Abstract
Cytosolic ferritin sequesters and stores iron and, consequently, protects cells against iron-mediated free radical damage. However, the function of the newly discovered mitochondrial ferritin (MtFt) is unknown. To examine the role of MtFt in cellular iron metabolism, we established a cell line that stably overexpresses mouse MtFt under the control of a tetracycline-responsive promoter. The overexpression of MtFt caused a dose-dependent iron deficiency in the cytosol that was revealed by increased RNA-binding activity of iron regulatory proteins (IRPs) along with an increase in transferrin receptor levels and decrease in cytosolic ferritin. Consequently, the induction of MtFt resulted in a dramatic increase in cellular iron uptake from transferrin, most of which was incorporated into MtFt. The induction of MtFt caused a shift of iron from cytosolic ferritin to MtFt. In addition, iron inserted into MtFt was less available for chelation than that in cytosolic ferritin and the expression of MtFt was associated with decreased mitochondrial and cytosolic aconitase activities, the latter being consistent with the increase in IRP-binding activity. In conclusion, our results indicate that overexpression of MtFt causes a dramatic change in intracellular iron homeostasis and that shunting iron to MtFt likely limits its availability for active iron proteins.
Introduction
Ferritins are ubiquitous, highly conserved iron storage proteins that play a critical role in cellular and organismal iron homeostasis. In mammalian cells, cytosolic ferritin consists of 2 subunits, the H and L chains. Twenty-four subunits assemble to form a spherical shell that can contain up to 4500 atoms of iron in a mineral and compact form.1,2 In ferritin shells, the H subunit has ferroxidase activity that converts soluble ferrous ions into inert ferric hydroxides. The L subunit lacks ferroxidase activity but is more efficient than the H subunit in inducing iron nucleation and mineralization within the shells.3,4 Ferritin H and L chain mRNAs have identical iron-responsive elements (IREs) at their 5′-untranslated regions (UTRs) that bind to cytosolic iron regulatory proteins IRP1 and IRP2 by an iron-dependent mechanism.2,5 When the level of iron in the so-called “labile iron pool” (LIP) is low, translation of ferritin mRNA is suppressed by the binding of IRPs to IREs, whereas iron supplementation up-regulates ferritin synthesis through the conversion of IRP1 to cytosolic aconitase and the degradation of IRP2.5-8
Mitochondria have a key role in iron metabolism because heme and various iron-sulfur (Fe-S) cluster-containing proteins are synthesized in them.9,10 The last step in heme biosynthesis, the insertion of Fe2+ into protoporphyrin IX by ferrochelatase, occurs in the mitochondrial matrix.9 Fe-S clusters are synthesized mainly, if not entirely, in mitochondria and are combined with mitochondrial apo-proteins to form mature proteins or are exported from mitochondria for use by cytosolic and nuclear proteins.10 Mitochondrial iron level must be well regulated because an inadequate supply of iron would impair the metabolic and respiratory activities of the organelle, whereas excess “free” iron in mitochondria would promote the generation of harmful reactive oxygen species, which are produced as a side reaction of mitochondrial electron transport.11,12
Defects in mitochondrial iron transport and utilization can result in mitochondrial iron overload. There is extensive iron accumulation in erythroblast mitochondria of patients with X-linked sideroblastic anemia due to defective erythroid-specific 5-aminolevulinic acid synthase (eALAS)9,13-15 and those with ringed sideroblasts associated with myelodysplastic syndrome.13,15 Mitochondrial iron overload has also been documented in patients with Friedreich ataxia with defective frataxin16-18 and in those with sideroblastic anemia with ataxia from defects in the Fe-S transporter ABC7.19 In addition, studies with yeast, the best-studied eukaryotic model of Fe-S cluster synthesis, showed that defects in any of the enzymes of the Fe-S cluster assembly pathway caused mitochondrial iron accumulation and lack of normal mitochondrial function.20
The recent discovery of mitochondrial ferritin (MtFt) has contributed to our understanding of mitochondrial iron metabolism21-24 and sideroblastic anemia. Although most cells, including normal erythroid cells, express extremely low levels of MtFt, it is highly expressed in erythroblast mitochondria (“ringed sideroblasts”) of patients with sideroblastic anemia21,24 and it was shown that the iron in ringed sideroblasts is in MtFt.24 In humans, MtFt is encoded by an intronless gene on chromosome 5q23.1. The protein is expressed as a 30-kDa precursor that is targeted to mitochondria. After intramitochondrial deletion of the N-terminal leader sequence, the 22-kDa MtFt subunits, which are highly homologous to H subunits, assemble into ferritin shells with ferroxidase activity.21,22 In HeLa cells, overexpression of human MtFt, which incorporates iron, results in decreased cytosolic ferritin and increased transferrin receptor levels, suggesting cytosolic iron deficiency.22
Although the regulation of MtFt expression and its functions are not understood, it is likely that it plays a role in cellular iron homeostasis. To investigate the role of MtFt in cellular iron uptake, distribution, and mobilization, we generated a human cell line with tetracycline (tet)–controlled expression of mouse MtFt with which we provide direct evidence that MtFt dramatically affects intracellular iron metabolism.
Materials and methods
Chemicals
EXPRE35S35S protein-labeling mix and 59FeCl3 were from Perkin Elmer (Boston, MA). All other reagents were purchased from Sigma (St Louis, MO) unless otherwise specified. The iron chelator salicylaldehyde isonicotinoyl hydrazone (SIH) was synthesized as described by Ponka et al.25
Plasmid constructs
The mouse MtFt cDNA encoding the full opening reading frame was amplified by polymerase chain reaction (PCR) using 5′-AGGGAATTCACCATGGGCCTGTCCTGCTTTTGGTTCTTCTC-3′ and 5′-GGCGGATCCTATTTAAGCGTAATCTGGAACATCGTATGGGTAGTGCTTGCTCTCGCTTCCAA-3′ primers with mouse testis QUICK-Clone cDNA library (Clontech, Mississauga, ON, Canada) and cloned into the pGEM-T vector (Promega, Madison, WI). This produced a plasmid containing a 768-bp fragment containing the entire mouse MtFt sequence, a C-terminal hemagglutinin (HA) epitope and an EcoRI site at the 5′ end and a BamHI site downstream of the stop codon at the 3′ end of MtFt. The 768-bp EcoRI/BamHI inserts were excised and subcloned into the EcoRI/BamHI site of pUHD10-3 (from Dr K. Pantopoulos, Lady Davis Institute, Montreal, QC, Canada) that contains a tetracycline-inducible hCMV minimal promoter26 and pcDNA3.1(-) (Invitrogen, Burlington, ON, Canada) to yield pUHD-MtFt and pcDNA3.1(-)–MtFt, respectively. Constructs with the correct sequences were confirmed by sequencing.
