Abstract
Interleukin-4 is implicated in the pathogenesis of HIV-induced AIDS and causes enhancement of replication of virus strains that use the CXCR4 (X4) coreceptor. In this study, we explored the effects of interleukin-4 (IL-4) antisense (AS) DNA on replication of X4, simian human immunodeficiency viruses, SHIVKU-2 and SHIV89.6P. AS IL-4 oligomer caused inhibition of virus replication in cultures of CD4+ T cells and macrophages derived from macaques. Plasmid expressing AS IL-4 DNA was also effective in abrogating virus replication in macrophage cultures. Relevance of these cell culture studies was confirmed in vivo by treating SHIV89.6P-infected macaques with AS IL-4 DNA. Six macaques were inoculated with the virus, and 4 were treated with AS IL-4 DNA. This resulted in a significant decrease in viral RNA concentrations in the liver, lungs, and spleen tissues that are all sites of virus replication in macrophages. This is the first demonstration of effective inhibition of an HIV-like virus in tissues by AS DNA of a cytokine. In the present era of increasing resistance of HIV to antiviral compounds, exploration of adjunct therapies directed at host responses in combination with antiretroviral drugs may be of value for the treatment of AIDS.
Introduction
The pathogenesis of HIV infection and the complex disease patterns caused by the virus are known to revolve around the dystrophic effects of the infection first, in helper CD4+ T cells, and later in cells of the macrophage lineage in organs such as the lungs, spleen, and brain.1-6 Loss of virus-infected CD4+ T cells heralds the loss of interferon-γ (IFN-γ)–mediated T-cell responses, and this creates an environment favorable for productive replication of opportunistic pathogens. Some of these agents are powerful inducers of T helper 2 (Th2) immune responses.7,8 Studies on coreceptor usage of HIV have shown that viruses that use the CCR5 (R5) coreceptor are usually associated with virus invasion across mucosal surfaces and highly productive phase of virus replication in activated memory CD4+ T lymphocytes. In many infected individuals, mutant virus strains that use the CXCR4 (X4) coreceptor evolve following the loss of Th1 cellular immune responses. Most of these viruses are capable of replicating only in naive CD4+ T lymphocytes, including T-cell lines,9,10 but some are also known to be dual tropic and capable of replicating productively in macrophages.11,12 Studies on replication of X4 HIV-1 in human macrophage cultures showed that interleukin-4 (IL-4) caused significant enhancement of virus replication.13 Whether IL-4 induced by opportunistic pathogens in immunocompromised HIV-infected persons causes enhancement of X4 virus replication is not known.
We have used the macaque simian–human immunodeficiency virus (SHIV) model of HIV disease to study the role of macrophage-tropic X4 viruses in the pathogenesis of AIDS and neurologic disease and, in particular, the interaction between the virus and macaque macrophages. Pathogenic SHIVs that have the X4 env gene of HIV-1 in a background of SIVmac239 such as SHIV89.6P14 and
Materials and methods
Viruses
SHIVKU-2, an X4 virus, was derived by sequential passage of the molecular clone, SHIV-4, which has the env, tat, rev, and vpu genes of HIV-1 HXBc2.19 SHIV89.6P, a dual tropic X4/R5 virus, was kindly provided by Dr N. Letvin (Harvard University, Cambridge, MA). Both viruses were propagated in macaque peripheral blood mononuclear cell (PBMC) cultures. Stock preparations had infectivity titers between 104 and 105 tissue culture infectious dose 50%. Cell cultures were inoculated with either virus at a multiplicity of 0.01.
Cell cultures
PBMCs from macaques were obtained by Ficoll-Hypaque (Sigma, St Louis, MO) gradient centrifugation as described earlier and prepared as suspension cultures grown in RPMI + 10% fetal bovine serum + 50 U/mL IL-2 (R10).18 Briefly, suspensions of PBMCs at a concentration of 1 × 106 cells/mL R10 medium were treated with staphylococcal enterotoxin A (Sigma) at 5 μg/mL for 24 hours. Cultures were then washed twice with RPMI and resuspended in R10 and inoculated with SHIV at a multiplicity (MOI) of 0.01 for 4 hours at 37°C. Cells were then extensively washed and replenished with fresh R10 and recombinant macaque IL-4 (rmIL-4) or AS IL-4 oligodeoxynucleotide (ODN). Every third day, 0.5 mL spent medium was removed from the cultures and replenished with fresh R10 and rmIL-4 or AS IL-4 ODN. Supernatant fluids were collected at regular intervals and monitored for viral p27 by enzyme-linked immunosorbent assay (ELISA). Monocyte-derived macrophage (MDM) cultures were obtained from PBMCs by incubation in macrophage differentiation medium consisting of RPMI medium supplemented with 20% heated human serum (56°C for 30 seconds), 5 U/mL macrophage colony-stimulating factor, 100 U/mL granulocyte-macrophage colony-stimulating factor, and 5% heat-inactivated rhesus monkey serum at 37°C for 7 days to allow adherent monocytes to differentiate into mature macrophages.18 Cells were plated in 96-well tissue culture dishes (Costar, Cambridge, MA) or as chamber slide preparations at a concentration of 8 × 105 cells/200 μL per well and incubated overnight at 37°C. Cultures were then rinsed to remove nonadherent cells, refed with the same medium, and maintained for 7 days. Macrophages were then inoculated with the virus at an MOI of 0.01 for 4 hours at 37°C. Cells were then extensively washed and replenished with fresh medium containing rmIL-4 or AS IL-4 ODN.
