Abstract
Hemophagocytic lymphohistiocytosis (HLH) is a rare disorder of immune dysregulation, characterized by end-organ damage from lymphocytic infiltration and macrophage activation. All known mutations associated with the HLH occur in genes critical in the perforin-granzyme pathway. Herein, we report HLH occurring in 2 female triplet infants who also had associated human herpesvirus type 8 (HHV-8) infections. The subjects had identical novel compound-heterozygous mutations in the Perforin alleles, resulting in undetectable perforin expression and NK-cell cytotoxicity. Both infants also had evidence of infection with HHV-8. These reports are, to our knowledge, the first cases of HLH in triplets and the first reported cases of HHV-8 infection associated with HLH in non–renal transplant and non–HIV-infected subjects.
Introduction
Hemophagocytic lymphohistiocytosis (HLH) is a rare disease in which the immune system becomes overactive due to its inability to effectively clear infections or regulate its response to infections or both.1-3 Despite aggressive chemotherapeutic interventions, patients with HLH have a high mortality rate as a result of end-organ damage. All of the known mutations associated with HLH are in genes involved in the granule exocytosis pathway (eg, PRF1, SH2D1A, UNC13D, RAB27A, CHS1, and others), implicating the critical role of this pathway in controlling immune responses to infections.4-7 We report 2 cases of HLH in triplets infected with human herpesvirus type 8 (HHV-8), both having novel compound heterozygous mutations in the perforin gene and absent natural killer (NK) cell function. To our knowledge, this is the first report of triplets presenting with HLH, and they are the first non-HIV and non–renal transplant subjects who have developed HLH in association with HHV-8 infection.
Study design
Subject samples
Subject samples and informed consents were collected in accordance with protocols approved by the University of Iowa Hospitals and Clinics Human Studies Committee and Internal Review Board. Informed consents and procedures on all subjects were performed in accordance with institutional ethical standards and with the Helsinki Declaration of 1975, as revised in 2000.
Light microscopy
Bone marrow aspirations were stained with standard May-Grünwald/Giemsa stain (Sigma, Saint Louis, MO). Light microscopy pictures were taken using a Nikon Microphot-SA microscope equipped with a 40× (original magnification, ×400) bright-field objective lens (Nikon, Garden City, NJ, Cytoseal XYL mounting medium (Richard-Allan Scientific, Kalamazoo, MI), an analysis camera with 10× magnification (Soft Imaging Systems, Lakewood, CA), and acquisition software (Soft Imaging Systems) were used.
Flow cytometry analysis
Peripheral blood mononuclear cells (PBMCs) were stained for surface and intracellular markers as previously described.8 Briefly, whole blood was stained with surface antibodies (CD4, CD8, and CD56; Becton Dickinson, Franklin Lakes, NJ), then fixed, permeabilized (Cytofix/Cytoperm; Pharmingen, San Jose, CA), and stained with primary-conjugated anti–granzyme A (CB9; Becton Dickinson-Pharmingen, San Diego, CA), anti–granzyme B (GB12; Caltag, San Francisco, CA), and antiperforin (δG9; Becton Dickinson-Pharmingen) antibodies. Samples were analyzed on a FACSCalibur (Becton Dickinson).
Flow-based killing assay
A flow-based killing assay was performed as previously described.8 Briefly, K562 target cells were labeled with 125 nM 5-(and-6-)-carboxyfluorescein diacetate succinimydyl ester (Molecular Probes, Eugene, OR), and then mixed and plated with isolated subject PBMCs at a 50:1 effector-target ratio. Duplicate samples were incubated for 6 hours. Next, 1 μg/mL 7-amino-actinomycin (Calbiochem, La Jolla, CA) was added to each sample before analysis. Negative (targets only) and positive (normal control) samples were performed in parallel.
Perforin mutational analysis
The Molecular Genetics Laboratory at Cincinnati's Children's Hospital (Dr Alexandra Filiopovich) analyzed genomic DNA samples from each subject for perforin gene mutations through polymerase chain reaction (PCR) and direct sequencing.
