The mechanism by which arsenic dramatically affects gene expression remains poorly understood. Here we report that prolonged exposure of acute promyelocytic leukemia NB4 cells to low levels of arsenic trioxide increased the expression of a set of genes responsible for reactive oxygen species (ROS) production. We hypothesize that arsenic-induced ROS in turn contribute partially to altered gene expression. To identify genes responsive to arsenic-induced ROS, we used microarray gene expression analysis and identified genes that responded to arsenic and hydrogen peroxide but whose response to arsenic was reversed by an ROS scavenger, N-acetyl-L-cysteine. We found that 26% of the genes significantly responsive to arsenic might have been directly altered by ROS. We further explored the mechanisms by which ROS affects gene regulation and found that the Sp1 transcription factor was oxidized by arsenic treatment, with a corresponding decrease in its in situ binding on the promoters of 3 genes, hTERT, C17, and c-Myc, whose expressions were significantly suppressed. We conclude that ROS contributed partly to arsenic-mediated gene regulation and that Sp1 oxidation contributed to gene suppression by arsenic-induced ROS.
Introduction
Arsenic trioxide has drawn broad attention in the past decade because of its dramatic therapeutic effect on acute promyelocytic leukemia (APL).1,2 Arsenic is among the oldest medicines used in human history; however, its clinical use was stalled for most of the past century because of its toxic and carcinogenic effects and because of the emergence of modern therapeutic modalities.2,3 Extensive studies indicate that arsenic has pleiotropic effects that make it proapoptotic, antiproliferative, and antiangiogenic.4 Its actions also appear to differ depending on the experimental conditions.4-7 Because of its pleiotropic effects, it is not surprising that arsenic alters the expression of many genes, such as those encoding cytokines and growth factors involved in inflammation,8 and of genes involved in reactive oxygen species (ROS) metabolism such as heme oxygenase 1.9
ROS induction by arsenic is well documented,7,10,11 and we recently reported that ROS is produced by arsenic-induced nicotinamide adenine dinucleotide phosphate (NADPH) oxidase in leukemic cells.12 We surmise that arsenic could exert its effects on gene regulation by the generation of ROS, which has been reported to activate Jun N-terminal kinase (JNK), p38, activator protein 1 (AP1), and nuclear factor kappa B (NFκB).4 We previously reported that arsenic suppresses hTERT expression,13 and we now hypothesize that arsenic-induced ROS alters gene expression. Not only is the full spectrum of genetic targets of arsenic-induced ROS undetermined, but most studies also identify arsenic-responsive genes with either sodium arsenite or sodium arsenate, a method not used in clinical practice. In the clinical setting, arsenic trioxide is continuously infused to achieve plasma trough levels of approximately 1 μM.14 In this study, we mimicked the clinical situation by treating human promyelocytic leukemia NB4 cells with arsenic trioxide (0.75 μM) for a prolonged period of time. Using microarray gene expression analysis, we have reported the identification of a set of arsenic-induced genes that encode components of NADPH oxidase, an enzyme complex responsible for the production of superoxide anions in myeloid cells. The induction of NADPH oxidase expression also corresponds with the enhanced production of ROS by arsenic-treated NB4 cells.12 In this study, we sought to identify the genes directly related to arsenic-induced ROS by identifying genes that are responsive to arsenic and hydrogen peroxide but whose altered expression by arsenic is reversed by an ROS scavenger, N-acetyl-L-cysteine (NAC). Among 26 highly arsenic-responsive genes, 6 could be directly linked to ROS production by arsenic. We further found that arsenic triggers the oxidation of the Sp1 transcription factor and attenuates its binding to the promoters of hTERT, MYC, and C17, perhaps contributing to their diminished expression.
Materials and methods
Cell culture and reagents
NB4 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum in a humidified incubator at 37°C. Arsenic trioxide was purchased from Sigma (St Louis, MO). The 100 mM stock solution in 1 N NaOH was then further diluted to 100 μM in phosphate-buffered saline (PBS), as previously described.13 Cells were passaged to keep a density less than 6 × 105/mL.
Microarray analysis
NB4 cells in log-phase growth with a density of 105/mL were treated with 0.75 μM arsenic trioxide for 10 days. Total RNA was isolated and subjected to Affymetrix oligonucleotide microarray analysis using HG_U133A chip, which contained 22 000 open reading frames from the human genome. Five replicates consisting of 2 controls and 3 arsenic-treated samples were tested. Raw signal intensities were normalized. Average signals of the control and treated samples were compared to obtain a ratio for each gene.