IL-4
rmIL-4 was provided by the Resource for Nonhuman Primate Immune Reagents, Emory University (Atlanta, GA) and was used at a concentration of 20 U/mL tissue culture medium. Cultures were replenished with fresh rmIL-4 every 3 days.
Antisense, sense, and scrambled ODNs
ODNs used in this study were custom-synthesized by Invitrogen Life Technologies (Carlsbad, CA). The sequences of the ODNs are as follows: antisense IL-4, 5′-GGGCTGCTGCTGGCTTTTTGCTT-3′; sense IL-4, 5′-AAGCAAAAAGCCAGCAGCAGCCC-3′; and scrambled IL-4, 5′-GGGCTGATGCGGCCTTTTTGCTT-3′. The ODNs were used at a concentration of 80 μM in tissue culture medium and replenished every 3 days. This concentration was not toxic as tested by trypan blue staining (data not shown).
Plasmids
Sense and AS IL-4 plasmids were constructed from the rhesus (Rh) IL-4 clone provided by the Resource for Nonhuman Primate Immune Reagents. The RhIL-4 plasmid was cut with either BstXI/ApaI (antisense) or NcoI/NotI (sense), and the restriction fragments obtained were then cloned into the EcoRV restriction site of pcDNA 3.1 (+) (Invitrogen, Carlsbad, CA) by blunt-end ligation to generate a cytomegalovirus–driven RhIL-4 AS and sense construct, respectively. A reporter plasmid gWIZ green fluorescent protein (GFP) supplied by GTS (San Diego, CA) was also used in our studies. Plasmids were amplified in Escherichia coli (DH10β from Gibco BRL, Carlsbad, CA) and extracted by endofree Mega/Giga plasmid purification kit from Qiagen (Valencia, CA). For in vitro experiments, primary macrophages grown in 24-well plates were transfected with 1 μg plasmid DNA per well complexed with the cationic lipid, in vitro jetPEI-Man (Qbiogene, Irvine, CA) as per the manufacturer's instructions.
Studies in macaques
Cynomolgus macaques, approximately 3 years of age, were used in these studies. One animal was used for determination of biodistribution of the gWIZ GFP, and 6 others were used for virus and AS DNA studies. The first animal was inoculated intravenously with 500 μg GFP plasmid complexed with the liposome in vivo MegaFectin (Qbiogene) by using the enhancer-1, according to the manufacturer's instructions. This animal was killed 2 days later, and lungs, liver, and spleen were harvested to assess localization of GFP. The other 6 animals were inoculated intravenously with 1 mL stock SHIV89.6P. Seven days later, 4 of the macaques were injected intravenously with 1 mg AS IL-4 DNA complexed with MegaFectin. Two of these animals were given a second injection of the AS DNA 2 days later. All 6 animals were killed 7 days later for assessment of virus replication in different tissues. The macaques were tranquilized with ketamine and then deeply anesthetized with pentobarbital. Laparotomies were performed, and the animals were exsanguinated from the abdominal aorta. A portion of the plasma was collected in ethylenediaminetetraacetic acid and used for determination of viral RNA content. The animals were then perfused via the left ventricle of the heart with 1 L saline, and perfusate from the right atrium was discarded. Tissues were then harvested rapidly. Portions of each organ were fixed in either 4% paraformaldehyde for histologic assays or in Streck fixative (Streck Laboratories, Omaha, NE) for immunofluorescence assays. Other portions of the tissues were snap-frozen in isomethyl butane chilled with liquid nitrogen. These tissues were used as sources of RNA.
