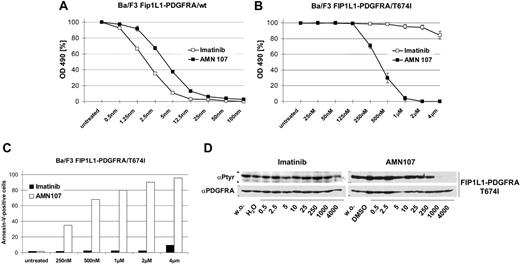
A fusion of the PDGFRA and FIP1L1 genes can be detected in cases of idiopathic hypereosinophilic syndrome (HES),1 and the resulting tyrosine kinase constitutes a drug target for the treatment of this disease with the tyrosine kinase inhibitor imatinib mesylate (Gleevec; Novartis, Basel, Switzerland).1,2 However, a mutation leading to threonine residue 674 in the FIP1L1-PDGFRA kinase domain being replaced by isoleucine is known to give rise to resistance to imatinib mesylate in patients with HES.1,3 This T674I exchange corresponds to T315I in BCR-ABL, which causes absolute resistance of chronic myelogenous leukemia (CML) to any available tyrosine kinase inhibitor, including imatinib mesylate,4 and the novel kinase inhibitor AMN107 (nilotinib; Novartis).5 To determine whether the T674I mutation in FIP1L1-PDGFRA causes resistance to AMN107 we used Ba/F3 cells transfected with both wild-type (wt) and the T674I mutant form of FIP1L1-PDGFRA. As expected, both inhibitors suppressed the growth of cells expressing FIP1L1-PDGFRA/wt at low nanomolar concentrations (Figure 1A). However, whereas FIP1L1-PDGFRAT674I caused a complete resistance to imatinib mesylate, AMN107 was capable of suppressing the growth of Ba/F3 cells transfected with FIP1L1-PDGFRA harboring this mutation with an inhibitory concentration at 50% (IC50) of 376 nM (Figure 1B). Also, AMN107, but not imatinib mesylate, induced apoptosis in FIP1L1-PDGFRA/T674I cells (Figure 1C). This was accompanied by suppression of FIP1L1-PDGFRA autophosphorylation in the presence of AMN107 (Figure 1D). Thus, AMN107 is active against FIP1L1-PDGFRA/T674I, a mutant that causes the resistance of HES to imatinib mesylate. This finding is in contrast to a recent report by Stover et al,6 describing resistance of FIP1L1-PDGFRA/T674I to AMN107. The conflicting findings may in part be explained by lower AMN107 concentrations used in some experiments in the report by Stover et al. It has to be emphasized that in our experiments, complete suppression of FIP1L1-PDGFRA/T674I was obtained at AMN107 concentrations that are achievable in patients where maximal and trough plasma drug concentrations of 3.6 and 1.7 μM were measured following treatment with AMN107 at 400 mg twice daily.7,8 As in patients with CML, where the corresponding mutation T315I constitutes the most frequent exchange leading to imatinib mesylate resistance, T674I may turn out to be a frequent cause of imatinib mesylate resistance in HES and is so far the only mutation identified in single cases of HES with imatinib mesylate resistance. Based on the presented data in such cases, a switch to AMN107 seems to be warranted.
FIP1L1-PDGFRA/T674I can be inhibited by AMN107 (nilotinib) but not imatinib mesylate. Growth inhibition of Ba/F3 cells expressing FIP1L1-PDGFRA/wt (A), and Ba/F3 cells expressing FIP1L1-PDGFRA/T674I (B) by imatinib mesylate and AMN107. Proliferation was measured using an MTT-based method by absorption of formazam at 490 nm. Measures were taken as triplicates after 48 hours of culture without and in the presence of inhibitor at the indicated concentrations. Values are expressed as mean values of growth inhibition from 3 independent experiments. For each experiment, measures were taken in triplicates. Bars indicate ± SE. OD indicates optical density. Induction of apoptosis by AMN107 in Ba/F3 cells expressing FIP1L1-PDGFRA/T674I (C). Annexin V–positive cells were measured after 48 hours of culture in the presence of AMN107 at the indicated concentrations. Two experiments were performed. Results of 1 representative experiment are shown. Inhibition of autophosphorylation in FIP1L1-PDGFRA/T674I transfected Ba/F3 cells (D). Cells were incubated without and in the presence of imatinib mesylate (left panel) or AMN107 (right panel) at the indicated concentrations. Cell lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Blots were probed for phosphotyrosine and PDGFRA.