SYBR green real-time PCR
For ID1, hTERT, and c-myc, the detection methods were described previously.13 Sequences of the primers and probe of ID1 are ID1-259F, 5-TCAAGGAGCTGGTGCCCA-3; ID1-326R, 5-GACGTGCTGGAGAATCTCCAC-3; and ID1-282, 5-CCCCAGAACCGCAAGGTGAGCA-3. Expression of other genes was determined by reverse transcription followed by SYBR green real-time polymerase chain reaction (PCR). cDNA was generated by heating a mixture of 15 μL containing 15 μg total RNA and 1 μg random primers (Promega, Madison, WI) to 70°C for 5 minutes. After immediate chilling on ice, 5 μLof 5 × reaction buffer, 5 μL dNTP (2.5 mM each), 40 U RNase inhibitor, and 200 U MMLV reverse transcriptase were added, and the mixture was incubated at 37°C for 1 hour. Ten nanograms RNA-equivalent of reverse transcription product was subjected to SYBR green quantitative real-time PCR. Each tube contained a 20-μL aliquot with 500 nM each primer, 200 μM dNTP, and PCR buffer with 1.75 mM MgCl2, 0.5 μL of 15 000-fold diluted SYBR green (Molecular Probes, Eugene, OR), and 0.5 U Platinum Taq (Invitrogen, Carlsbad, CA), unless stated otherwise. Primer sequences are available on request. All PCR reactions were performed at 95°C for 10 minutes, followed by 40 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute. Fail-safe buffers (Epicenter, Madison, WI) were used occasionally, as indicated. Multicomponent analysis (dissociation curve) was performed to ensure a single PCR product by heating the PCR products to 95°C for 20 seconds, 50°C for 20 seconds, and 98°C for 20 seconds with ramping time for 20 minutes. The signals were detected using the ABI 7700 sequence detection system (Applied Biosystems, Foster City, CA). All signals were normalized by the expression of large acidic ribosomal protein (RPLP0).
Detection of ROS by flow cytometry
NB4 cells treated with 0.75 (μM arsenic with or without NAC (American Regent Laboratories, Shirley, NY) for 10 days were washed with PBS and resuspended in complete medium with original concentrations of arsenic, followed by incubation with 0.5 μM dihydrorhodamine 123 (DHR123) (Sigma, St Louis, MO) for 30 minutes at 37°C. ROS fluorescence intensity was determined by flow cytometry with excitation at 490 nm and emission at 520 nm.
Detection of ROS by chemiluminescence
To detect extracellular and intracellular hydrogen peroxide, 10 μM luminol was added to 1 × 106 cells in 2 mL aerated complete PBS (PBS with 0.5 mM MgCl2, 0.7 mM CaCl2, and 0.1% glucose) supplemented with 10 μg/mL horseradish peroxidase (HRP). Chemiluminescence was measured continuously in a Berthold LB9505 6-channel luminometer at 37°C for 30 minutes, as described.12
Chromatin immunoprecipitation
Twenty million cells to be immunoprecipitated per antibody were crosslinked with 1% formaldehyde for 10 minutes; this was followed by the addition of 0.125 M glycine to quench the cross-linking. The pellet was washed twice with 1 × PBS with protease inhibitor and then lysed sequentially with cell lysis buffer (5 mM PIPES [piperazine diethanesulfonic acid], pH 8.0, 85 mM KCl, 0.5% NP40, and protease inhibitor) and 2 mL nuclear lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane], pH 8.1, 10 mM EDTA [ethylenediaminetetraacetic acid], 1% SDS [sodium dodecyl sulfate], and protease inhibitor). The lysate was sonicated to yield a DNA fragment length of approximately 1 kb. The soluble chromatin was precleared with protein A-coated Staphylococcus cells (Roche, Basel, Switzerland). The precleared chromatin was collected, and then 4 vol dilution buffer (0.01% SDS, 1.1% Triton X 100, 1.2 mM EDTA, 16.7 mM Tris pH 8.1, and 167 mM NaCl) was added. Four micrograms Sp1 antibody (Pep2; Santa Cruz Biotechnology, Santa Cruz, CA), c-Myc antibody (sc764; Santa Cruz Biotechnology), or hepatocyte growth factor antibody (HGF Ab; Santa Cruz Biotechnology) was added, and the mixture was rotated in a cold room overnight. Antibody-bound chromatin was pulled down by protein A-coated Staphylococcus cells. After extensive washing twice with dialysis buffer (2 mM EDTA, 50 mM Tris, pH 8.0, 0.2% sarkosyl) and 6 times with wash buffer (100 mM Tris, pH 9, 500 mM LiCl, 1% NP 40, 1% deoxycholic acid), the chromatin was eluted by 50 mM sodium bicarbonate plus 1% SDS. The cross-linking was reversed in 0.3 M NaCl plus 10 μg RNase A at 65°C for 4 to 5 hours, and the DNA was purified with phenol/chloroform and dissolved in 30 μL water. DNA fragments 5 to 6 kb away from the Sp1 binding sites were tested as negative controls. Primer sequences are listed in Table 1. For each 20-μL reaction mixture, there were 1 μL DNA, 500 μM primers, 0.5 μL 15 000-fold diluted SYBR green, 0.5 U Platinum Taq, and 10 μL fail-safe buffer (G buffer for hTERT, A buffer for c-myc, L buffer for C17). PCR was performed with 40 cycles at 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute. Signals were detected using the ABI 7700 sequence detection system.
Fractionation of cytoplasmic and nuclear protein
Cytoplasmic and nuclear proteins were fractionated using the NE-PER kit from Pierce Biotechnology (Rockford, IL) according to the manufacturer's instructions.