Virus assays
Supernatant fluids from virus-inoculated cultures were examined for virus content by using the reverse transcriptase assay or an ELISA measuring viral Gag p27. Viral RNA concentrations in tissues were assessed by using real-time reverse transcription (RT)–PCR as previously described.20 Briefly, total RNA isolated from frozen tissues was treated with DNAase and subjected to real-time RT-PCR (ABI, Foster City, CA) using gag primers and a Taqman Probe with thermal cycler conditions as described. Levels of hypoxanthine phosphoribosyl transferase (HPRT) mRNA, a housekeeping gene, were also measured by real-time RT-PCR to normalize the viral load. The amplification efficiencies of the gag and HPRT targets can be considered essentially equal, as the difference in their slopes (ΔS) of the standard curves was within 0.2.
Immunocytochemistry and histochemistry
Immunocytochemical analysis was performed on chamber slide preparations of macrophage cultures or on sections from paraffin-embedded tissues. Slides were treated with murine monoclonal antibody to p27, the Gag protein of simian immunodeficiency virus (SIV; ABI, Columbia, MD) followed by treatment with biotinylated goat anti-mouse immunoglobulin G (DAKO, Carpenteria, CA), peroxidase-conjugated streptavidin (DAKO), and NovaRed substrate (Vector Laboratories, Burlingame, CA), which yields a reddish reaction product.21 For fluorescence microscopy, Streck-fixed tissues were frozen, embedded at –25°C in ornithine carbamoyltransferase compound (OCT) (Miles, Elkhart, IN), and cut into 5- to 10-μm thick sections. Micrographs were captured on a Nikon Eclipse E600 microscope equipped with a 4 ×/0.13 or 10 ×/0.30 objective lens, optem 1 × DC10 NN camera and analysis image processing acquisition software (Nikon Instruments, Melville, NY). Confocal images were captured on LSM 510 Laser Scanning Microscope with a Plan-Neo 25 ×/0.80 IK DIC objective lens and LSM software (Carl Zeiss Microscopy, Jena, Germany).
Semiquantitative RT-PCR analyses
RNA was extracted from tissue samples by using Trizol reagent (Life Technologies, Grand Island, NY). Semiquantitative RT-PCR analyses were performed on the RNA by using the Access RT-PCR kit (Promega, Madison, WI) in a Perkin-Elmer (Emeryville, CA) DNA Thermal Cycler 480 as described earlier by using macaque IL-4 primers19 or CXCR4 primers.22
Real-time RT-PCR analysis for IL-4
For quantification of cellular IL-4 mRNA levels, total RNA isolated from macrophages treated with rmIL-4 or AS IL-4 was subjected to real-time RT-PCR by using IL-4 primers (5′-TTT CAC AGG CAC AAG CAG CT-3′, 5′-GCC AGG CCC CAG AGG TT-3′) and Taqman probe (5′-6FAM-CCG ATT CCT GAA ACG GCT CGA CAG-TAMRA) labeled at the 5′ end with the reporter dye FAM (6-carboxyfluorescein) and at the 3′ end with the quencher dye TAMRA (6-carboxytetramethyl-rhodamine) using ABI Prism7700. Primers and the Taqman probe were designed by using the Primer Express software (ABI). Thermal cycling conditions consisted of 50°C for 2 minutes for uracil-N-glycosylase, 60°C for 30 minutes and 95°C for 10 minutes, followed by 44 cycles of 95°C for 15 seconds and 60°C for 30 seconds. Prime RNAse inhibitor was used in the reactions (7.5 U; Brinkmann-Eppendorf, Westbury, NY), and reaction volumes were 25 μL. Standard curves were performed by using 6- to 10-fold dilutions of nuclear runoff IL-4 RNA. Samples were analyzed twice, in duplicate. As a measure of cellular mRNA levels, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA copy numbers in the RNA samples were also determined by a real-time RT-PCR Taqman assay over 40 cycles (ABI). The IL-4 mRNA levels were normalized to the cellular GAPDH mRNA number to express the results from an equivalent cell number.
Results
IL-4 enhanced X4 virus replication in macaque PBMCs and macrophages
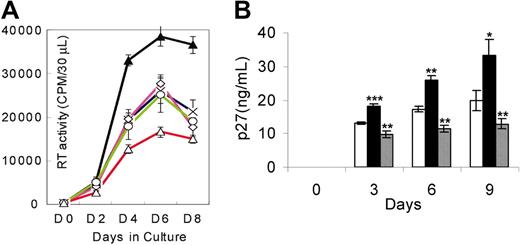
Since the pathogenesis of HIV-1 infection in humans and SHIV infection in macaques is associated with infection in CD4+ T cells and macrophages, we investigated whether IL-4 would modulate SHIV replication in these 2 cell lineages. Infection with either SHIVKU-2 or SHIV89.6P yielded similar results in cell culture systems. As seen in Figure 1, IL-4 caused significant increases in viral titers of SHIVKU-2 in PBMCs (Figure 1A) and macrophage cultures (Figure 1B), compared with untreated cultures.