Generation of stable MtFt-expressing cell line
The pUHD-MtFt plasmid was cotransfected with the puromycin-resistant pBabe plasmid into tTA-H1299 cells27 by Lipofectamine Plus (Invitrogen). Stable transfectants were selected in medium containing 2 μg/mL puromycin, 250 μg/mL G418, and 2 μg/mL tetracycline. Representative clones were isolated for further characterization. The cells were maintained in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) tetracycline-free fetal bovine serum (Clontech), 2 mM glutamine, and 100 U/mL penicillin, and 0.1 ng/mL streptomycin. MtFt expression was induced by removal of tetracycline (tet-off system).27
Generation of mitochondrial leader deletion mutation and transient transfection
The MtFt leader sequence was predicted using MitoProt II software28 and the report by Corsi et al.22 A truncated MtFt sequence (deletion of the first encoded 56 amino acid residues, Δ56-MtFt) was PCR-amplified by the primer pair of 5′-GGCGGATCCTATTTAAGCGTAATCTGGAACATCGTATGGGTAGTGCTTGCTCTCGCTTCCAA-3′ and 5′-AGGGAATTCACCATGGGCGACTCCACTAGACCTTCCAG-3′. Then the 603-bp fragment was subcloned into pcDNA3.1(-) and pUHD10-3 as described (see “Plasmid constructs”) for the full-length MtFt sequence. Transient transfection with pcDNA3.1(-)–MtFt and pcDNA3.1(-)–Δ56-MtFt were performed with the Lipofectamine Plus reagent. Protein expression was examined using Western blot analysis.
Western blotting
Cells were lysed with RIPA buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) or Munro buffer (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.5, 3 mM MgCl2, 40 mM NaCl, 5% glycerol, 1 mM dithiothreitol, and 0.2% Nonidet P-40) and the lysates were immediately boiled in Laemmli loading buffer for 10 minutes. Protein content was determined by the Bradford assay (Bio-Rad, Hercules, CA). Equal amounts of protein were loaded, resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to a nitrocellulose blotting membrane (Pall, Pensacola, FL). The blots were blocked by incubating them for 1 hour with 5% nonfat milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBST) and hybridized with anti-HA rabbit polyclonal IgG (Y11; Santa Cruz Biotechnology, Santa Cruz, CA), antitransferrin receptor (anti-TfR; Zymed, South San Francisco, CA), antiferritin (Dako, Carpinteria, CA), or anti–β-actin (Sigma, Oakville, ON, Canada). After washing 3 times for 15 minutes each with PBST, the blots were incubated for 1 hour with horseradish peroxidase-linked secondary antibody. Dilutions were 1:1000 for primary antibodies and 1:10 000 for antirabbit or antimouse IgG secondary antibodies. Peroxidase-coupled secondary antibodies were detected with Western lighting (Amersham, Buckinghamshire, United Kingdom).
IRP activity
A gel retardation assay, as described previously,29 was used to measure the interaction between IRPs and IREs. Briefly, 6 × 106 cells were lysed at 4°C in 100 μL Munro buffer and the lysates were centrifuged for 10 minutes at 10 000g to remove nuclei and unlysed cells. Samples of cytoplasmic extracts were diluted with Munro buffer without 0.2% Nonidet P-40 to a protein concentration of 1 μg/μL, and 10-μg aliquots were incubated with an excess amount of 32P-labeled pST18-fer RNA transcript (containing one IRE).30 This RNA was transcribed in vitro from a linearized plasmid template using T7 RNA polymerase in the presence of α-32P] UTP. To form RNA-protein complexes, cytoplasmic extracts were incubated for 10 minutes at room temperature with this probe. Heparin (5 μg) was added and the incubation was continued for another 10 minutes to prevent nonspecific binding. RNA-protein complexes were analyzed in 6% nondenaturing polyacrylamide gels. In parallel experiments, samples were treated with 2% β-mercaptoethanol (β-ME) before adding the RNA probe to maximally activate IRP1-binding activity.
Iron uptake
Human apo-transferrin (Tf) was labeled with 59Fe as described previously.31 Equal numbers of cells (3 × 106 cells) were seeded without or with tetracycline, cultured for 48 hours, and then incubated with 1 μM 59Fe-Tf at 37°C for 24 hours. Cells were washed 3 times with ice-cold PBS, and radioactivity in the cell pellet and culture medium was measured in a γ counter (Packard Cobra II auto-gamma; Perkin-Elmer, Wellesley, MA).
Cellular iron distribution
After labeling with 59Fe-Tf at 37°C for the indicated times, the cells were harvested and lysed at 4°C with lysis buffer (0.14 M NaCl, 0.1 M HEPES, pH 7.4, and 1.5% Triton X-100), and the lysates were analyzed by electrophoresis followed by autoradiography as described previously.32,33 Briefly, following centrifugation at 10 000g for 10 minutes at 4°C, equal amounts of lysate samples were loaded onto native gradient gels (3%-20% polyacrylamide). After 4 hours of electrophoresis, the gels were dried and exposed to x-ray film.
Metabolic labeling and immunoprecipitation
Cells were incubated for 1 hour with 100 μCi/mL (3.7 MBq/mL) 35S-methionine and 35S-cysteine and lysed with RIPA buffer. Lysates containing equal amounts of protein were first treated with 30 μL protein A-Sepharose (Amersham, Piscataway, NJ) to remove proteins that bind nonspecifically to the beads. Then, 5 μg antiferritin antibody (Roche, Penzberg, Germany) was added to the supernatants, which were then incubated for 3 hours at 4°C. Protein A-Sepharose suspension (60 μL; 50% beads in RIPA buffer) was added to immunoprecipitate the ferritin-IgG complexes. The beads were washed 3 times with cold RIPA buffer (10 minutes each) and then boiled for 5 minutes with Laemmli loading buffer. Immunoprecipitated proteins were separated by 12.5% SDS-PAGE. The gels were treated with Amplify (Amersham), dried, and analyzed by autoradiography with a Phosphor Imaging Screen and a Storm (840) Scanner (Molecular Dynamics, San Diego, CA).
Aconitase (EC 4.2.1.3) assay
Both cytosolic and mitochondrial aconitase activities were measured by isocitric dehydrogenase-coupled nicotinamide adenine dinucleotide phosphate (NADPH) formation as previously described.34 Briefly, 100 μL extracts containing equal amounts of protein was added to 800 μL of a solution containing 330 mM Tris-HCl, pH 7.4, 1.5 mM manganese sulfate, 0.6 mM NADP, and 1 U/mL isocitrate dehydrogenase. The reaction was started by adding 100 μL of 20 mM sodium citrate, and production of NADPH was monitored by measuring the increase in absorbance at 340 nm. One unit of aconitase is defined as the amount that converts 1 μmol citrate per hour to isocitrate at pH 7.4, at 25°C.
Other methods
Mitochondria were isolated by an established method.35 Briefly, 1 × 108 cells were homogenized in a glass-Teflon homogenizer containing isolation medium (32 mM sucrose, 1 mM EDTA [ethylenediaminetetraacetic acid], and 10 mM Tris-HCl, pH 7.4). The homogenate was centrifuged at 1500g for 10 minutes to remove nuclei and unlysed cells. The supernatant was centrifuged at 17 000g for 10 minutes at 4°C and the pelleted mitochondria were washed twice with the isolation buffer. The postmitochondrial fraction was used as cytosolic fraction. Cross-contamination of cytosol and mitochondria was determined on the basis of the amount of activity of marker enzymes for cytosol (lactate dehydrogenase [LDH], EC 1.1.1.27)36 and mitochondria (succinate dehydrogenase [SDH], EC 1.3.99.1).37 Mitochondria contamination of cytosol was less than 0.5% (percent LDH activity in mitochondria/total cell homogenate) and cytosolic contamination of mitochondria was less than 1% (percent SDH activity in postmitochondrial fraction/total cell homogenate in RIPA buffer). Total nonheme iron was measured using the ferrozine method described by Fish38 in cells grown for 72 hours without or with tetracycline. Cellular heme and nonheme 59Fe were measured by an acid precipitation method.39 Briefly, uninduced and induced clone B9 cells were grown for 2 days, 59Fe-Tf was added, and the cells were cultured for an additional day. The cells were washed 3 times in PBS and lysed by adding 0.2 M HCl, and the lysates were boiled for 10 minutes to dissociate nonheme iron from proteins. 59Fe-heme was precipitated by adding trichloroacetic acid to a final concentration of 7% and incubated at 4°C for 15 minutes. After centrifugation at 2000g for 5 minutes, the pellets were washed 3 times with 7% trichloroacetic acid and radioactivity in the pellets (heme 59Fe) and supernatants (nonheme 59Fe) was measured in a γ counter.