Sp1 immunoprecipitation
After immunoprecipitation of Sp1, 2 × 107 NB4 cells were lysed with 1.5 mL RIPA buffer (10 mM Tris, pH 8.5, 50 mM NaCl, 0.05% NP40, 0.1% SDS, and 0.5% deoxycholic acid) with protease inhibitor and then sheared with a 23-gauge needle. The soluble part was precleared with 50 μL protein A Sepharose (4 Fast Flow; Amersham Biosciences, Piscataway, NJ), followed by the addition of 4 μg Sp1 antibody (Pep 2) and by incubation at 4°C for 1 hour. Fifty microliters protein A Sepharose was then added, and the mixture was rotated at 4°C for 1 hour. The beads were washed extensively 3 times with RIPA buffer containing protease inhibitors.
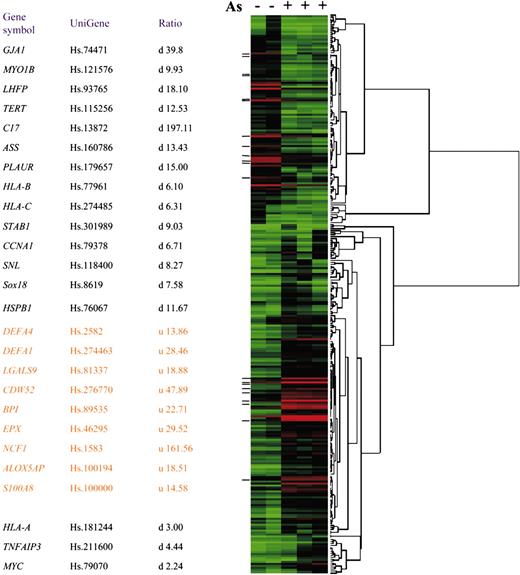
Expression profile of arsenic-responsive genes. (Left) Gene symbols, UniGene numbers, and change ratios of expression as revealed by microarray analysis of the 26 highly responsive genes for RT-PCR after hydrogen peroxide treatment or NAC rescue experiment. Three genes, denoted with asterisks, were not changed more than 6-fold; hence, they are omitted from the heat map. u indicates up-regulated; d, down-regulated. (Right) Heat map demonstrating the arsenicinduced changes in expression (at least 6-fold) of 106 down-regulated and 177 up-regulated genes.
Expression profile of arsenic-responsive genes. (Left) Gene symbols, UniGene numbers, and change ratios of expression as revealed by microarray analysis of the 26 highly responsive genes for RT-PCR after hydrogen peroxide treatment or NAC rescue experiment. Three genes, denoted with asterisks, were not changed more than 6-fold; hence, they are omitted from the heat map. u indicates up-regulated; d, down-regulated. (Right) Heat map demonstrating the arsenicinduced changes in expression (at least 6-fold) of 106 down-regulated and 177 up-regulated genes.
Detection of Sp1 glycosylation
Sp1-bound protein A Sepharose was boiled in 1 × complete Laemmli buffer for 5 minutes and resolved in SDS-polyacrylamide gel electrophoresis (SDS-PAGE). After transfer to nitrocellulose membrane, total Sp1 and glycosylated Sp1 were detected with the antibody of Sp1 (Pep2) and 110.6 antibody (a gift from Dr Gerald Hart, Johns Hopkins University), respectively. To induce deglycosylation, cells were treated with 40 μM 6-diazo-5-oxo-L-norleucine (Sigma) or were deprived of glucose with glucose-free medium for 18 hours.
Detection of Sp1 oxidation
Procedures for the detection of Sp1 oxidation have been described.15,16 Sp1-bound protein A Sepharose was incubated with 20 μM BIAM (N-[biotinoyl]-N'-[iodoacetyl]ethylenediamine) (Molecular Probes) in de-oxygenated buffer containing 50 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.5, 100 mM NaCl, 0.5% Triton X-100, and protease inhibitors for 15 minutes at room temperature. The reaction was stopped on the addition of 100 μL of 1 × Laemmli buffer and boiling for 5 minutes. The mixture was then resolved on 8% SDS-PAGE. After transfer to nitrocellulose, the BIAM-incorporated Sp1 was detected with streptavidin-conjugated HRP. Band intensities were quantified with UVP gel box (Epi Chemi II Darkroom, UVP, Upland, CA).
Results
Genetic targets of arsenic in NB4 cells
To determine the effect of arsenic on gene expression in acute promyelocytic leukemia cells, we exposed NB4 cells (105/mL) to 0.75 μM arsenic for 10 days, which slowed their growth without significant apoptosis (data not shown). Given that we previously observed that arsenic dramatically diminishes hTERT expression,13 we verified the effect of arsenic by the suppression of hTERT expression to less than 1% before we subjected the RNA samples for microarray analysis (data not shown). To determine the genes regulated by this low-dose, prolonged arsenic treatment, we compared the normalized and average signals of each gene in the control and arsenic-treated samples. Using a array of 22 000 genes, we found 106 down-regulated genes and 177 up-regulated genes, altered by at least 6-fold after arsenic treatment (Figure 1; Table S1) (for Table S1, see the Supplemental Table link at the top of the online article on the Blood website).