Replication of SHIVKU-2 in response to rmIL-4 and AS IL-4 in macaque PBMCs and monocyte-derived macrophages. (A) PBMCs were treated with staphylococcal enterotoxin A for 24 hours, following which the cells were inoculated with the virus in the medium containing IL-2 and incubated for 4 hours at 37°C. Cells were then washed and treated with either rmIL-4 or AS IL-4 ODN, which was replenished every third day (described in “Materials and methods”). × indicates no IL-4 treatment; ▴, treatment with IL-4; ⋄, with IL-4 sense ODN; ○, with IL-4 scrambled ODN; and ▵, with IL-4 antisense ODN. Sequence-specific inhibition of SHIVKU-2 replication was seen by AS IL-4 ODN in macaque PBMCs. (B) Macrophage cultures were inoculated with SHIVKU-2 and incubated at 37°C for 4 hours, after which they were rinsed 3 times and replenished with medium containing rmIL-4 (▪) or AS IL-4 ODN (▦). □ indicates no treatment with IL-4. Every third day, fresh rmIL-4 or AS IL-4 DNA was added to the cultures. Aliquots of the culture supernatant fluid were assayed for SHIV Gag p27 content by ELISA or by RT assays at different time intervals. The experiment reported is representative of a set of 3 different experiments. Two-tailed t tests were performed. *P < .05; **P < .005; ***P < .001 as compared with control infected cultures. The data are presented as the mean ± standard deviation of 3 independent experiments.
Replication of SHIVKU-2 in response to rmIL-4 and AS IL-4 in macaque PBMCs and monocyte-derived macrophages. (A) PBMCs were treated with staphylococcal enterotoxin A for 24 hours, following which the cells were inoculated with the virus in the medium containing IL-2 and incubated for 4 hours at 37°C. Cells were then washed and treated with either rmIL-4 or AS IL-4 ODN, which was replenished every third day (described in “Materials and methods”). × indicates no IL-4 treatment; ▴, treatment with IL-4; ⋄, with IL-4 sense ODN; ○, with IL-4 scrambled ODN; and ▵, with IL-4 antisense ODN. Sequence-specific inhibition of SHIVKU-2 replication was seen by AS IL-4 ODN in macaque PBMCs. (B) Macrophage cultures were inoculated with SHIVKU-2 and incubated at 37°C for 4 hours, after which they were rinsed 3 times and replenished with medium containing rmIL-4 (▪) or AS IL-4 ODN (▦). □ indicates no treatment with IL-4. Every third day, fresh rmIL-4 or AS IL-4 DNA was added to the cultures. Aliquots of the culture supernatant fluid were assayed for SHIV Gag p27 content by ELISA or by RT assays at different time intervals. The experiment reported is representative of a set of 3 different experiments. Two-tailed t tests were performed. *P < .05; **P < .005; ***P < .001 as compared with control infected cultures. The data are presented as the mean ± standard deviation of 3 independent experiments.
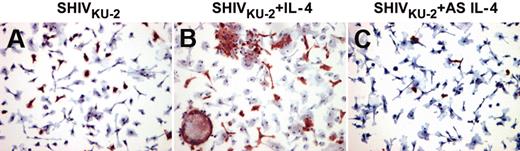
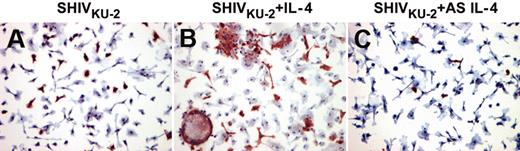
Furthermore, IL-4 also mediated enhancement of viral replication in a macaque CD4+ T-cell line immortalized by herpes virus Saimiri (data not shown). This enhanced viral replication was also demonstrated by immunocytochemical analyses, showing that IL-4 increased the numbers of virus-positive macrophages and enhanced viral cytopathic effects characterized by multinucleated giant cell formation (Figure 2).
Viral protein staining in SHIVKU-2–infected rhesus MDMs after treatment with IL-4 protein and AS IL-4 ODN. MDM cultures were grown on chamber slides, inoculated with SHIVKU-2 for 4 hours at 37°C, washed, and untreated (A) or then treated with either rmIL-4 (B) or AS IL-4 (C) for 48 hours. Cells were then fixed and immunostained using anti-Gag p27 antibody.
Viral protein staining in SHIVKU-2–infected rhesus MDMs after treatment with IL-4 protein and AS IL-4 ODN. MDM cultures were grown on chamber slides, inoculated with SHIVKU-2 for 4 hours at 37°C, washed, and untreated (A) or then treated with either rmIL-4 (B) or AS IL-4 (C) for 48 hours. Cells were then fixed and immunostained using anti-Gag p27 antibody.
