Abstract
Hepcidin, a key regulator of iron metabolism, is synthesized by the liver. Hepcidin binds to the iron exporter ferroportin to regulate the release of iron into plasma from macrophages, hepatocytes, and enterocytes. We analyzed liver samples from patients undergoing hepatic surgery for cancer or receiving liver transplants and analyzed correlations between clinical parameters and liver hepcidin mRNA and urinary hepcidin concentrations. Despite the many potential confounding influences, urinary hepcidin concentrations significantly correlated with hepatic hepcidin mRNA concentrations, indicating that hepcidin quantification in urine is a valid approach to evaluate hepcidin expression. Moreover, we found in humans that hepcidin levels correlated with hepatic iron stores and hemoglobin levels and may also be affected by hepatic dysfunction. (Blood. 2005;106:746-748)
Introduction
Hepcidin is a peptide found in human plasma and urine1,2 and synthesized in the liver.1-3 Studies in mice, humans, and cell cultures demonstrated that hepcidin mRNA levels were regulated by iron stores,3 inflammation,3,4-6 anemia, and hypoxia,4 and influenced by the mouse strain and sex and transcription factors involved in hepatocyte differentiation.7,8 Hepcidin-deficient mice accumulated iron in the liver and pancreas9 and mice engineered to overexpress hepcidin were born with severe iron deficiency that was not corrected by iron supplementation.10 Taken together these data suggested that hepcidin was a hormone involved in the regulation of iron metabolism.9 Studies of patients with iron disorders demonstrated the involvement of hepcidin in regulation of iron metabolism during juvenile hemochromatosis,11 classical HFE-related hemochromatosis,12,13 and chronic inflammatory diseases.5,14 Hepcidin appears to act by inhibiting cellular iron export through binding directly to the iron exporter ferroportin and inducing its internalization and degradation.15
Recent understanding of iron metabolism in mice is based on measurements of hepatic hepcidin mRNA, whereas the corresponding human studies examine urinary hepcidin concentrations. We examined the correlation between hepcidin mRNA and urinary hepcidin concentrations in the context of human liver disease.
Study design
Patients
Thirty-six patients, who were operated on for primary or secondary liver carcinoma or received transplants for cirrhosis, were included in this study. The patients with known HFE genetic hemochromatosis, expected to exhibit abnormal hepcidin regulation,12,16 were excluded. The study was approved by the local ethics committee (CCPPRB) and informed consent was obtained from the patients.
From each patient we collected clinical data and serum, urine, and nontumoral liver samples. Liver tissues were frozen in liquid nitrogen or fixed in formalin for histologic studies. The hepatic fibrosis status was evaluated according to the Metavir score17 ; 20 samples had no or slight fibrosis, 7 samples were moderately fibrotic, and 9 samples were strongly fibrotic or cirrhotic.
Methods
Clinical laboratory studies. Clinical laboratory assays were performed at the Rennes University Hospital. The liver iron concentration (LIC) was evaluated as described previously.18
Quantitative RT-PCR. Total RNAs were extracted using the SV Total RNA Isolation System (Promega, Madison, WI). Quality-checked RNA (1 μg) was used for reverse transcription (RT) following the manufacturer's protocol (Advantage RT-for-PCR Kit, Clontech, Palo Alto, CA). We performed polymerase chain reactions (PCRs) in triplicate to evaluate the hepcidin expression in each sample compared with the RNA 18S expression and with a standard set using the qPCR-Core-kit according to the manufacturer's instructions (Eurogentec, Seraing, Belgium). Primer and probe sequences used for the hepcidin amplification were: forward primer 5′-TCCCACAACAGACGGGACAA-3′, reverse 5′-AGCAGCCGCAGCAGAAAAT-3′ and FAM/TAMRA probe 5′-CCATGTTCCAGAGGCGAAGGAGGC-3′. The amplified 137-bp PCR fragment was checked by sequencing. For RNA 18S amplification we used the 18S genomic control kit (Eurogentec). The PCR was run on ABI PRISM 7000 sequence detection system (Applied Bioscience, London, United Kingdom): 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
Data analysis. For each sample, hepcidin mRNA threshold cycle (Ct) value was normalized with 18S RNA Ct value and compared to value of a standard sample (mix of 4 liver samples exhibiting readily detectable level of hepcidin mRNA as previously analyzed by Northern blot). Results were expressed in log 2 of ratio sample versus standard and noted for convenience in arbitrary units (AU).