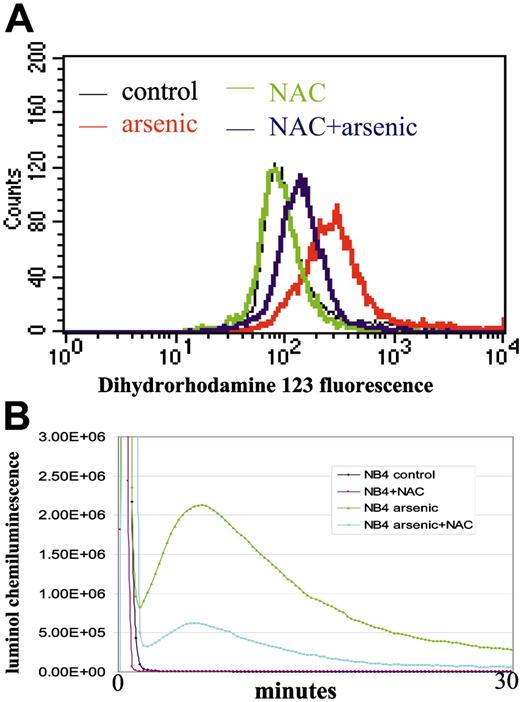
With the use of EASE software, gene ontology analysis showed that many of the up-regulated genes and those involved in immune and host defense responses were related to ROS production (Figure 1).17 For example, neutrophil cytosolic factor 1 (NCF1, p47PHOX, a p47-kD subunit of NADPH oxidase complex)18 and S100 calciumbinding factor A8 (S100A8)19 are related to the activation of NADPH oxidase, an enzyme complex responsible for the production of superoxide anion (respiratory burst) by phagocytes challenged with microbes. Other arsenic-induced alterations included 5-lipoxygenase activator (ALOX5AP), which involves arachidonate metabolism,20,21 Galectin 9,22 and eosinophil peroxidase (EPX)23-25 (Figure 1). Concurrent with the up-regulation of these genes, we observed arsenic-induced ROS production using flow cytometry and chemiluminescence (Figure 2A-B). Other genes such as bactericidal/permeability-increasing protein (BPI), defensin, Charcot-Leyden crystal protein (CLC), and interleukin-8 receptor α (IL-8Rα) are related to tissue inflammation, chemotaxis, and host defense (Figure 1; Table S1).26-29 There were, however, no specific patterns of genes down-regulated by arsenic, as determined by the EASE software. C17, a gene with a putative cytokinelike sequence but without known function,30 is the most dramatically suppressed. As previously reported,13 hTERT was also markedly down-regulated, as determined by microarray analysis, providing quality control for the effects of arsenic on NB4 cells (Figure 1). Changes in the protein levels of some of these genes were confirmed in previous studies.12,13
Arsenic induces ROS production. (A) ROS detection by flow cytometry using dihydrorhodamine 123 as a probe. Pretreatment with NAC significantly blunts ROS production after arsenic treatment. (B) ROS detection by luminol chemiluminescence. Cotreatment with NAC significantly blunts arsenic-induced ROS. Experiments were repeated twice with similar results. The y-axis represents arbitrary units of signal intensity, and the x-axis represents duration of measurement in minutes.
Arsenic induces ROS production. (A) ROS detection by flow cytometry using dihydrorhodamine 123 as a probe. Pretreatment with NAC significantly blunts ROS production after arsenic treatment. (B) ROS detection by luminol chemiluminescence. Cotreatment with NAC significantly blunts arsenic-induced ROS. Experiments were repeated twice with similar results. The y-axis represents arbitrary units of signal intensity, and the x-axis represents duration of measurement in minutes.
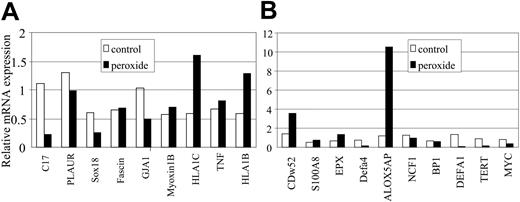
Hydrogen peroxide treatment revealed a subset of genes regulated in concordance with arsenic. Because arsenic induces ROS,7,10,11 we hypothesized that ROS, through the action of hydrogen peroxide, participates in and contributes to altering gene expression. To test this hypothesis, we sought to identify genes whose expressions are altered by hydrogen peroxide among the 26 genes that are significantly altered by arsenic (Figure 1). We treated NB4 cells with 20 μM hydrogen peroxide for 1 day, a regimen that did not cause obvious cell death, and analyzed the gene expression level by SYBR RT-PCR. Among the 17 genes down-regulated by arsenic, we found that 7 (C17, TERT, MYC, GJA1, SOX18, CCNA1, STAB1) were suppressed by hydrogen peroxide. Among 9 genes up-regulated by arsenic, 2 (CDw52, ALOX5AP) were also stimulated by hydrogen peroxide (Figure 3A-B).