FIP1L1-PDGFRA/T674I can be inhibited by AMN107 (nilotinib) but not imatinib mesylate. Growth inhibition of Ba/F3 cells expressing FIP1L1-PDGFRA/wt (A), and Ba/F3 cells expressing FIP1L1-PDGFRA/T674I (B) by imatinib mesylate and AMN107. Proliferation was measured using an MTT-based method by absorption of formazam at 490 nm. Measures were taken as triplicates after 48 hours of culture without and in the presence of inhibitor at the indicated concentrations. Values are expressed as mean values of growth inhibition from 3 independent experiments. For each experiment, measures were taken in triplicates. Bars indicate ± SE. OD indicates optical density. Induction of apoptosis by AMN107 in Ba/F3 cells expressing FIP1L1-PDGFRA/T674I (C). Annexin V–positive cells were measured after 48 hours of culture in the presence of AMN107 at the indicated concentrations. Two experiments were performed. Results of 1 representative experiment are shown. Inhibition of autophosphorylation in FIP1L1-PDGFRA/T674I transfected Ba/F3 cells (D). Cells were incubated without and in the presence of imatinib mesylate (left panel) or AMN107 (right panel) at the indicated concentrations. Cell lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Blots were probed for phosphotyrosine and PDGFRA.
The definition of resistance is in the eye of the beholder
We appreciate the observations made by Duyster et al that relate to sensitivity of clinical alleles of FIP1L1-PDGFRA associated with hypereosinophilic syndrome to inhibition with AMN107. Our cellular and biochemical experiments are generally in agreement in showing that the FIP1L1-PDGFRA T674I imatinib mesylate resistance allele is also highly resistant to inhibition with AMN107, with more than 100-fold higher IC50 than for FIP1L1-PDGFRA.1 There are several potential explanations for why we observed modest effects on cell growth and tyrosine phosphorylation content of FIP1L1-PDGFRA T674I transformed cells at 1 μM AMN107 compared with the results annotated here, but at most there may be a several fold difference in our estimates of cellular IC50.
Several other points merit consideration. First, we have previously reported that PKC412 is a potent inhibitor of the FIP1L1-PDGFRA T674I allele,2 with an IC50 of approximately 100 nM vs approximately 376 nM reported here for AMN107. From this perspective, PKC412 may be as appropriate an alternative for FIP1L1-PDGFRA patients who develop resistance due to acquired T674I as AMN107. In this context, it is also worth noting the importance of including controls for nonspecific or off-target toxicities for tyrosine kinase inhibitors such as AMN107 or PKC412, as they are selective rather than specific. Parental Ba/F3 cells would have been 1 such control in these experiments, but in our view the most compelling controls for nonspecific toxicity or off-target effects are kinase mutations that are selected for resistance to the kinase inhibitor in question. For example, we generated a PKC412 resistance allele in the context of FIP1L1-PDGFRA with an N659D substitution.2 We observed that cells stably transduced with FIP1L1-PDGFRA N659D were resistant to the effects of PKC412, establishing FIP1L1-PDGFRA as the critical target for cellular cytotoxicity mediated by PKC412 in this context, rather than off-target or nonspecific effects. Indeed, the finding of the FIP1L1-PDGFRA T674I imatinib mesylate resistance mutation in a patient with clinically resistant HES provides the most compelling evidence that FIP1L1-PDGFRA causes HES and is the target of imatinib mesylate. It would be useful to have comparable controls for AMN107 as well, although it seems most likely that the cytotoxic effects of AMN107 treatment are attributable to inhibition of the T674I allele.
Second, we completely agree that relative resistance of FIP1L1-PDGFRA T674I to AMN107 does not exclude potential clinical applicability and efficacy if adequate plasma concentrations can be achieved. It may be of value to consider either AMN107 or PKC412 as second-line agents. For the time being, the point is moot, as neither of these drugs is Food and Drug Administration (FDA) approved for any indication, but could potentially be accessed through compassionate use mechanisms.
It may be, as suggested by the authors, that FIP1L1-PDGFRA T674I will emerge as an important clinical problem in treatment of HES, but so far it has not. Over the last several years increasing numbers of patients with HES have been treated with imatinib mesylate, but there are only 2 reported cases of resistance to imatinib mesylate.3,4 Both of these developed in the context of acute eosinophilic leukemias rather than in HES. It is certainly possible that more cases will ultimately surface, and proactive development of inhibitors such as PKC412 and AMN107 that address this problem seems a prudent course of action. But it is possible that these will be no more effective in the setting of acute eosinophilic leukemia than imatinib mesylate is in the context of CML in blast crisis.
However, it is also possible that resistance will develop less frequently in FIP1L1-PDGFRA–positive HES due to the remarkable potency of imatinib mesylate in this context. Imatinib mesylate is several hundred–fold more potent as a FIP1L1-PDGFRA inhibitor than as a BCR-ABL inhibitor. Only time will tell, but one might hope that highly potent kinase inhibitors such as imatinib mesylate in the context of FIP1L1-PDGFRA, or AMN107 or dasatinib in the context of BCR-ABL–positive disease, will more effectively suppress emergence of resistant clones due to point mutations in the target kinases, than lower potency inhibitors.
Correspondence: D. Gary Gilliland, Brigham and Women's Hospital, Division of Hematology/Oncology, Karp Family Research Building, 1 Blackfan Circle, 5th Floor, Boston, MA 02115; e-mail: ggilliland@rics.bwh.harvard.edu.