Specific inhibition of SHIV replication by AS IL-4 ODNs
We next inquired whether AS IL-4 ODN treatment would abrogate virus replication in macaque PBMC and macrophage cultures. The cultures were inoculated at an MOI of 0.01 and 4 hours later treated with either AS DNA of IL-4 or various control ODNs. In these experiments, antisense IL-4 DNA was added to virus-inoculated cultures, and supernatant fluids collected on days 3, 6, and 9 after infection were assayed for SIV Gag. The cultures were replenished with fresh AS IL-4 ODN every third day. As seen in Figure 1A-B, AS IL-4 DNA specifically and markedly inhibited virus replication (80%-90%) as measured by reverse transcriptase assays or ELISA. In contrast to cultures treated with IL-4, AS IL-4 ODN decreased the numbers of virus-positive macrophages and the extent of viral cytopathogenicity characterized by multinucleated giant cell formation (Figure 2). This effect was again confirmed also in the CD4+ T-cell line (data not shown). The effect of AS IL-4 ODN on virus replication was sequence-specific since sense and scrambled IL-4 oligos used at similar concentrations did not inhibit virus replication (Figure 1A). To further rule out nonspecific effects of IL-4, we selected an irrelevant cytokine, IL-8, as a control. AS IL-8 ODNs when used at the same concentration as AS IL-4 failed to inhibit virus replication (data not shown).
Modulation of IL-4 and CXCR4 RNAs by exogenous IL-4 and AS IL-4 ODN
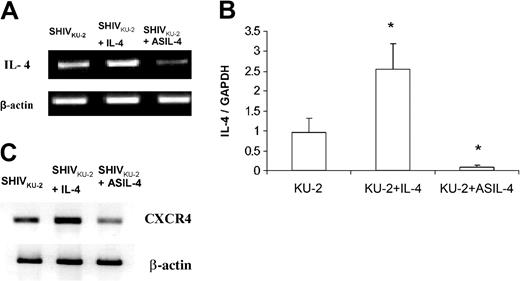
To explore whether the enhancing effect of IL-4 was associated with modulation of endogenous IL-4 production and whether AS ODN designed against IL-4 truly inhibited endogenously produced IL-4 in treated cultures, we performed an IL-4–specific RT-PCR analysis of RNA extracted from infected cultures that were treated with IL-4 and AS IL-4 DNA. As shown in Figure 3A, infected macrophages treated with IL-4 showed a marked increase in IL-4 mRNA, while cultures treated with AS DNA showed significantly reduced levels of IL-4 mRNA. These findings were confirmed by real-time RT-PCR (Figure 3B). We and others had shown previously that the virus-enhancing effect of IL-4 correlated with increased expression of the viral coreceptor, CXCR4, in treated cultures.13,21,22 Analysis of CXCR4 message by RT-PCR demonstrated a down-regulation of CXCR4 mRNA in SHIV-infected CD4+ T cells treated with AS IL-4 ODN as opposed to an up-regulation in IL-4–treated cultures, emphasizing the specificity of the AS IL-4 effect (Figure 3C).
Levels of IL-4 mRNA in SHIVKU-2–infected CD4+ T cells treated with rmIL-4 or AS IL-4 as seen by RT-PCR and quantitative real-time RT-PCR. Following 24 hours of treatment, cellular RNA was extracted by Trizol, DNAse treated, and assessed for IL-4 mRNA levels by RT-PCR (A) and quantitative real-time RT-PCR (B). Panel B represents a ratio of IL-4 and GAPDH RNA transcripts. *P < .05 compared with SHIV-infected cells. (C) RT-PCR analysis of CXCR4 mRNA in SHIVKU-2–infected MDM in the presence of rmIL-4 or AS IL-4 ODN by using macaque CXCR4 primers. The data are presented as the mean ± standard deviation of 3 independent experiments.
Levels of IL-4 mRNA in SHIVKU-2–infected CD4+ T cells treated with rmIL-4 or AS IL-4 as seen by RT-PCR and quantitative real-time RT-PCR. Following 24 hours of treatment, cellular RNA was extracted by Trizol, DNAse treated, and assessed for IL-4 mRNA levels by RT-PCR (A) and quantitative real-time RT-PCR (B). Panel B represents a ratio of IL-4 and GAPDH RNA transcripts. *P < .05 compared with SHIV-infected cells. (C) RT-PCR analysis of CXCR4 mRNA in SHIVKU-2–infected MDM in the presence of rmIL-4 or AS IL-4 ODN by using macaque CXCR4 primers. The data are presented as the mean ± standard deviation of 3 independent experiments.