Results
Characterization of transfected mouse MtFt and establishment of a tetracycline-inducible stable expression cell line
The sequence of the cloned mouse MtFt was aligned with the published sequence of mouse MtFt (NM_026286) and 2 variations were found. At nucleotide 37 there was an A>G substitution in the mitochondrial leader sequence and at nucleotide 488 there was a G>A substitution in the ferroxidase center (numbered from ATG start codon). The first resulted in a Ser → Gly change and the second to a Gly → Glu change (G107E; Glu is well conserved in human H, L, and mitochondrial ferritins, and many animals, plants, and bacteria ferritins1,21 ). MitoProt II software28 predicted that, although it would be preferred in the mitochondrial target sequence, the Ser replacement by Gly would not apparently decrease mitochondrial import efficiency.
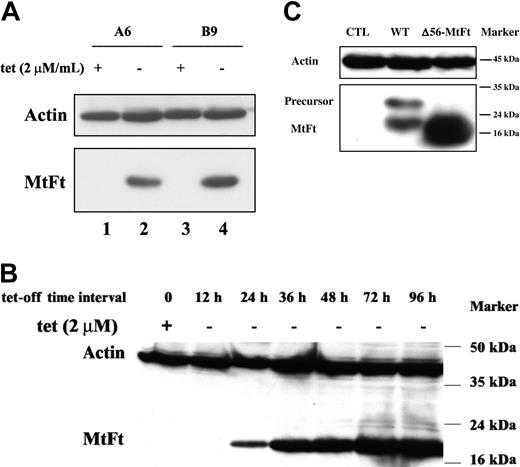
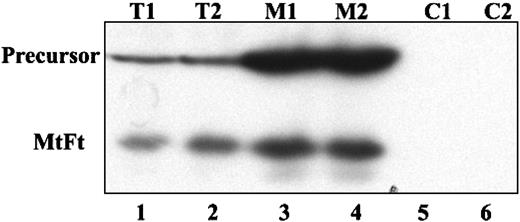
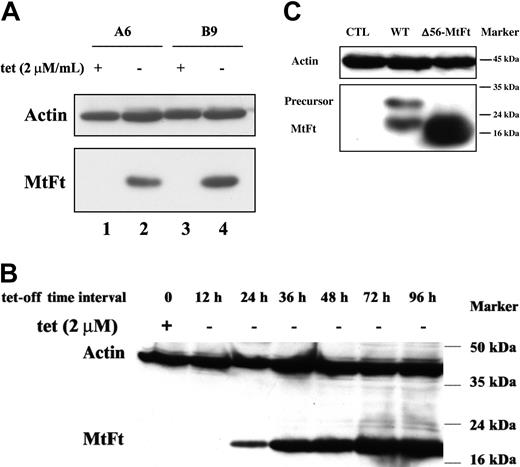
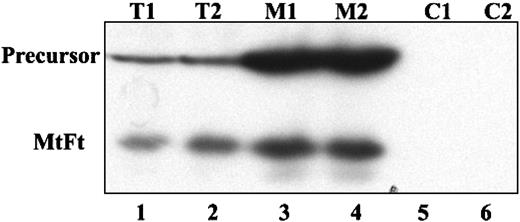
Human H1299 cells expressing the transactivator tTA were transfected with cDNAs encoding HA-tagged MtFt under the control of a tetracycline-inducible promoter (tet-off system).27 MtFt protein expression was analyzed by Western blotting using anti-HA tag antibody. Stable clones were screened and representative clones were selected (Figure 1A). Clone B9 was used for further characterization. Removal of tetracycline from the medium for 12 to 96 hours resulted in a profound induction of MtFt expression (Figure 1B). Expression of MtFt protein was time dependent and reached a maximum after 72 hours of induction. In agreement, a previous report about human MtFt,22 mouse MtFt from stable transfectants migrated as a single band with a molecular mass of approximately 22 kDa on SDS-PAGE. On the other hand, from transient transfection with pcDNA 3.1-MtFt, 2 bands, the precursor and the mature protein, were detected by Western blotting. The precursor was about 28 kDa and was processed to the 22-kDa mature protein (Figure 1C). Transient transfection of cDNA with Δ56-MtFt resulted in nearly the same molecular size band corresponding to mature MtFt (Figure 1C). This is consistent with removal of the predicted leader sequence eliminated at the predicted position. Figure 2 illustrates the distribution of MtFt in clone B9 cells. Most of the MtFt, if not all, localized to the mitochondrial fraction (M1 and M2), but not in the postmitochondrial fraction (C1 and C2). The MtFt was concentrated about 10 times in the mitochondrial fraction compared with whole-cell homogenates (T1 and T2). This result suggests that the precursor of MtFt is targeted to mitochondria where it is processed to the mature protein.
Tetracycline-inducible expression of mouse MtFt in H1299 cells. (A) Human tTA-H1299 lung cancer cells were stably transfected with HA-epitope–tagged wild-type mouse MtFt. Two representative MtFt transfectants (clones A6 and B9) were grown for 48 hours with 2 μg tetracycline/mL (+) or without tetracycline (-) and analyzed for expression of HA-tagged the MtFt by Western blotting. The hybridization membrane was probed with antibodies to HA (bottom) and stripped and reprobed with β-actin (top). The apparent size of the MtFt band was 22 kDa. (B) Time course of MtFt expression in clone B9. The figure shows that the amount of MtFt expression increased with time from 24 to 96 hours. (C) Expression of MtFt mutant without mitochondrial leader sequence. H1299 cells were transiently transfected (48 hours) with either full-length MtFt (WT) or a mutant lacking the leader sequence (Δ56-MtFt). Full-length MtFt transfection yielded an unprocessed 28-kDa band in addition to the mature 22-kDa form. The mutant protein migrated as a single band of almost identical size to the mature WT.
Tetracycline-inducible expression of mouse MtFt in H1299 cells. (A) Human tTA-H1299 lung cancer cells were stably transfected with HA-epitope–tagged wild-type mouse MtFt. Two representative MtFt transfectants (clones A6 and B9) were grown for 48 hours with 2 μg tetracycline/mL (+) or without tetracycline (-) and analyzed for expression of HA-tagged the MtFt by Western blotting. The hybridization membrane was probed with antibodies to HA (bottom) and stripped and reprobed with β-actin (top). The apparent size of the MtFt band was 22 kDa. (B) Time course of MtFt expression in clone B9. The figure shows that the amount of MtFt expression increased with time from 24 to 96 hours. (C) Expression of MtFt mutant without mitochondrial leader sequence. H1299 cells were transiently transfected (48 hours) with either full-length MtFt (WT) or a mutant lacking the leader sequence (Δ56-MtFt). Full-length MtFt transfection yielded an unprocessed 28-kDa band in addition to the mature 22-kDa form. The mutant protein migrated as a single band of almost identical size to the mature WT.