Nested PCR for HHV-8
Nested PCR for HHV-8 was performed as previously described.9,10 Briefly, DNA was isolated from each subject sample (DNeasy; Qiagen, Valencia, CA), then the HHV-8–specific 336–base pair (bp) open reading frame 26 (ORF-26) region (nucleotide position 890 to 1226) was amplified by nested PCR using an outside primer pair (5′-ATCTATCCAAGTGCACACTGC-3′ and 5′-CTGGGAACCAAGGCTGATAGG-3′) and an inside primer pair (5′-GATGATCCCTCTGACAACCT-3′ and 5′-GGATCCGTGTTGTCTACG-3′). The product of the nested PCR was resolved on a 1.5% agarose gel. The BC-3 and Mewo cell lines were used for positive and negative controls, respectively.9-11
Results and discussion
Case report A
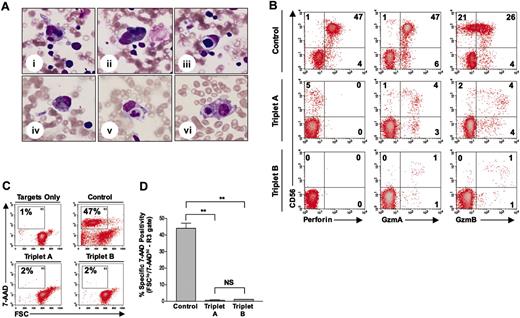
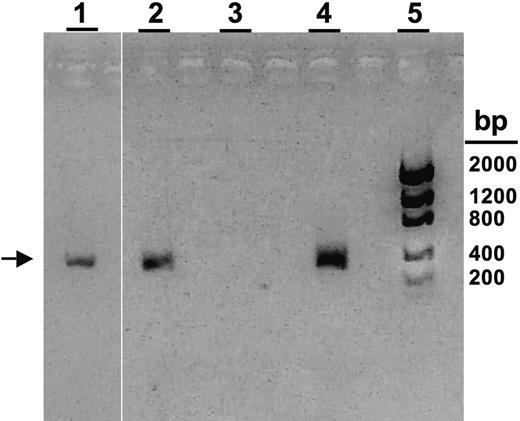
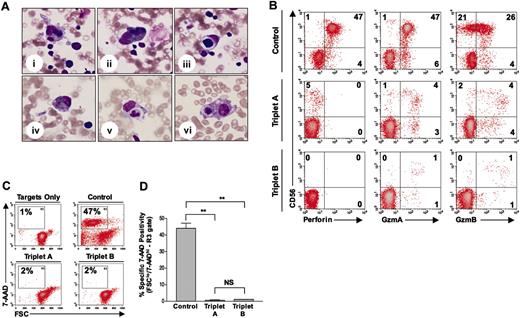
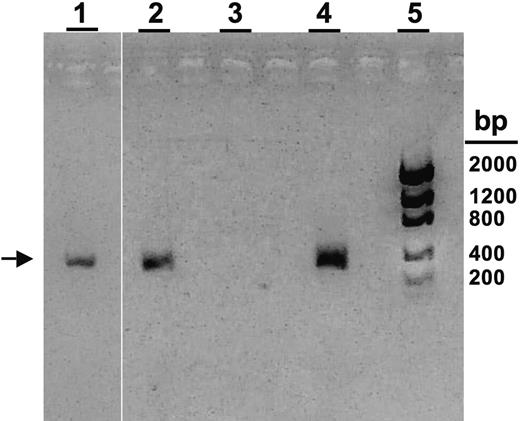
A 4-month-old multiethnic girl (triplet A; sister to triplets B and C) was transferred to our institution for persistent fevers and pancytopenia. Birth history was remarkable for delivery at 29 weeks, and Group B streptococcus septicemia at 4 weeks of age. Family history was unremarkable, and the parents were not related. Physical examination upon presentation revealed hepatosplenomegaly. Peripheral blood examination revealed a white blood cell (WBC) count of 3.4 × 109/L (3400 cells/μL), hemoglobin (Hgb) of 52 g/L (5.2 g/dL), platelets of 37 × 109/L (37 000/μL), with an absolute neutrophil count (ANC) of 210 cells/μL, absolute lymphocyte count (ALC) of 2500 cells/μL, and an absolute monocyte count (AMC) of 370 cells/μL. Abnormal chemistries included a triglyceride level of 1.7 mmol/L (151 mg/dL; normal, < 1.7 mmol/L [< 150 mg/dL]), ferritin of 2529 ng/dL (normal, 10-290 ng/dL), and fibrinogen level of 2.22 g/L (222 mg/dL; normal, 2.3-3.9 g/L [230-390 mg/dL]). Cerebral spinal fluid (CSF) evaluation revealed 5 WBCs/mm3. Bone marrow evaluation revealed hemophagocytosis (Figure 1Ai-iii), without evidence of leukemia. An extensive infectious disease assessment was notable for a positive PCR test for HHV-8 from peripheral blood but not from her CSF sample (Figure 2). A clinical diagnosis of HLH was made based on Histiocyte Society Criteria, and she was started on the HLH-94 protocol consisting of etoposide, dexamethasone, and cyclosporine A.12 Immunophenotyping revealed a normal NK-cell (CD56+CD8–) percentage (5%; normal: 2%-14% for age) (Figure 1B), although in vitro NK-cell functional analysis showed no significant killing of K562 target cells (Figure 1C-D).3,8 Normal expression of granzymes A and B was demonstrated in both NK and CD8+ T cells (Figure 1B; data not shown). Flow cytometry revealed no detectable perforin antibody reactivity (Figure 1B). Genetic analysis demonstrated novel compound heterozygous mutations in the perforin gene (Table 1). Evaluation of her parents demonstrated their carrier status for the same perforin gene mutations (Table 1). Triplet A responded to HLH-94 therapy with normalization of WBC counts and chemistries and with decreased bone marrow hemophagocytosis.