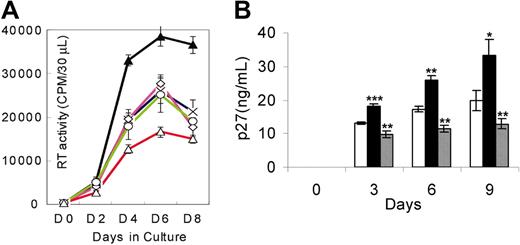
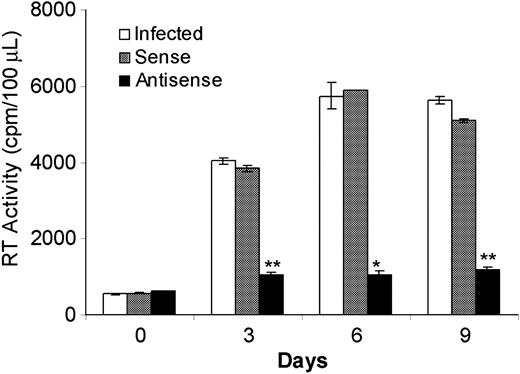
SHIV replication in the presence of sense or AS IL-4 plasmid DNA
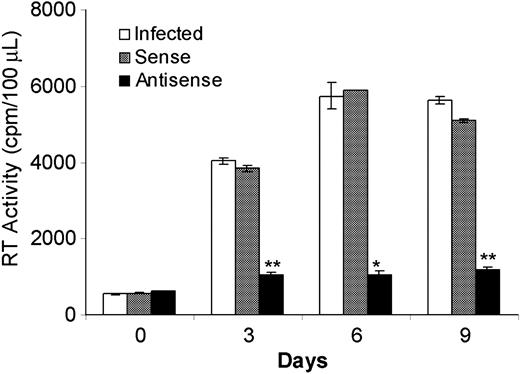
AS IL-4 ODNs provided a tool to assess the role of endogenously produced IL-4 on SHIV replication. The observed inhibition of virus replication in virus-infected cultures of lymphocytes and macrophages by AS IL-4 ODNs suggested that this mechanism may provide a novel therapeutic approach. However, the prohibitive cost of ODNs prompted us to explore whether recombinant AS IL-4 DNA could be substituted with synthetic ODNs for the purpose of inhibiting endogenous IL-4 production. Plasmids expressing AS or sense IL-4 were constructed and transfected into virus-infected macrophages with the use of the cationic lipid, in vitro jet-PEI-Man (Qbiogene). A single treatment of the infected cultures with AS IL-4 resulted in marked and sustained reduction of virus replication by as much as 85% for the next 9 days. (Figure 4; P ≤ .001). In contrast, no significant inhibition of virus replication was observed in control cultures administered a sense IL-4 plasmid. These results confirmed that the AS IL-4–expressing vector had the same effect as the ODNs and, therefore, could be substituted for the latter in therapeutic attempts.
SHIV89.6P replication in macrophages in the presence of sense or AS IL-4 plasmid. Plasmids expressing AS (▪) or sense IL-4 (▦) were transfected into infected macrophages with the use of the cationic lipid, in vitro jetPEI-Man. □ indicates no IL-4. SHIV infection was then monitored by RT activity at different time intervals. *P < .01; ** P ≤ .001 compared with control-infected cultures.
SHIV89.6P replication in macrophages in the presence of sense or AS IL-4 plasmid. Plasmids expressing AS (▪) or sense IL-4 (▦) were transfected into infected macrophages with the use of the cationic lipid, in vitro jetPEI-Man. □ indicates no IL-4. SHIV infection was then monitored by RT activity at different time intervals. *P < .01; ** P ≤ .001 compared with control-infected cultures.
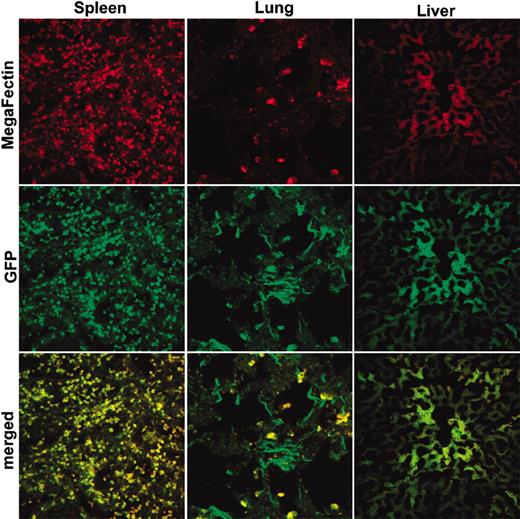
Biodistribution of GFP in macaque tissues
As a preliminary investigation prior to intravenous delivery of AS DNA to infected macaques, we examined the biodistribution of a reporter gene, GFP, in 1 macaque. The animal was injected intravenously with the GFP plasmid and killed 2 days later. As shown in Figure 5, direct visualization of fixed tissue sections by using confocal microscopy revealed colocalization of MegaFectin (red fluorescence) and GFP (green fluorescence) in lungs, liver, and spleen. These 3 organs are primarily involved in the clearance of particulate foreign matter from the blood. This experiment was a prerequisite for new follow-up experiments that used liposomes containing AS IL-4 DNA as a therapy for infected macaques.