Overexpressed MtFt is targeted to mitochondria in B9 cells. H1299 cells were transiently transfected with full-length MtFt (48 hours) and the transfected cells were divided into 2 parts. One half of the cells were lysed and the cell extract was used directly for analysis of MtFt expression by Western blotting; the other half of the cells was used to isolate mitochondrial and postmitochondrial fractions. Then, 20 mg of the total cell extracts (T1 and T2, duplicates), 10 mg of the mitochondrial fractions (M1 and M2, duplicates), and 20 mg of the postmitochondrial fractions (C1 and C2, duplicates) were separated by SDS-PAGE and analyzed for the expression of MtFt by Western blotting. The figure shows that in mitochondrial fractions MtFt was concentrated, whereas in postmitochondrial fractions, no MtFt is observed.
Overexpressed MtFt is targeted to mitochondria in B9 cells. H1299 cells were transiently transfected with full-length MtFt (48 hours) and the transfected cells were divided into 2 parts. One half of the cells were lysed and the cell extract was used directly for analysis of MtFt expression by Western blotting; the other half of the cells was used to isolate mitochondrial and postmitochondrial fractions. Then, 20 mg of the total cell extracts (T1 and T2, duplicates), 10 mg of the mitochondrial fractions (M1 and M2, duplicates), and 20 mg of the postmitochondrial fractions (C1 and C2, duplicates) were separated by SDS-PAGE and analyzed for the expression of MtFt by Western blotting. The figure shows that in mitochondrial fractions MtFt was concentrated, whereas in postmitochondrial fractions, no MtFt is observed.
MtFt-induced cellular iron starvation
IRPs sense the level of intracellular iron in the LIP and coordinate the expression of ferritin and Tf receptor (TfR) through binding to IREs located in the 5′-UTR of ferritin mRNA and the 3′-UTR of TfR mRNA.2,5-8 We examined the effect of MtFt expression on IRP/IRE-binding activity. Cells were grown in the presence or absence of tetracycline for 24 to 72 hours, and lysates were then analyzed by gel retardation assays using an IRE probe. Removal of tetracycline led to a significant increase in IRE-binding activity in cells expressing MtFt, about 2.8- and 1.8-fold (quantified by densitometry) after 48 or 72 hours of induction, respectively (Figure 3A, lanes 1-6). As expected, there was no change in total IRP activity that was revealed when cell extracts were treated with β-ME (Figure 3A, lanes 7-12). Because in human tissues and cells, both IRP1/IRE and IRP2/IRE complexes comigrate in gel retardation assays, here they are referred to as “IRP.”
Effects of MtFt overexpression on the IRE-IRP system, TfR levels, and ferritin synthesis. (A) MtFt overexpression increases IRP-binding activity. Stably transfected H1299 cells (clone B9) were seeded in 100-mm dishes and grown for 24 hours (lanes 1-2, 7-8), 48 hours (lanes 3-4, 9-10), and 72 hours (lanes 5-6, 11-12) without (-) or with (+) 2 μg/mL tetracycline. Cytoplasmic extracts (10 μg) were assayed for their ability to retard the migration of a 32P-labeled IRE probe in the absence (lanes 1-6) or presence (lanes 7-12) of 2% β-ME. (B) Stimulation of TfR expression by MtFt overexpression. Clone B9 cells were seeded (25% confluence) in 100-mm dishes and grown for 24, 48, and 72 hours without (-) or with (+) tetracycline. The cell lysates were analyzed for expression of MtFt, TfR, and actin by Western blotting. The filters were probed with antibodies against HA-tag (MtFt, upper panel), stripped, and reprobed with TfR (middle panel) and actin (lower panel), respectively. (C-D) Effect of MtFt overexpression on cytosolic ferritin synthesis. In panel C, cells treated without or with tetracycline for 24 hours (lanes 1-2), 48 hours (lanes 3-4), and 72 hours (lanes 5-6) were metabolically labeled with [35S] methionine for 1 hour and cell extracts were subjected to immunoprecipitation with an antiferritin antibody. The immunoprecipitated proteins were separated by SDS-PAGE on 12.5% gels and autoradiographed. In panel D, the same treatment of the cells was used to measure the steady-state amount of ferritins (lower bands) by Western blotting (top band is MtFt that is recognized by the antiferritin antibody).
Effects of MtFt overexpression on the IRE-IRP system, TfR levels, and ferritin synthesis. (A) MtFt overexpression increases IRP-binding activity. Stably transfected H1299 cells (clone B9) were seeded in 100-mm dishes and grown for 24 hours (lanes 1-2, 7-8), 48 hours (lanes 3-4, 9-10), and 72 hours (lanes 5-6, 11-12) without (-) or with (+) 2 μg/mL tetracycline. Cytoplasmic extracts (10 μg) were assayed for their ability to retard the migration of a 32P-labeled IRE probe in the absence (lanes 1-6) or presence (lanes 7-12) of 2% β-ME. (B) Stimulation of TfR expression by MtFt overexpression. Clone B9 cells were seeded (25% confluence) in 100-mm dishes and grown for 24, 48, and 72 hours without (-) or with (+) tetracycline. The cell lysates were analyzed for expression of MtFt, TfR, and actin by Western blotting. The filters were probed with antibodies against HA-tag (MtFt, upper panel), stripped, and reprobed with TfR (middle panel) and actin (lower panel), respectively. (C-D) Effect of MtFt overexpression on cytosolic ferritin synthesis. In panel C, cells treated without or with tetracycline for 24 hours (lanes 1-2), 48 hours (lanes 3-4), and 72 hours (lanes 5-6) were metabolically labeled with [35S] methionine for 1 hour and cell extracts were subjected to immunoprecipitation with an antiferritin antibody. The immunoprecipitated proteins were separated by SDS-PAGE on 12.5% gels and autoradiographed. In panel D, the same treatment of the cells was used to measure the steady-state amount of ferritins (lower bands) by Western blotting (top band is MtFt that is recognized by the antiferritin antibody).
Next, we examined the effect of overexpression of MtFt on the expression of TfR. Clone B9 cells were grown in the presence or absence of tetracycline for 24 to 72 hours and TfR levels were analyzed by Western blotting. The increased expression of MtFt correlated with a time- and dose-dependent increase in TfR levels (2.3- and 2.8-fold after 2 and 3 days of induction, respectively; Figure 3B), whereas the levels of β-actin did not change. This increase in the levels of TfR after expression of MtFt is consistent with a cellular response to iron deficiency. To explore this, we examined the role of MtFt in the expression of cytosolic ferritin, which is regulated at the posttranscriptional (translational) level by IRE-IRP interactions.5-9,40 After cells were labeled metabolically with 35S-methionine and 35S-cysteine, ferritin synthesis was analyzed by immunoprecipitation (Figure 3C). Expression of MtFt after the removal of tetracycline clearly inhibited ferritin synthesis (Figure 3C, lanes 2, 4, and 6) even after 24 hours of induction. We also measured the steady-state level of ferritins (Figure 3D) in the cells with the same treatment. A decrease in cytosolic ferritin levels was found in the cells with 24-hour induction of MtFt expression; after 48-hour induction of MtFt, the cytosolic ferritin was barely detectable. It appears that MtFt promoted an apparent iron-deficient phenotype in H1299 cells, namely, increased IRP-binding activity, increased TfR levels, and inhibited cytosolic ferritin synthesis.