Hemophagocytosis and lack of perforin detection and NK-cell cytotoxicity in case reports A and B. (A) Bone marrow evaluation of triplet A (case report A; i-iii) and triplet B (case report B; iv-vi), showing hemophagocytosis of bone marrow elements (eg, red blood cells and neutrophils) by histiocytes. All pictures are from May-Grünwald/Giemsa–stained slides and are shown at × 400 magnification. (B) Perforin and granzymes A and B antibody reactivity in CD56+CD8– NK cells from control (top row), triplet A (middle row), and triplet B (bottom row). Percentage of positive cells is shown in each quadrant. GzmA indicates granzyme A; GzmB, granzyme B. (C) Representative flow-based cytotoxicity results, demonstrating NK-cell–induced apoptosis (FSClo/7-AAD+hi) of major histocompatibility complex (MHC) class I–negative target cells (K562) in control, triplet A, and triplet B. Percentage of apoptotic target cells is shown in the R3 gate of each panel. FSC indicates forward scatter; 7-AAD, 7-aminoactinomycin D. (D) Summary of NK-cell killing, demonstrating the average percentage of apoptotic target cells detected in the R3 gate for each subject. Effector-target (E / T) ratio was 50:1 for all samples. Values shown are the mean ± SD. **P < .01, NS = not significant.
Hemophagocytosis and lack of perforin detection and NK-cell cytotoxicity in case reports A and B. (A) Bone marrow evaluation of triplet A (case report A; i-iii) and triplet B (case report B; iv-vi), showing hemophagocytosis of bone marrow elements (eg, red blood cells and neutrophils) by histiocytes. All pictures are from May-Grünwald/Giemsa–stained slides and are shown at × 400 magnification. (B) Perforin and granzymes A and B antibody reactivity in CD56+CD8– NK cells from control (top row), triplet A (middle row), and triplet B (bottom row). Percentage of positive cells is shown in each quadrant. GzmA indicates granzyme A; GzmB, granzyme B. (C) Representative flow-based cytotoxicity results, demonstrating NK-cell–induced apoptosis (FSClo/7-AAD+hi) of major histocompatibility complex (MHC) class I–negative target cells (K562) in control, triplet A, and triplet B. Percentage of apoptotic target cells is shown in the R3 gate of each panel. FSC indicates forward scatter; 7-AAD, 7-aminoactinomycin D. (D) Summary of NK-cell killing, demonstrating the average percentage of apoptotic target cells detected in the R3 gate for each subject. Effector-target (E / T) ratio was 50:1 for all samples. Values shown are the mean ± SD. **P < .01, NS = not significant.
PCR analysis of buffy coat and CSF samples from affected triplets for HHV-8. Specific PCR product for ORF-26 of HHV-8 is shown (336-bp product; arrow).9 Lane 1, triplet B CSF sample; lane 2, triplet A buffy coat sample; lane 3, Mewo cell line (negative control); lane 4, BC-3 cell line (positive control); lane 5: DNA bp ladder.
PCR analysis of buffy coat and CSF samples from affected triplets for HHV-8. Specific PCR product for ORF-26 of HHV-8 is shown (336-bp product; arrow).9 Lane 1, triplet B CSF sample; lane 2, triplet A buffy coat sample; lane 3, Mewo cell line (negative control); lane 4, BC-3 cell line (positive control); lane 5: DNA bp ladder.