Colocalization of GFP (green) and in vivo MegaFectin complexes (red) in spleen, lungs, and liver of a macaque. Two days after intravenous injection of GFP/MegaFectin complex, the animal was killed, tissues were collected, and fixed or frozen sections were examined to visualize distribution with use of confocal microscopy of spleen (left column), lungs (middle column), and liver (right column).
Colocalization of GFP (green) and in vivo MegaFectin complexes (red) in spleen, lungs, and liver of a macaque. Two days after intravenous injection of GFP/MegaFectin complex, the animal was killed, tissues were collected, and fixed or frozen sections were examined to visualize distribution with use of confocal microscopy of spleen (left column), lungs (middle column), and liver (right column).
Inhibition of virus replication in macaques treated with liposomes containing AS IL-4 DNA
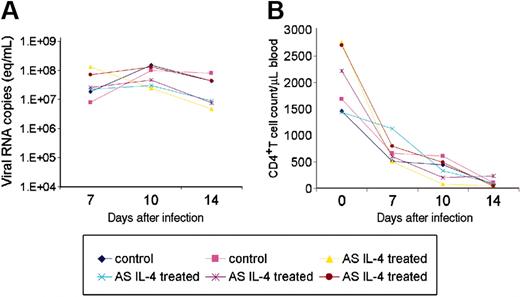
Next, we chose to treat monkeys with AS IL-4 DNA during the acute phase of SHIV infection, because significant levels of systemic viral replication are observed during the first 2 to 4 weeks after infection. The animals were, therefore, inoculated with SHIV89.6P; 2 animals were given AS IL-4 on day 7 after infection, and 2 others received the AS IL-4 on days 7 and 9 after infection. The 2 virus control animals were not treated. All 6 animals were killed at 2 weeks after infection. Blood from the 4 treated and 2 untreated animals was analyzed on days 7, 10, and 14 for viral RNA content in plasma and CD4+ T-cell counts in the mononuclear cells in peripheral blood. Figure 6A shows that by day 7 after inoculation, all 6 animals had viral RNA concentrations varying from 107 to 108 copies/mL plasma. Titers remained essentially at this level by day 10 and declined slightly by day 14. In 3 of the 4 AS IL-4–treated animals there was a 1 log10 reduction in the plasma viral RNA concentration compared with the control animals. CD4+ T-cell numbers declined precipitously in both the treated and untreated animals following virus inoculation, as shown in Figure 6B. There was no significant difference between the CD4+ T-cell counts from the control or treated animals.
Plasma viral load and CD4+ T cell counts in the peripheral blood of ASIL-4 treated and untreated macaques. (A) Cell-free plasma viral RNA load of the treated and untreated macaques at multiple time points was determined by real-time RT-PCR by using standards covering 6 orders of magnitude. Viral loads are shown as viral RNA copies per milliliter plasma. (B) CD4+ T-cell profile in the control and AS IL-4–treated SHIV89.6P–infected macaques. The absolute number of CD4+ T cells per microliter blood was calculated by multiplying the percentage of lymphocyte subset with the absolute number of lymphocytes per microliter from complete blood count.
Plasma viral load and CD4+ T cell counts in the peripheral blood of ASIL-4 treated and untreated macaques. (A) Cell-free plasma viral RNA load of the treated and untreated macaques at multiple time points was determined by real-time RT-PCR by using standards covering 6 orders of magnitude. Viral loads are shown as viral RNA copies per milliliter plasma. (B) CD4+ T-cell profile in the control and AS IL-4–treated SHIV89.6P–infected macaques. The absolute number of CD4+ T cells per microliter blood was calculated by multiplying the percentage of lymphocyte subset with the absolute number of lymphocytes per microliter from complete blood count.