Effect of MtFt expression on uptake of 59Fe-Tf and mitochondrial iron content
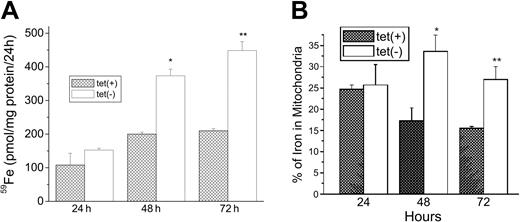
We next examined the role of MtFt expression in the uptake of 59Fe from 59Fe-Tf and the distribution of 59Fe between cytosol and mitochondria. Cells grown for 0, 24, or 48 hours with or without tetracycline were incubated with 59Fe-Tf for another 24 hours and cell-associated radioactivity was measured. Expression of MtFt on removal of tetracycline resulted in a significant (P < .01) increase in 59Fe uptake from 59Fe-Tf by approximately 180% and 250% at 48 hours and 72 hours of induction, respectively (Figure 4A). Analysis of the distribution of radioactivity between cytosol and mitochondria showed that MtFt-expressing cells had a significantly higher percentage of the 59Fe taken up in mitochondria. At 48 hours of induction, it increased from 17.5% in uninduced cells to 33.0% in cells expressing MtFt. After 72 hours of induction, the percentage of 59Fe in mitochondria was about 27% in cells expressing MtFt, still significantly higher than in the control cells (16.4%; Figure 4B). Most, if not all, of this increased 59Fe was likely in MtFt. In another experiment, it was found that MtFt increased total cellular nonheme iron from 0.264 ± 0.009 nM/106 cells to 0.358 ± 0.012 nM/106 cells after 72 hours of induction of MtFt expression. Thus, MtFt expression led to cytosolic iron deficiency and increased total iron uptake with more iron shunted into the mitochondria.
Increase of total uptake of 59Fe and localization of 59Fe into mitochondria by MtFt overexpression. (A) Total iron uptake. Clone B cells (30% confluence), grown for 24, 48, or 72 hours without (□) or with tetracycline ( ), were incubated for 24 hours with 1 μM 59Fe-Tf and washed 3 times in PBS, and their radioactivity was measured. (B) Distribution of iron between cytosol and mitochondria. The cells treated as described in panel A were homogenized in lysis buffer (sucrose 0.25 M, 10 mM HEPES, pH 7.4, 0.15% bovine serum albumin). The cytosolic and mitochondrial fractions were prepared by differential centrifugation and their radioactivities were measured. The amount of iron in mitochondria is expressed as a percentage of total 59Fe taken up. *P < .01 compared with 48-hour tet-on cells; **P < .01 compared with 72 hour tet-on cells. Data shown are the means ± SD of triplicate determinations from a typical experiment that was performed 3 times.
), were incubated for 24 hours with 1 μM 59Fe-Tf and washed 3 times in PBS, and their radioactivity was measured. (B) Distribution of iron between cytosol and mitochondria. The cells treated as described in panel A were homogenized in lysis buffer (sucrose 0.25 M, 10 mM HEPES, pH 7.4, 0.15% bovine serum albumin). The cytosolic and mitochondrial fractions were prepared by differential centrifugation and their radioactivities were measured. The amount of iron in mitochondria is expressed as a percentage of total 59Fe taken up. *P < .01 compared with 48-hour tet-on cells; **P < .01 compared with 72 hour tet-on cells. Data shown are the means ± SD of triplicate determinations from a typical experiment that was performed 3 times.
Increase of total uptake of 59Fe and localization of 59Fe into mitochondria by MtFt overexpression. (A) Total iron uptake. Clone B cells (30% confluence), grown for 24, 48, or 72 hours without (□) or with tetracycline ( ), were incubated for 24 hours with 1 μM 59Fe-Tf and washed 3 times in PBS, and their radioactivity was measured. (B) Distribution of iron between cytosol and mitochondria. The cells treated as described in panel A were homogenized in lysis buffer (sucrose 0.25 M, 10 mM HEPES, pH 7.4, 0.15% bovine serum albumin). The cytosolic and mitochondrial fractions were prepared by differential centrifugation and their radioactivities were measured. The amount of iron in mitochondria is expressed as a percentage of total 59Fe taken up. *P < .01 compared with 48-hour tet-on cells; **P < .01 compared with 72 hour tet-on cells. Data shown are the means ± SD of triplicate determinations from a typical experiment that was performed 3 times.
), were incubated for 24 hours with 1 μM 59Fe-Tf and washed 3 times in PBS, and their radioactivity was measured. (B) Distribution of iron between cytosol and mitochondria. The cells treated as described in panel A were homogenized in lysis buffer (sucrose 0.25 M, 10 mM HEPES, pH 7.4, 0.15% bovine serum albumin). The cytosolic and mitochondrial fractions were prepared by differential centrifugation and their radioactivities were measured. The amount of iron in mitochondria is expressed as a percentage of total 59Fe taken up. *P < .01 compared with 48-hour tet-on cells; **P < .01 compared with 72 hour tet-on cells. Data shown are the means ± SD of triplicate determinations from a typical experiment that was performed 3 times.
Cellular iron distribution after induction of MtFt expression and SIH mobilization of intracellular iron
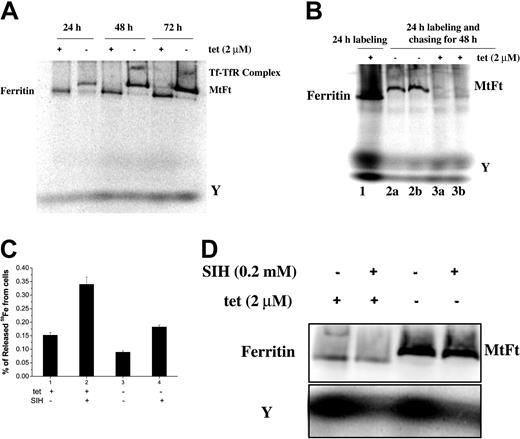
To further examine the effect of MtFt on cellular iron distribution, we used native PAGE combined with 59Fe autoradiography to evaluate the distribution of 59Fe in the major cellular iron-containing molecules. Figure 5A shows that MtFt (confirmed by supershift assay; data not shown) migrated more slowly than cytosolic ferritin but their positions were not far apart, suggesting that MtFt probably forms multimeric shells similar to cytosolic ferritin. Induction of MtFt expression almost totally blocked iron from being incorporated into cytosolic ferritin, most of it being found in MtFt, whereas in uninduced cells the majority of the 59Fe was taken up by cytosolic ferritin (Figure 5A). These results suggest that MtFt forms functional ferritin-like shells and competes with cytosolic ferritin for iron. There also was an increase in 59Fe-Tf-TfR complexes after 48 and 72 hours of induction of MtFt expression, whereas the amount of 59Fe in the low-molecular-weight fraction, referred to as fraction “Y” in a previous study,32 did not apparently change (Figure 5A). The positions of the Tf and Tf-TfR bands was verified by supershifts using anti-Tf antibodies (data not shown). This, together with the low incorporation of 59Fe into cytosolic ferritin, suggests that most of the iron from Tf shunted into mitochondria may bypass the cytosol. Next, the cells were incubated with 59Fe-Tf for 24 hours, washed, and cultured for 48 hours without 59Fe and without tetracycline to induce MtFt expression. Figure 5B clearly shows that the induction of MtFt leads to the mobilization of iron from both cytosolic ferritin and fraction Y, indicating that cytosolic iron can also be incorporated into MtFt.