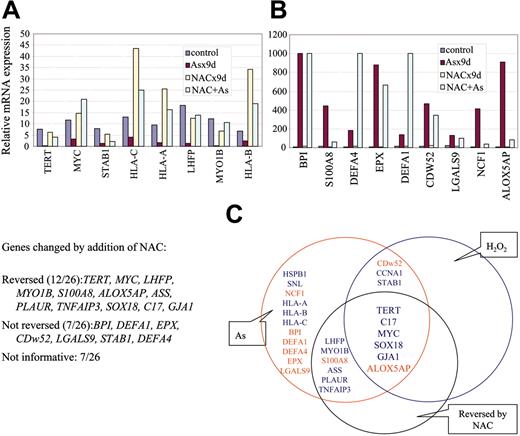
If gene expression is altered by ROS, we surmised that the alteration by arsenic would be rescued by NAC, an ROS scavenger. Therefore, we treated NB4 cells with 0.75 μM arsenic for 10 days with or without coincubation with 10 mM NAC. ROS production was significantly decreased with NAC, as measured by flow cytometry (Figure 2A) and chemiluminescence (Figure 2B). Among 26 highly responsive genes tested, the expression of 12 was reversed by NAC (Figure 4A-B). Combining the genes responsive to hydrogen peroxide and those with expression reversed by NAC, we found that 5 genes down-regulated by arsenic (C17, MYC, TERT, SOX18, and GJA1) and 1 gene up-regulated by arsenic (ALOX5AP) are ROS responsive (Figure 4c). These observations suggest that ROS is responsible for the altered expression of a subset (6 of 26; 26%) of genes triggered by arsenic.
Decreased in vivo Sp1 DNA binding after arsenic treatment
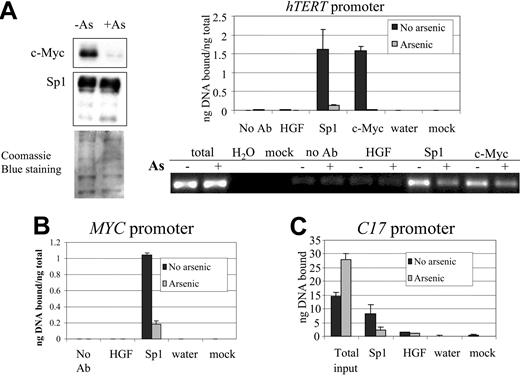
Although our findings suggest that ROS accounts for approximately one fourth of genes responsive to arsenic, the ROS-induced molecular mechanisms that cause altered gene expression are likely to be complex. In particular, many transcription factors are modulated by ROS, including NFκB, AP1, and Sp1. We focus on the Sp1 transcription factor because we have previously demonstrated that Sp1 activity on the hTERT promoter is attenuated by arsenic, as determined by electrophoretic mobility shift assay (EMSA) and luciferase reporter assays.13 Moreover, Sp1 is redox sensitive.31,32 We first tested the Sp1 in vivo binding capacity on the hTERT promoter by chromatin immunoprecipitation (ChIP). We found that after 10 days of 0.75 μM arsenic treatment, Sp1 protein binding decreased by 80% of the control at the Sp1-binding sites in the hTERT promoter (Figure 5A, right panel), without a commensurate decrease in Sp1 protein level in the nuclear extracts of cells treated with arsenic (Figure 5A, left panel). In contrast, c-Myc protein levels decreased significantly after arsenic treatment (Figure 5A, left panel), as previously reported.13 We then tested Sp1 in vivo binding to consensus Sp1 sites in the promoters of the other 2 targets, c-Myc and C17. ChIP assays confirmed Sp1 binding on some but not all of these consensus sites, and the binding decreased in arsenic-treated cells (Figure 5B-C; data not shown). The control genomic regions located 5 to 6 kb away from the consensus Sp1-binding sites revealed no significant Sp1 binding (data not shown). ChIP with a control immunoglobulin G (IgG) (hepatocyte growth factor antibody) or without antibody did not yield a significant amount of bound DNA (Figure 5A-C). These findings suggest that posttranslational modification of Sp1, rather than its level, could be responsible for its decreased DNA-binding activity on arsenic exposure.
Genes responsive to hydrogen peroxide and arsenic. Nineteen of 26 genes are represented in this figure. Among 9 arsenic up-regulated genes, 2 (CDw52 and ALOX5AP) are also up-regulated by hydrogen peroxide. Among 17 arsenic down-regulated genes, 7 (C17, TERT, MYC, GJA1, SOX18, CCNA1, and STAB1) are also down-regulated by hydrogen peroxide (only 5 were presented in this figure); see Figure 4. Values are averages of triplicates. Independent experiments were performed at least twice with similar results.
Genes responsive to hydrogen peroxide and arsenic. Nineteen of 26 genes are represented in this figure. Among 9 arsenic up-regulated genes, 2 (CDw52 and ALOX5AP) are also up-regulated by hydrogen peroxide. Among 17 arsenic down-regulated genes, 7 (C17, TERT, MYC, GJA1, SOX18, CCNA1, and STAB1) are also down-regulated by hydrogen peroxide (only 5 were presented in this figure); see Figure 4. Values are averages of triplicates. Independent experiments were performed at least twice with similar results.