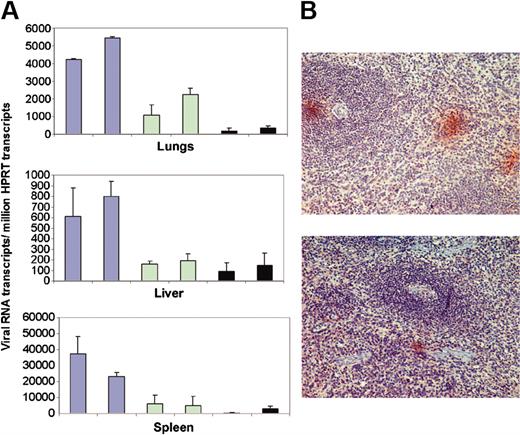
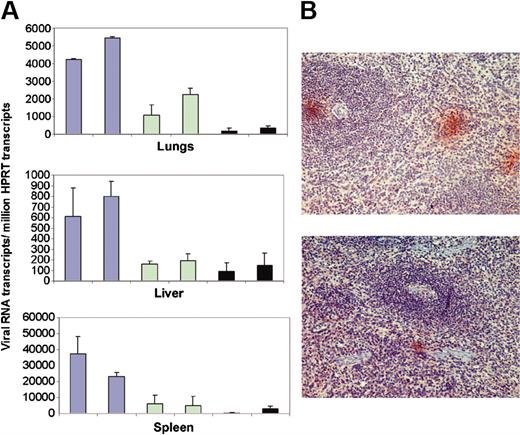
Having determined that intravenous administration of liposomes would be cleared from blood by cells in the liver, lungs, and spleen, these tissues coincidentally being the site of virus replication predominantly in macrophages,4,6 we then investigated whether AS DNA of IL-4 would inhibit virus replication in these organs in vivo. Tissue samples of organs in which the AS IL-4 DNA was expected to accumulate were examined for virus content. Liver, lung, and splenic tissues from the 4 treated and the 2 untreated infected macaques were examined for presence of full-length viral gag mRNA by real-time RT-PCR. Gag RNA copy numbers were normalized to the copy number of the cellular mRNA for HPRT, also measured by real time RT-PCR. The gag/HPRT ratio showed that the tissues from treated animals exhibited markedly less viral RNA than those of the 2 control animals. Further, the tissues from the 2 animals that received 2 injections of liposome/AS IL-4 DNA had even less viral RNA than the 2 that received only a single treatment (Figure 7A). Specifically, animals receiving a single injection of AS DNA had 60% less viral RNA, whereas the 2 that were treated twice had about 80% to 90% reduction in the viral RNA in spleen and lungs compared with the 2 untreated controls. Immunohistochemical studies on spleen sections that used a Gag-specific antibody corroborated the viral RNA quantification data since only a few small foci of viral-infected cells were found in the tissue sections of the AS-treated animals compared with many large foci of viral antigen-positive cells in splenic sections of the untreated animals (Figure 7B). The virus-positive cells in the spleen were primarily macrophages in the red pulp area of the spleen (data not shown).
Viral RNA load in AS IL-4 DNA treated and untreated SHIV-infected macaques as seen by real-time RT-PCR and immunohistochemistry. (A) Viral RNA levels in the lungs (top), liver (middle), and spleen (bottom) in SHIV89.6P-infected macaques (blue bars); viral RNA in infected macaques given a single dose of AS IL-4 (green bars); and viral RNA in the animals receiving 2 injections of the AS DNA (black bars). Viral gag mRNA/million HPRT ratios obtained after real-time RT-PCR are presented. RNA was extracted from 3 different regions of each tissue and analyzed individually. (B) p27 Staining in the spleen sections of control (top) and treated animals (bottom).
Viral RNA load in AS IL-4 DNA treated and untreated SHIV-infected macaques as seen by real-time RT-PCR and immunohistochemistry. (A) Viral RNA levels in the lungs (top), liver (middle), and spleen (bottom) in SHIV89.6P-infected macaques (blue bars); viral RNA in infected macaques given a single dose of AS IL-4 (green bars); and viral RNA in the animals receiving 2 injections of the AS DNA (black bars). Viral gag mRNA/million HPRT ratios obtained after real-time RT-PCR are presented. RNA was extracted from 3 different regions of each tissue and analyzed individually. (B) p27 Staining in the spleen sections of control (top) and treated animals (bottom).
Discussion
The pathogenesis of X4 SHIV infection in macaques reproduces the late phase of HIV pathogenesis in which development of X4 virus mutants is associated with rapid loss of CD4+ T cells23 and, in some cases, intense replication of these viruses in tissue macrophages. In the macaque model, pathogenic X4 SHIVs, SHIVKU,15 SHIV89.6P,14 and
Prepublished online as Blood First Edition Paper, December 23, 2004; DOI 10.1182/blood-2004-09-3515.
Supported by grants from the National Institutes of Health (MH-62969-01, AI-29382, NS-32203, RR-16443, MH068212, MH072355) and Center of Biomedical Research Excellence (COBRE; P20-RR16443).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Mingzhao Huang and Fenglan Jia for their technical assistance. We also thank Eileen Roach for assistance with the preparation of the final figures used in this article and the NIH-funded Resource for Nonhuman Immune Reagents for providing rmIL-4 reagents.