The effect of MtFt on distribution and trafficking of intracellular 59Fe and on the availability of 59Fe for chelation by SIH. (A) Distribution of 59Fe-containing molecules. Induced and uninduced clone B9 cells were incubated with 59Fe-Tf for 1, 2, or 3 days and lysed with Triton X-100 lysis buffer (1.5% Triton X-100, 0.14 M NaCl, 0.1 M HEPES, pH 7.4, and proteinases cocktail). The lysates were centrifuged at 10 000g for 10 minutes and the soluble fractions (40 μg) were separated by native gradient gel electrophoresis, which was followed by 59Fe autoradiography. (B) Mobilization of iron from endogenous ferritin by MtFt overexpression. Uninduced clone B9 cells (80% confluence) were incubated with 1 μM 59Fe-Tf and after 24 hours some of the cells were processed immediately and the others were reseeded (50% confluence), and incubated for another 48 hours without (2a and 2b) or with (3a and 3b; a and b represent duplicates) tetracycline. After washing, lysing and centrifugation, the lysates were subjected to native PAGE, which was followed by 59Fe autoradiography. (C) 59Fe mobilization from wild-type and MtFt-expressing cells by the iron chelator SIH. Induced and uninduced clone B9 cells were cultured for 48 hours without (-) or with (+) tetracycline, and then for another 24 hours with 1 μM 59Fe-Tf. After washing 3 times with cold PBS, the cells were incubated without or with 200 μM SIH. After 5 hours of incubation, 59Fe in the culture medium and the cells was measured by a γ counter. Data shown are the means ± SD of triplicate determinations from a typical experiment that was performed 3 times. (D) 59Fe mobilization from cytosolic ferritin and MtFt by the iron chelator SIH. Cells were treated as described in panel C. After the pellets were lysed, the extracts were subjected to native PAGE, which was followed by 59Fe autoradiography. The intensities of the bands, corresponding to cytosolic ferritin, MtFt, and fraction Y, were quantified by densitometry. (Note: exposure time for fraction Y was 3-fold longer that that for ferritins.) For panel D, a representative of 5 experiments that gave similar results is shown.
The effect of MtFt on distribution and trafficking of intracellular 59Fe and on the availability of 59Fe for chelation by SIH. (A) Distribution of 59Fe-containing molecules. Induced and uninduced clone B9 cells were incubated with 59Fe-Tf for 1, 2, or 3 days and lysed with Triton X-100 lysis buffer (1.5% Triton X-100, 0.14 M NaCl, 0.1 M HEPES, pH 7.4, and proteinases cocktail). The lysates were centrifuged at 10 000g for 10 minutes and the soluble fractions (40 μg) were separated by native gradient gel electrophoresis, which was followed by 59Fe autoradiography. (B) Mobilization of iron from endogenous ferritin by MtFt overexpression. Uninduced clone B9 cells (80% confluence) were incubated with 1 μM 59Fe-Tf and after 24 hours some of the cells were processed immediately and the others were reseeded (50% confluence), and incubated for another 48 hours without (2a and 2b) or with (3a and 3b; a and b represent duplicates) tetracycline. After washing, lysing and centrifugation, the lysates were subjected to native PAGE, which was followed by 59Fe autoradiography. (C) 59Fe mobilization from wild-type and MtFt-expressing cells by the iron chelator SIH. Induced and uninduced clone B9 cells were cultured for 48 hours without (-) or with (+) tetracycline, and then for another 24 hours with 1 μM 59Fe-Tf. After washing 3 times with cold PBS, the cells were incubated without or with 200 μM SIH. After 5 hours of incubation, 59Fe in the culture medium and the cells was measured by a γ counter. Data shown are the means ± SD of triplicate determinations from a typical experiment that was performed 3 times. (D) 59Fe mobilization from cytosolic ferritin and MtFt by the iron chelator SIH. Cells were treated as described in panel C. After the pellets were lysed, the extracts were subjected to native PAGE, which was followed by 59Fe autoradiography. The intensities of the bands, corresponding to cytosolic ferritin, MtFt, and fraction Y, were quantified by densitometry. (Note: exposure time for fraction Y was 3-fold longer that that for ferritins.) For panel D, a representative of 5 experiments that gave similar results is shown.
SIH, which mobilizes iron from reticulocytes whose mitochondria are loaded with nonheme iron,25 was used to examine whether iron in MtFt can be chelated (Figure 5C-D). Uninduced and induced (48 hours) cells were incubated with 59Fe-Tf for 24 hours and with SIH for another 5 hours, after which 59Fe in the medium and cells was measured. Figure 5C shows that SIH released 33% of the total cellular 59Fe from uninduced cells, whereas only 18% of 59Fe was released from MtFt-expressing cells. Similarly, SIH released about 31% (quantified by densitometry) of 59Fe from cytosolic ferritin, whereas only 21% was released from MtFt in induced cells (Figure 5D).
MtFt induction inhibits both mitochondrial and cytosolic aconitase activities and 59Fe incorporation into heme
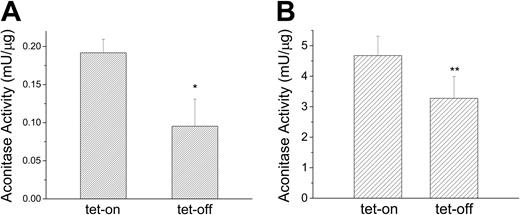
Because IRP1 and cytosolic aconitase are the same protein with 2 different functions and because mitochondrial aconitase is an Fe-S cluster protein whose mRNA contains a functional IRE in its 5′-UTR,41,42 we examined whether MtFt-induced cytosolic iron depletion directly affects the activities of these 2 Fe-S cluster proteins. MtFt expression was induced for 72 hours and isocitric dehydrogenase-coupled NADPH formation was used to measure aconitase activities of both proteins. Consistent with the previous result showing increased IRP-binding activity on MtFt induction (Figure 3A), cytosolic aconitase activity decreased about 50% (P < .01) in induced cells (Figure 6A). Interestingly, mitochondrial aconitase also responded to MtFt induction and was decreased by about 30% (P < .05) in induced cells (Figure 6B).
MtFt overexpression reduces both cytosolic and mitochondrial aconitase activities. Clone B9 cells (30% confluence), grown for 72 hours without or with tetracycline, were harvested and homogenized in lysis buffer (sucrose 0.25 M, 10 mM HEPES, pH 7.4, 0.15% bovine serum albumin). Cytosolic (A) and mitochondrial (B) fractions separated by differential centrifugation and assayed for aconitase activity. t test *P < .01 compared with tet-on cells; **P < .05 compared with tet-on cells. Data shown are the means ± SD of triplicate determinations from a typical experiment that was performed 3 times.