Effect of NAC on arsenic-induced gene expression. (A-B) Certain arsenic-induced gene expression changes can be blunted by at least 50% with NAC cotreatment (reversed). There are also genes whose expression is altered by arsenic but that remain unaffected by NAC (not reversed). Some genes were responsive (more than 2-fold) to NAC alone without arsenic and hence were categorized as being “not informative.” Values are averages of triplicates. Independent experiments were performed at least twice with similar results. (C) Schematic diagram showing that 6 of 26 genes tested are directly related to ROS production (common area of the 3 circles). Blue highlights down-regulated genes, and red highlights up-regulated genes by arsenic treatment. The 9 genes in the area common to the red and blue circles denote those with consensus change of expression after treatment with arsenic or hydrogen peroxide. The 12 genes in the area common to the red and black circles are those whose expression change after arsenic treatment could be reversed by NAC.
Effect of NAC on arsenic-induced gene expression. (A-B) Certain arsenic-induced gene expression changes can be blunted by at least 50% with NAC cotreatment (reversed). There are also genes whose expression is altered by arsenic but that remain unaffected by NAC (not reversed). Some genes were responsive (more than 2-fold) to NAC alone without arsenic and hence were categorized as being “not informative.” Values are averages of triplicates. Independent experiments were performed at least twice with similar results. (C) Schematic diagram showing that 6 of 26 genes tested are directly related to ROS production (common area of the 3 circles). Blue highlights down-regulated genes, and red highlights up-regulated genes by arsenic treatment. The 9 genes in the area common to the red and blue circles denote those with consensus change of expression after treatment with arsenic or hydrogen peroxide. The 12 genes in the area common to the red and black circles are those whose expression change after arsenic treatment could be reversed by NAC.
In vivo Sp1 binding to the promoters of hTERT, c-Myc, and C17, as determined by ChIP assay. (A, left) Nuclear extracts from the same samples used in ChIP were immunoblotted with c-Myc and Sp1 antibodies. Coomassie blue staining served as loading control. (Right) SYBR green real-time PCR shows diminished Sp1 and c-Myc binding on the hTERT promoter after arsenic (0.75 μM for 12 days) exposure. Mock indicates no nuclear extract in the reaction; No Ab, omission of antibody in the immunoprecipitation procedures. HGF antibody was used as a control IgG. PCR products obtained at 35 cycles and resolved on agarose gel are shown below the graph. (B-C) SYBR green real-time PCR quantification of ChIP using anti-Sp1 antibody shows a significant decrease in Sp1 binding in the promoters of MYC (B) and C17 (C) after arsenic treatment. Error bars represent standard deviations from triplicate experiments.
In vivo Sp1 binding to the promoters of hTERT, c-Myc, and C17, as determined by ChIP assay. (A, left) Nuclear extracts from the same samples used in ChIP were immunoblotted with c-Myc and Sp1 antibodies. Coomassie blue staining served as loading control. (Right) SYBR green real-time PCR shows diminished Sp1 and c-Myc binding on the hTERT promoter after arsenic (0.75 μM for 12 days) exposure. Mock indicates no nuclear extract in the reaction; No Ab, omission of antibody in the immunoprecipitation procedures. HGF antibody was used as a control IgG. PCR products obtained at 35 cycles and resolved on agarose gel are shown below the graph. (B-C) SYBR green real-time PCR quantification of ChIP using anti-Sp1 antibody shows a significant decrease in Sp1 binding in the promoters of MYC (B) and C17 (C) after arsenic treatment. Error bars represent standard deviations from triplicate experiments.
Deglycosylation of Sp1 by arsenic
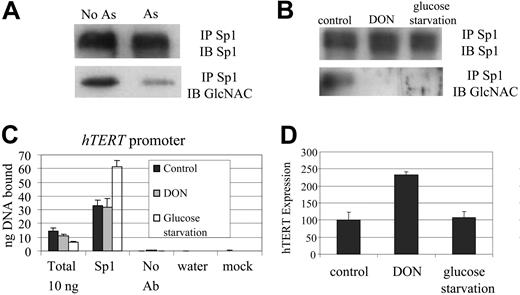
Because Sp1 protein is O-linked glycosylated,33,34 we sought to determine whether arsenic changes the glycosylation status of Sp1 with resultant alteration of its DNA binding in NB4 cells. After exposing NB4 cells to arsenic, we immunoprecipitated Sp1 protein and immunoblotted with an antibody (110.6) that recognizes O-linked glycosylated proteins. We found that arsenic treatment decreased the glycosylation of Sp1 protein (Figure 6A). However, the deglycosylation of Sp1, as induced by glucose deprivation or treatment of cells with 6-diazo-5-oxo-L-norleucine, did not decrease hTERT expression or in vivo Sp1 binding to the hTERT promoter (Figure 6B-D). Hence, deglycosylation, though associated with arsenic treatment, is insufficient to diminish the DNA-binding activity of Sp1 to the hTERT promoter or to inhibit hTERT expression.