Hepcidin assay. Urinary hepcidin assay was performed as previously described5 and hepcidin concentration in urine expressed as nanograms hepcidin per milligrams creatinine. Because the lower detection limit of the urinary hepcidin assay is 1 ng/mg creatinine, we attributed a value of 1 ng/mg creatinine to samples with lower value.
Statistical analysis. The statistical analysis was performed on Statview software (SAS institute, Cary, NC) using nonparametric tests. P less than .05 was considered significant.
Results and discussion
Hepcidin mRNA expression is correlated with the urinary hepcidin level
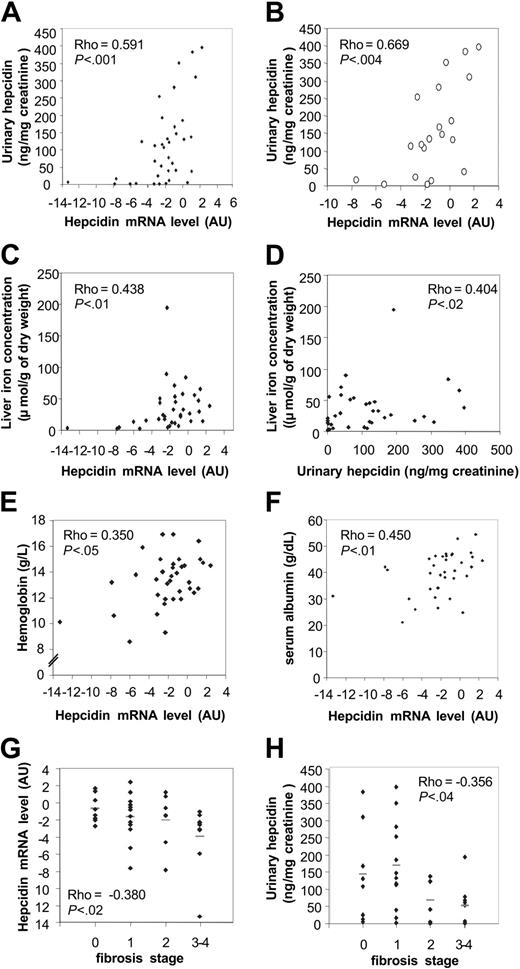
Hepcidin mRNA and urinary hepcidin levels appeared to be positively correlated (Figure 1A), with levels of both parameters increasing in parallel. This result demonstrates that the urinary hepcidin assay can be considered a valid reflection of hepatic hepcidin production. The correlation, however, was not ideal, possibly related to the inclusion of patients with fibrosis, which could have modified the relationship due to the heterogeneity of the liver in such cases. This interpretation is supported by improved correlation between hepcidin mRNA and urinary hepcidin (Figure 1B) when patients with more than slight fibrosis were excluded. In the nonfibrotic group, the correlation could be partly obscured by (1) additional regulatory mechanisms during translation, secretion, and urinary excretion processes; (2) detection limits of the present urinary hepcidin assay; and (3) the acute effects of anesthesia and surgery that could selectively influence hepatic hepcidin mRNA concentrations but not urinary hepcidin concentrations.
Hepatic hepcidin mRNA level, urinary hepcidin level, and clinical parameters
As judged by laboratory studies, our patients displayed a broad range of inflammation status and iron loading (Table 1). The analysis of the relationship between hepcidin mRNA, urinary hepcidin, and other clinical data led us to identify significant correlations, involving iron status, hematologic parameters, and hepatic functional status (Table 1).