Case report B
Genetic testing on triplet B (sister to triplets A and C) revealed that she carried the same perforin gene mutations as triplet A (Table 1). She remained asymptomatic with normal laboratory evaluations until 6 months of age, when she developed respiratory symptoms with persistent fevers. Physical examination revealed an irritable infant with hepatomegaly. Peripheral blood examination showed a WBC count of 45.0 × 109/L (4500 cell/μL), Hgb of 100.3 g/L (10.3 g/dL), platelets of 72 × 109/L (72 000/μL), an ANC of 1300 cells/μL, an ALC of 2600 cells/μL, and an AMC of 90 cells/μL. Abnormal chemistries included a ferritin level of 2105 ng/dL, prothrombin time of 13 seconds (normal, 9-12 seconds), alanine aminotransferase of 423 IU/L (normal, 3-30 IU/L), γ-glutamyl transpeptidase of 200 IU/L (normal, 1-39 IU/L), total bilirubin of 25.65 μmol/L (1.5 mg/dL; normal, 3.42-17.1 μmol/L [0.2-1 mg/dL]), and a fibrinogen of 1.75 g/L (175 mg/dL). CSF evaluation showed a total of 1 WBC/mm3. Bone marrow evaluation revealed hemophagocytosis (Figure 1Aiv-vi). Virology assessment revealed a positive PCR test for HHV-8 DNA from her CSF (Figure 2). Peripheral blood analysis revealed no circulating NK cells and few granzyme A– or granzyme B–expressing cells (≤1%; Figure 1B). Similar to triplet A, she had no detectable perforin antibody reactivity and minimal NK-cell killing (Figure 1B-D).8 The clinical diagnosis of HLH was made, and she was started on HLH-94 therapy with subsequent improvement in her blood counts and serum chemistries.12
Case report C
Triplet C (sister to triplets A and B) presented for medical evaluation upon recognition that her 2 siblings (triplets A and B) had HLH. Physical examination was unremarkable. All laboratory tests were within normal limits. Genetic analysis showed that she was a carrier of a single mutated perforin allele, similar to her father's mutation (Table 1). She has required no further medical intervention.
These case reports are to our knowledge the first cases of HLH occurring in a set of triplets. There have been 2 previous reports of perforin gene mutations in twins.13,14 Twins reported by Lipton et al14 presented in the newborn period with symptoms consistent with HLH. Genetic analysis demonstrated a single missense mutation in the perforin gene (PRF1) in only one of the twins and their mother. It was presumed that each twin inherited an additional genetic mutation in the perforin gene from their father; however, this point was not documented.14 Busiello et al13 reported on 2 fraternal twins having the same homozygous perforin mutations. Despite both twins having the same perforin mutations, only one reportedly developed HLH. The unaffected twin reportedly demonstrated normal NK-cell activity.13
In our study, the affected triplets presented within 12 weeks of each other following a viral prodrome. HHV-8 infection was documented by PCR amplification in both subjects using a previously published technique.10 HHV-8 was noted in the peripheral blood of triplet A, but not her CSF, while triplet B only had HHV-8 detected in her CSF. These differences are probably related to the timing of sample collections from each of the 2 subjects. The source of the HHV-8 infection is difficult to discern; however, since a small percentage of healthy American adults are HHV-8 seropositive, one possibility is transmission via a blood product while in the nursery.15 Although HLH has been previously associated with HHV-8 infection, all of these reports have been in HIV-infected individuals with associated Kaposi sarcoma,16-18 or patients who received a renal transplant.19,20
Genetic analysis demonstrated that triplets A and B had the same compound heterozygous mutations in the perforin gene. Neither subject demonstrated intracellular perforin expression by flow cytometry. Although certain perforin mutations may not allow for antibody reactivity, functional analysis of both subjects also demonstrated no significant NK-cell cytotoxicity, indicating that the compound heterozygous mutations result in nonexpressed or nonfunctional perforin. Similar to Busiello et al,13 there were also phenotypic differences between each affected triplet in our study, including time of onset of disease, as well as differences in the percentage of circulating NK cells, and expression of cellular granzymes A and B. These results suggest that, despite having the same genetic mutations, there are significant polygenetic influences in the development of HLH.
This report and others demonstrate the critical importance of the perforin-granzyme pathway in NK-cell function and viral clearance. Moreover, it stresses the importance of this pathway in regulating immune responses to viral infections. We have recently reported that human T regulatory cells also use the perforin-granzyme pathway. We demonstrated that both adaptive and natural human T regulatory cells are able to “suppress” autologous naive T-cell proliferation by killing them through the perforin-granzyme pathway.3,8 These reports support the important role that this pathway plays in the immune dysregulation found in HLH. Specifically, they demonstrate that the perforin-granzyme pathway is not only important in the initial immune response to viral infections, but that it is also a pathway used in regulating or “suppressing” the immune response during the contraction phase via the CD4+ T regulatory compartment.
Prepublished online as Blood First Edition Paper, April 19, 2005; DOI 10.1182/blood-2005-03-0950.
Supported by the Children's Miracle Network (F.D.G.) and the National Institutes of Health (grant AI22795) (C.G.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Cincinnati Children's Research Foundation Molecular Genetics Laboratory (moleculargenetics@cchmc.org) performed all perforin gene mutation analyses.