Hydrogen peroxide decreases Sp1 DNA binding
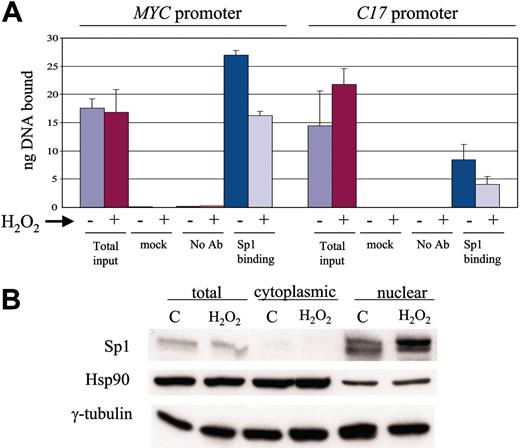
Given that changes in glycosylation could not account for the effects of arsenic, we sought to determine whether the oxidation of Sp1 could lead to its diminished activity. We found that after 1 day of hydrogen peroxide (20 μM) treatment, in vivo Sp1 binding on the promoters of MYC and C17 decreased significantly (Figure 7A). However, the amount of nuclear Sp1 protein was not changed after hydrogen peroxide treatment (Figure 7B). Thus, the oxidation of Sp1 can decrease its DNA binding activity.
Arsenic and hydrogen peroxide treatment induces oxidation of Sp1
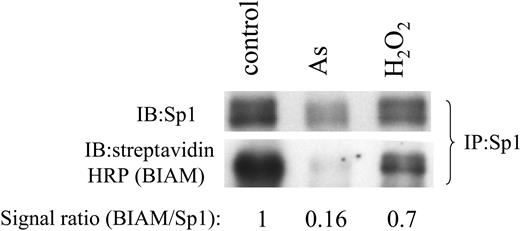
Observations that arsenic induces ROS and that arsenic and hydrogen peroxide treatment leads to diminished Sp1 function support the hypothesis that arsenic causes Sp1 oxidation and inhibits its DNA-binding activity. To confirm this hypothesis, we used the BIAM assay, which detects free sulfhydryl moieties on proteins, such that oxidation of the sulfhydryl groups would diminish BIAM incorporation. We found that sulfhydryl moieties of Sp1 are oxidized in the presence of hydrogen peroxide or arsenic (Figure 8). These observations suggest that arsenic induces ROS, which in turn oxidizes Sp1 and inhibits its function.
Sp1 deglycosylation after arsenic treatment. (A) Sp1 glycosylation was decreased after arsenic treatment, as detected by immunoblotting (IB) with O-linked GlcNAc-specific antibody (110.6 Ab) after immunoprecipitation (IP) with an anti-Sp1 antibody. The Sp1 antibody-detected signals serve as loading controls. (B) Sp1 deglycosylation was induced by glucose starvation (24 hours) or treatment with DON (40 μM for 24 hours). (C) Decrease in Sp1 glycosylation did not diminish in vivo Sp1 binding on the hTERT promoter, as determined by chromatin immunoprecipitation. (D) Deglycosylation of Sp1 did not result in decreased hTERT expression, as determined by quantitative RT-PCR. Error bars represent standard deviations from triplicate experiments.
Sp1 deglycosylation after arsenic treatment. (A) Sp1 glycosylation was decreased after arsenic treatment, as detected by immunoblotting (IB) with O-linked GlcNAc-specific antibody (110.6 Ab) after immunoprecipitation (IP) with an anti-Sp1 antibody. The Sp1 antibody-detected signals serve as loading controls. (B) Sp1 deglycosylation was induced by glucose starvation (24 hours) or treatment with DON (40 μM for 24 hours). (C) Decrease in Sp1 glycosylation did not diminish in vivo Sp1 binding on the hTERT promoter, as determined by chromatin immunoprecipitation. (D) Deglycosylation of Sp1 did not result in decreased hTERT expression, as determined by quantitative RT-PCR. Error bars represent standard deviations from triplicate experiments.
Hydrogen peroxide treatment decreases in vivo Sp1 binding to MYC and C17 promoters. (A) ChIP assay showed decrease of Sp1 binding on the promoters of c-Myc and C17 after oxidation by hydrogen peroxide treatment. (B) Cell fractionation after hydrogen peroxide treatment showed no decrease in the level of Sp1, which was exclusively nuclear. Cytosolic heat shock protein 90 (Hsp90) was used as a control for fractionation. γ-Tubulin was used as a loading control.
Hydrogen peroxide treatment decreases in vivo Sp1 binding to MYC and C17 promoters. (A) ChIP assay showed decrease of Sp1 binding on the promoters of c-Myc and C17 after oxidation by hydrogen peroxide treatment. (B) Cell fractionation after hydrogen peroxide treatment showed no decrease in the level of Sp1, which was exclusively nuclear. Cytosolic heat shock protein 90 (Hsp90) was used as a control for fractionation. γ-Tubulin was used as a loading control.
Discussion
Previous studies and this report suggest that arsenic has multiple mechanisms of altering gene expression. We observed that many genes induced by arsenic are involved in ROS production and that ROS can inhibit gene expression, in part by the oxidation of Sp1. Oxidized Sp1 loses DNA-binding activity and contributes to the suppression of genes such as MYC, hTERT, and C17.