Thus, LIC appeared to be significantly correlated not only with hepcidin mRNA, as previously reported in untreated hemochromatotic patients,12 but also with urinary hepcidin levels (Figure 1D). In addition, urinary hepcidin was correlated with ferritinemia as previously documented.5 In a group of patients not selected for diseases of iron metabolism12 or inflammatory disorders,5 these observations document a relationship between hepatic iron and urinary hepcidin levels. However, in our population the 2 hepcidin parameters did not significantly correlate with either serum iron concentration or with transferrin saturation.
Our study also demonstrates a relationship between hemoglobin level and hepcidin mRNA expression (Figure 1E), supporting the hypothesis of an impact of anemia or hypoxia (or both) on hepcidin mRNA expression, as reported in mice.4 However, we did not find a significant correlation between erythroid parameters and urinary hepcidin concentration, suggesting again additional regulatory mechanisms.
Correlations between hepcidin mRNA levels, urinary hepcidin concentrations, and clinical parameters. Urinary hepcidin levels expressed in nanogram per milligram urinary creatinine were correlated with hepcidin mRNA levels expressed in arbitrary units (log 2 scale) in the whole population of the 36 patients (♦; A) and within the subgroup of 20 patients with no or slight fibrosis (○; B). In our total population (n = 36) LIC was correlated with hepcidin mRNA (C) and urinary hepcidin (D) levels. Hepcidin mRNA level was correlated with blood hemoglobin value (E) as well as serum albumin level (F). Inverse correlations were found for hepatic fibrosis status and hepcidin mRNA (G) and urinary hepcidin levels (H). Horizontal bars correspond to the mean values.
Correlations between hepcidin mRNA levels, urinary hepcidin concentrations, and clinical parameters. Urinary hepcidin levels expressed in nanogram per milligram urinary creatinine were correlated with hepcidin mRNA levels expressed in arbitrary units (log 2 scale) in the whole population of the 36 patients (♦; A) and within the subgroup of 20 patients with no or slight fibrosis (○; B). In our total population (n = 36) LIC was correlated with hepcidin mRNA (C) and urinary hepcidin (D) levels. Hepcidin mRNA level was correlated with blood hemoglobin value (E) as well as serum albumin level (F). Inverse correlations were found for hepatic fibrosis status and hepcidin mRNA (G) and urinary hepcidin levels (H). Horizontal bars correspond to the mean values.
Surprisingly, we did not find a relationship between hepcidin expression and C-reactive protein (CRP) levels, an indicator of inflammatory state. The heterogeneity of our population, the small number of patients exhibiting high CRP levels, the possible lack of CRP sensitivity for expressing hepatic inflammatory status, and the presence of confounding regulatory factors could partly explain this result.
Finally, we found that parameters reflecting hepatic function were correlated with hepcidin levels (Figure 1F-H). Thus, serum albumin was positively correlated with hepcidin mRNA levels, whereas fibrosis status was negatively correlated with hepcidin mRNA and urinary hepcidin levels. Taken together, these results suggest that hepatic function and the effects of disease processes on hepatocytes could modulate iron metabolism. This hypothesis is reinforced within the subgroup of 20 patients with no or slight fibrosis, which exhibits an improvement of the correlations, especially those involving urinary hepcidin with LIC (rho = 0.632; P < .01) and ferritin (rho = 0.755; P < .001). In addition, a relationship between hepcidin mRNA and ferritinemia appears in this group (rho = 0.515; P < .03), as previously described.13 Conversely, all these correlations disappeared in the patient subgroup exhibiting high levels of fibrosis or cirrhosis.
In conclusion, our results demonstrate that urinary hepcidin levels reflect hepatic hepcidin mRNA expression. Moreover, despite the heterogeneity of our patients, they show in humans a relationship between hepcidin levels, hepatic iron stores, hemoglobin level, and hepatic function. With further refinement, hepcidin assays could contribute to the diagnosis and improved understanding of the pathophysiology of iron disorders.
Prepublished online as Blood First Edition Paper, March 29, 2005; DOI 10.1182/blood-2004-12-4855.
Supported by a PHRC regional grant (R09-01) and by the QLRT-2001-00444 EC contract.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the Biological Resource Centre (BRC) of Rennes for the supplying of human biologic samples and Medhi Alizadeh for helpful discussion.