Abstract
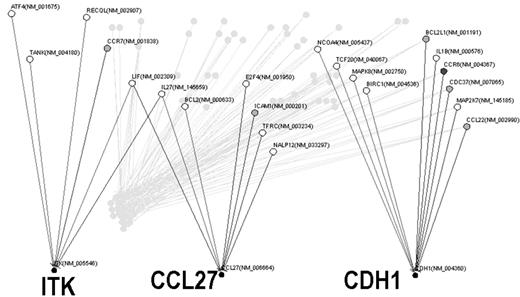
The immune response to viral infection involves complex network of dynamic gene and protein interactions. Comprehensive gene expression analysis of the host immune response against viruses has been extensively studied, however, the mechanism of virus-induced immune response is not completely understood. This might be due in part to the difficulty of finding pathologically relevant genes, despite the fact that DNA microarray technology can simultaneously monitor the expression of thousands of genes. Likewise, it is hard to estimate how each gene interferes during the viral infection. Thus, construction of gene networks from microarray gene expression data is becoming an important challenge in the post-genome era. Human herpesvirus 6 (HHV-6) is a β-herpesvirus that is closely related to human cytomegalovirus. HHV-6 shows a predominant tropism for CD4 T lymphocytes, on which it exerts marked cytopathic effects. Understanding of the clinical spectrum of HHV-6 is still evolving, however, in vitro interactions between HHV-6 and other viruses, such as the human immunodeficiency virus (HIV), and their relevance to the in vivo situation has become increasingly apparent. We present here the dynamic gene network of the host immune response during human herpesvirus type 6 (HHV-6) infection in an adult T cell leukemia (ATL) cell line. Using a pathway-focused oligonucleotide DNA microarray, we found a possible association between chemokine genes regulating Th1/Th2 balance and genes regulating T-cell proliferation during HHV-6B infection. Gene network analysis using an integrated comprehensive workbench, VoyaGene® revealed that a gene encoding a TEC-family kinase, ITK, might be a putative modulator in the host immune response against HHV-6B infection. We conclude that Th2-dominated inflammatory reaction in host cells may play an important role in HHV-6B infected T cells, thereby suggesting the possibility that ITK might be a therapeutic target in diseases related to dysregulation of Th1/Th2 balance. This study describes a novel approach to find genes related with the complex host-virus interaction using microarray data employing the Bayesian statistical framework.
Author notes
Corresponding author