNA damage and potentially oncogenic rearrangements. This system can be used to determine the relative importance of repair proteins in the maintenance of genome stability following exposure of CD34+ cells to chemotherapeutic regimens that include etoposide.
Prepublished online as Blood First Edition Paper, November 4, 2004; DOI 10.1182/blood-2004-07-2683.
Supported in part by R01CA100159 and the Stewart Trust Foundation (C.R.); and RO1CA77683, RO1CA85469, and Leukemia and Lymphoma Society Specialized Center of Research (SCOR) grant (C.A.F.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Anna Vitebsky for technical assistance, Vladin Miljkovic and Eric Rappaport for sequencing, and Dorothy Warburton for FISH.