MtFt overexpression reduces both cytosolic and mitochondrial aconitase activities. Clone B9 cells (30% confluence), grown for 72 hours without or with tetracycline, were harvested and homogenized in lysis buffer (sucrose 0.25 M, 10 mM HEPES, pH 7.4, 0.15% bovine serum albumin). Cytosolic (A) and mitochondrial (B) fractions separated by differential centrifugation and assayed for aconitase activity. t test *P < .01 compared with tet-on cells; **P < .05 compared with tet-on cells. Data shown are the means ± SD of triplicate determinations from a typical experiment that was performed 3 times.
In another experiment, uninduced and induced clone B9 cells were grown for 2 days, 59Fe-Tf was added, and the cells were cultured for an additional day. After washing, 59Fe was measured in the heme and nonheme iron fractions. The ratios of heme-59Fe to nonheme 59Fe were 0.110 ± 0.013 and 0.086 ± 0.003 (P = .03) in uninduced and induced cells, respectively, suggesting that in cells expressing MtFt less iron is available for heme synthesis.
Discussion
Iron is a precious metal for organisms and is involved in a broad spectrum of essential biologic functions. However, some aspects of iron chemistry diminish its appeal. Unless bound to specific ligands, at pH 7.4 and physiologic oxygen tension, iron would exist only in the forms of virtually insoluble ferric hydroxides. Moreover, the catalytic action of iron in one-electron redox reactions can play an important role in the formation of toxic free radicals that ultimately cause oxidative damage to vital cell structures. Therefore, organisms are equipped with specific proteins designed for this metal's acquisition, transport, and storage in a soluble nontoxic form.
Levi et al21 recently identified a new intronless gene that encodes an H-ferritin–like protein, MtFt, that is expressed only in mitochondria. Most cells and tissues, including normal erythroblasts, express very low levels of MtFt.21 However, MtFt levels increase dramatically in ringed sideroblasts from patients with sideroblastic anemia.21,24 It is well known that a ringed sideroblast is a pathologic erythroid precursor containing excessive deposits of nonheme iron with characteristic perinuclear distribution accounting for the ring appearance.13-15 However, neither the pathogenesis of mitochondrial iron accumulation in ringed sideroblasts nor the function of MtFt is known.
Previous work by Corsi et al22 has documented that the overexpression of human MtFt in HeLa cells leads to a decrease in cytosolic ferritin levels and enhances TfR expression. Our results confirm and expand on their work. In this study we cloned the mouse MtFt gene, which was then transfected into a cell line under the control of a tetracycline-responsive promoter.27 We showed that removal of tetracycline in clone B9 cells caused a time-dependent induction of MtFt, starting as early as 12 hours and reaching maximum at 72 hours (Figure 1B). MtFt expressed in stable transfectants migrated as a single band with a molecular mass of about 22 kDa, corresponding to the mature mitochondrial protein. However, transient transfectants expressed not only mature MtFt but also a 28-kDa protein, likely a precursor of MtFt expressed in mitochondria (Figure 1C). As expected, transient transfection with cDNA lacking mitochondrial leader sequence generated protein with a molecular size corresponding to that of mature MtFt (Figure 1C).
Exploiting the “tet-off” system in stable transfectants, we confirmed that the expression of MtFt leads to an increase in TfR levels (Figure 3B) and a total suppression of cytosolic ferritin synthesis (Figure 3C-D) that confirms a decrease in cytosolic ferritin levels described earlier.22 However, in addition to this we have also demonstrated, for the first time, that the overexpression of MtFt causes a dramatic increase in IRP binding to IREs (Figure 3A), a finding that provides direct evidence of cytosolic iron deficiency. In another set of experiments we documented that MtFt-expressing cells, as compared with their wild-type counterparts, took up Tf-borne 59Fe with significantly higher rates (Figure 4A) which is likely the result of increased TfR expression. Moreover, mitochondria from induced cells contained about twice as much radioactive iron at 48 hours (Figure 4B) and their total nonheme iron content increased by approximately 36% at 72 hours compared with controls. In contrast to uninduced cells in which the majority of the radioactive iron was in cytosolic ferritin, induced cells had virtually no 59Fe in cytosolic ferritin and contained virtually all their radioactive iron in MtFt (Figure 5A). Moreover, the induction of MtFt very effectively mobilized 59Fe from the cytosolic ferritin of uninduced cells that had previously been incubated with 59Fe-Tf; in fact, most cellular 59Fe following the induction ended up in MtFt (Figure 5B). These experimental results demonstrate not only a remarkable capacity of MtFt to acquire iron from cytosolic sources, but also a very efficient translocation of Tf-derived iron into MtFt. Although the mechanism underlying this is unknown, the MtFt-inducible system described here could be exploited to characterize, at least in part, a yet unknown pathway involved in mitochondrial iron use. In addition, the rapid mobilization of iron from cytosolic ferritin to MtFt in our inducible system will also be a useful tool in investigating the nature of the release of iron from ferritins in situ, a mechanism that has been one of iron metabolism's enduring dilemmas.
It was previously shown that a 1-hour incubation of reticulocytes with 59Fe-Tf in the presence of porphyrin synthesis inhibitors leads to a dramatic accumulation of nonheme 59Fe in mitochondria39,43 that is available for chelation by SIH; this chelator, at 200 μM, removed approximately 45% of 59Fe per hour from reticulocytes with mitochondrial nonheme 59Fe accumulation.44 It has also been shown that certain 2-pyridylcarboxaldehyde isonicotinoyl hydrazone analogues can effectively mobilize this nonheme iron accumulated in reticulocyte mitochondria.45 The chemical form in which 59Fe accumulates in reticulocyte mitochondria is unknown, but the radioiron is unlikely inserted in MtFt because this protein is not expressed in normal erythroid cells. The efficacy with which SIH mobilizes 59Fe from reticulocytes suggests that mitochondrial membranes do not represent much of a barrier for this chelator. We deemed it important to examine the capacity of SIH to mobilize 59Fe from cells that contain most of their radioactive iron either in cytosolic or mitochondrial ferritin. We found that SIH mobilized 59Fe less efficiently from cells that contained 59Fe in MtFt than from those having 59Fe stored in cytosolic ferritin (Figure 5C). Similarly, SIH removed 59Fe much more readily from cytosolic than mitochondrial ferritin (Figure 5D). This decreased availability to iron chelation was also observed in HeLa cells overexpressing human MtFt, when using desferrioxamine to mobilize the iron stored in MtFt.22
We also investigated whether or not the expression of MtFt affects some proteins in which iron is essential for their function. Our data clearly demonstrate that the induction of MtFt significantly inhibited the activity of both cytosolic and mitochondrial aconitase (Figure 6), enzymes that require an intact [4Fe-4S] cluster for their function. It needs to be pointed out that a lower activity of mitochondrial aconitase activity may, at least in part, be due to decreased synthesis of this enzyme because its mRNA contains an IRE in its 5′-UTR.41,42 Moreover, our data suggest that in MtFt-expressing cells iron is less available for heme synthesis than in wild-type cells.
In summary, this study has demonstrated that MtFt is capable of very effectively acquiring endosomal, Tf-borne iron as well as cytosolic iron that is inserted in ferritin. Moreover, the induction of MtFt compromises the function of at least some nonheme iron proteins and also appears to interfere with iron delivery to heme. Furthermore, compared with cytosolic ferritin, iron inserted into MtFt is less available for chelation by the strong chelator SIH. Collectively, our data suggest that the avidity of MtFt for, and its association with, iron is stronger than those of its cytosolic counterpart. These properties of MtFt could be potentially useful in diminishing oxidative damage in conditions associated with mitochondrial iron overload, such as Friedreich ataxia16,17 or X-linked sideroblastic anemia with ataxia.19
Prepublished online as Blood First Edition Paper, November 2, 2004; DOI 10.1182/blood-2004-07-2722.