Arsenic induces targets that produce ROS, such as NADPH oxidase and arachidonate metabolic enzymes. Eosinophil peroxidase, which is also induced by arsenic, uses hydrogen peroxide as a substrate to generate hypochlorous acid (HOCl), leading to macromolecular chlorination and oxidation.35 These targets have not been previously reported as arsenic-responsive genes perhaps because of the different experimental conditions and cell lines used. EPX is specific to myeloid cells, and NADPH oxidase is a well-characterized ROS-producing complex that is largely confined to myeloid cells. Other notable targets of arsenic identified in our study include BPI, defensin, CLC, and IL-8Rα. These proteins are involved in neutrophil chemotaxis, inflammatory responses, and antimicrobial functions. Decreased EPX, BPI, or defensin levels are associated with a rare congenital disease, neutrophil-specific granule (NSG) deficiency, which is linked to susceptibility to infections.36 Mutations in one of the subunits of NADPH oxidase result in immunocompromised congenital chronic granulomatous disease (CGD), underscoring the importance of NADPH oxidase in host immunity.37,38 Because NADPH oxidase, BPI, EPX, and defensin are all related to the host neutrophil immune response to pathogens, it is intriguing to speculate that immunity in NSG-deficient patients might be restored by arsenic treatment.
Arsenic and hydrogen peroxide treatments result in oxidation of Sp1 protein. Immunoprecipitated (IP) Sp1 from control, arsenic, or hydrogen peroxide-treated samples was incubated with BIAM, and this was followed by detection by immunoblotting (IB) with streptavidin-conjugated HRP. The same immunoprecipitate was subjected to detection by Sp1 antibody as a loading control. BIAM/Sp1 signal ratios represent relative BIAM incorporation.
Arsenic and hydrogen peroxide treatments result in oxidation of Sp1 protein. Immunoprecipitated (IP) Sp1 from control, arsenic, or hydrogen peroxide-treated samples was incubated with BIAM, and this was followed by detection by immunoblotting (IB) with streptavidin-conjugated HRP. The same immunoprecipitate was subjected to detection by Sp1 antibody as a loading control. BIAM/Sp1 signal ratios represent relative BIAM incorporation.
Previous studies have implicated ROS in arsenic-induced cell death and NADPH oxidase in arsenic-induced ROS production in leukemia cell lines.12 In our study, these observations did not seem to correlate with cell differentiation because we could not see cell differentiation by morphology or immunophenotyping. In this report, we define the significance of ROS in the arsenic-induced alteration of gene expression. Among a group of highly arsenicresponsive genes, one quarter of the genes appears to be affected by arsenic-induced ROS, with MYC, hTERT, and C17 the most dramatically inhibited. Our earlier study suggested that Sp1 DNA-binding ability was inactivated by arsenic treatment, accounting for the suppression of transcription of hTERT and c-Myc by arsenic.13 Sp1 protein has been shown to be phosphorylated,39,40 O-linked glycosylated,33 or oxidized.31,32 We observe that the deglycosylation of Sp1 is insufficient to inhibit its association with target genes as determined by ChIP. Because the direct addition of arsenic to nuclear extracts did not decrease Sp1 activity, as assayed by EMSA (data not shown), we surmise that Sp1 is oxidized indirectly by arsenic through the induction of ROS. The sulfhydryl groups in proteins are prime targets of ROS modification,41,42 and our biochemical study using BIAM revealed the oxidation of Sp1 sulfhydryl groups in cells exposed to arsenic. These observations are compatible with the oxidation and inactivation of Sp1 through arsenic-induced ROS. Sp1 protein was originally reported as a positive regulator of gene transcription,43,44 and subsequent studies demonstrated that Sp1 could transactivate numerous genes. However, recent reports indicate that Sp1 could also serve as a negative regulator of gene transcription.45,46 Although we demonstrate in this report that arsenic-induced ROS compromises Sp1 binding, leading to the suppression or induction of gene expression, it is difficult to gauge the quantitative impact of Sp1 oxidation on the change of gene expression in cells treated with arsenic because transcriptional regulation is complex and usually beyond the influence of a single transcription factor. Moreover, we cannot rule out that cells might have overcome detrimental effects of prolonged arsenic exposure through adaptive mechanisms that may also affect the expression of some vital genes independently of Sp1 oxidation. Given the prolonged and low-dose treatment, the present study may also provide insight into the toxicologic and therapeutic effects of arsenic trioxide. Notwithstanding these caveats, our study nevertheless provides firm evidence that arsenic-mediated oxidation of Sp1 correlates with the responses of several genes to arsenic.
In aggregate, our observations suggest that arsenic suppression of gene expression, particularly MYC, hTERT, and C17, is mediated in part by ROS produced by arsenic-induced genes. Although this specific mechanism contributes to our understanding of the biologic effects of arsenic, the relatively low percentage of genes whose expression was altered by ROS suggests that this specific pathway may only play a minor role in the pleiotropic effects of arsenic. Other transcription factors and mechanisms could also be involved.
Prepublished online as Blood First Edition Paper, March 10, 2005; DOI 10.1182/blood-2005-01-0241.
Supported by National Institutes of Health grant R37CA51497 (C.V.D.) and T32GM07819.
C.V.D. is Johns Hopkins Family Professor in Oncology Research.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Michael Trush for assistance with the chemiluminescence assay. We appreciate comments from L. Gardner, M. McDevitt, and K. Zeller.