Supported by grants (P.P.) and a scholarship (A.D.S.) from the Canadian Institutes of Health Research.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Kostas Pantopoulos and Jian Wang for the tTA-H1299 cells and Dr Lukas Kuhn for pST18-fer plasmid.

![Figure 3. Effects of MtFt overexpression on the IRE-IRP system, TfR levels, and ferritin synthesis. (A) MtFt overexpression increases IRP-binding activity. Stably transfected H1299 cells (clone B9) were seeded in 100-mm dishes and grown for 24 hours (lanes 1-2, 7-8), 48 hours (lanes 3-4, 9-10), and 72 hours (lanes 5-6, 11-12) without (-) or with (+) 2 μg/mL tetracycline. Cytoplasmic extracts (10 μg) were assayed for their ability to retard the migration of a 32P-labeled IRE probe in the absence (lanes 1-6) or presence (lanes 7-12) of 2% β-ME. (B) Stimulation of TfR expression by MtFt overexpression. Clone B9 cells were seeded (25% confluence) in 100-mm dishes and grown for 24, 48, and 72 hours without (-) or with (+) tetracycline. The cell lysates were analyzed for expression of MtFt, TfR, and actin by Western blotting. The filters were probed with antibodies against HA-tag (MtFt, upper panel), stripped, and reprobed with TfR (middle panel) and actin (lower panel), respectively. (C-D) Effect of MtFt overexpression on cytosolic ferritin synthesis. In panel C, cells treated without or with tetracycline for 24 hours (lanes 1-2), 48 hours (lanes 3-4), and 72 hours (lanes 5-6) were metabolically labeled with [35S] methionine for 1 hour and cell extracts were subjected to immunoprecipitation with an antiferritin antibody. The immunoprecipitated proteins were separated by SDS-PAGE on 12.5% gels and autoradiographed. In panel D, the same treatment of the cells was used to measure the steady-state amount of ferritins (lower bands) by Western blotting (top band is MtFt that is recognized by the antiferritin antibody).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/5/10.1182_blood-2004-07-2722/6/m_zh80050575040003.jpeg?Expires=1766311969&Signature=Dt-KFnMM0PVqtKUuCmBCJib5B5D1mQweft--3lEcANe6wPu1M6Ki3EgzpMBO5ImEA3ZZ~Xd9~dUzupteeZfy2PeZcj~f~5vZ0YZBh32YUuohx7nqB8rsdgEN5YnbzgP2jJTTR46OzqD9y-Kx3YpK3SXpZ1MpZdiG~-X5qDEVeJeDSZ2K0TfHHZFHJHPurrJS7QvPqnyUvG3X9RFk5zs6aYuwQir-AWYXWtF3-dSmPj3GjJsSA4ucGB3uBKFcFSjrQWSM4b2HD2fqBc2iiRMxrpDsMRo18MRg~RFjXfynH7jBsOiGYwRQjklmtksXIo8pyv4eds6re0OJw~RAmxiWDg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



![Figure 3. Effects of MtFt overexpression on the IRE-IRP system, TfR levels, and ferritin synthesis. (A) MtFt overexpression increases IRP-binding activity. Stably transfected H1299 cells (clone B9) were seeded in 100-mm dishes and grown for 24 hours (lanes 1-2, 7-8), 48 hours (lanes 3-4, 9-10), and 72 hours (lanes 5-6, 11-12) without (-) or with (+) 2 μg/mL tetracycline. Cytoplasmic extracts (10 μg) were assayed for their ability to retard the migration of a 32P-labeled IRE probe in the absence (lanes 1-6) or presence (lanes 7-12) of 2% β-ME. (B) Stimulation of TfR expression by MtFt overexpression. Clone B9 cells were seeded (25% confluence) in 100-mm dishes and grown for 24, 48, and 72 hours without (-) or with (+) tetracycline. The cell lysates were analyzed for expression of MtFt, TfR, and actin by Western blotting. The filters were probed with antibodies against HA-tag (MtFt, upper panel), stripped, and reprobed with TfR (middle panel) and actin (lower panel), respectively. (C-D) Effect of MtFt overexpression on cytosolic ferritin synthesis. In panel C, cells treated without or with tetracycline for 24 hours (lanes 1-2), 48 hours (lanes 3-4), and 72 hours (lanes 5-6) were metabolically labeled with [35S] methionine for 1 hour and cell extracts were subjected to immunoprecipitation with an antiferritin antibody. The immunoprecipitated proteins were separated by SDS-PAGE on 12.5% gels and autoradiographed. In panel D, the same treatment of the cells was used to measure the steady-state amount of ferritins (lower bands) by Western blotting (top band is MtFt that is recognized by the antiferritin antibody).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/5/10.1182_blood-2004-07-2722/6/m_zh80050575040003.jpeg?Expires=1766311970&Signature=Lq43jqXG6UahYqelj3j-frPIU2jInyYd6R5yOXmIg~oy3fO7kYuxjGpINHvifqB30QNcgXxyd3-PTK~LXW1-zWgzac-XqftH1ewKjew0G672lWFazFUkuBZPlhPAmw4vVi-b3YjBOlOsU7QiIxrBsqfWjJlTdQtN0lD9a4XOVQgPR3d1hR2x4LizW8FmDhMpF68fpLTFNg633VRav-8ZYvFr9yIimOfogU8aehE1V6~14iXnGxmTw0NNd0GTDtGZ99~Kt134Ynw0P~H9zTawo6UYt1FwUplKVs-Jh3oiH3xHUJFTXwyD0q1a~EMpe6fiBtjoFK7fAfpYlyw3vNcEpw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

 ), were incubated for 24 hours with 1 μM 59Fe-Tf and washed 3 times in PBS, and their radioactivity was measured. (B) Distribution of iron between cytosol and mitochondria. The cells treated as described in panel A were homogenized in lysis buffer (sucrose 0.25 M, 10 mM HEPES, pH 7.4, 0.15% bovine serum albumin). The cytosolic and mitochondrial fractions were prepared by differential centrifugation and their radioactivities were measured. The amount of iron in mitochondria is expressed as a percentage of total 59Fe taken up. *P < .01 compared with 48-hour tet-on cells; **P < .01 compared with 72 hour tet-on cells. Data shown are the means ± SD of triplicate determinations from a typical experiment that was performed 3 times.
), were incubated for 24 hours with 1 μM 59Fe-Tf and washed 3 times in PBS, and their radioactivity was measured. (B) Distribution of iron between cytosol and mitochondria. The cells treated as described in panel A were homogenized in lysis buffer (sucrose 0.25 M, 10 mM HEPES, pH 7.4, 0.15% bovine serum albumin). The cytosolic and mitochondrial fractions were prepared by differential centrifugation and their radioactivities were measured. The amount of iron in mitochondria is expressed as a percentage of total 59Fe taken up. *P < .01 compared with 48-hour tet-on cells; **P < .01 compared with 72 hour tet-on cells. Data shown are the means ± SD of triplicate determinations from a typical experiment that was performed 3 times.