Abstract
Childhood acute lymphoblastic leukemia (ALL) consists of various subtypes that respond differently to cytotoxic drugs and therefore have a markedly different clinical outcome. We used microarrays to investigate, in 190 children with ALL at initial diagnosis, whether 70 key apoptosis genes were differentially expressed between leukemic subgroups defined by lineage, genetic subtype, in vitro drug resistance, and clinical outcome. The expression of 44 of 70 genes was significantly different in T-versus B-lineage ALL, 22 genes differed in hyperdiploid versus nonhyperdiploid, 16 in TEL-AML1-positive versus-negative, and 13 in E2A-rearranged versus germ-line B-lineage ALL. Expression of MCL1 and DAPK1 was significantly associated with prednisolone sensitivity, whereas BCL2L13, HRK, and TNF were related to L-asparaginase resistance. BCL2L13 overexpression was also associated with unfavorable clinical outcome (P < .001). Multivariate analysis including known risk factors revealed that BCL2L13 expression was an independent prognostic factor (P = .011).
The same trend was observed in a validation group of 92 children with ALL treated on a different protocol at St Jude (P = .051). In conclusion, ALL subtypes have a unique expression pattern of apoptosis genes and our data suggest that selective genes are linked to cellular drug resistance and prognosis in childhood B-lineage ALL. (Blood. 2006; 107:769-776)
Introduction
The treatment of pediatric acute lymphoblastic leukemia (ALL) has greatly improved over the past 3 decades, resulting in long-term disease-free survival (DFS) of approximately 80%.1 Despite this progress, therapy resistance still forms a major obstacle to successful treatment in a significant number of children. Childhood ALL is a heterogeneous disease consisting of various genetic subtypes such as t(9;22)/BCR-ABL, t(12;21)/TEL-AML1, hyperdiploid (> 50 chromosomes), 11q23/MLL rearranged, t(1;19)/E2A-PBX1, and T-lineage ALL, which differ markedly in their treatment response.2 The in vitro response to chemotherapy can be studied by exposure of primary patient samples to cytostatic drugs in a cell-kill assay such as the methyl-thiazol-tetrazolium (MTT) assay. We and others have previously demonstrated that children with ALL whose leukemia cells exhibit in vitro resistance to single drugs or a combination of drugs (ie, prednisolone, vincristine, and L-asparaginase [PVA]) have a significantly worse prognosis than patients with sensitive leukemic cells.3-7 In addition, leukemia subtypes with a relatively unfavorable prognosis have been associated with in vitro drug resistance8-10 and subtypes with a favorable prognosis with in vitro drug sensitivity.11,12
Apoptosis is the predominant form of cell death triggered in vivo and in vitro by drugs in hematologic malignancies.13 There are 2 major routes by which apoptosis can be induced: (1) the mitochondrial or intrinsic apoptosis pathway; and (2) the death receptor-mediated or extrinsic apoptosis pathway. Both apoptotic pathways have been extensively reviewed elsewhere.14-17 Briefly, the intrinsic route is initiated by mitochondrial damage that leads to release of apoptogenic factors, such as cytochrome c, Smac/Diablo, and apoptosis-inducing factor (AIF), from the mitochondrial intermembrane space.17 The release of these factors is mediated by Bcl-2 family proteins, a group of key regulators of the intrinsic apoptosis pathway that consists of proapoptotic and antiapoptotic members. Upon its release into the cytoplasm, cytochrome c forms a complex known as the apoptosome consisting of apoptotic protease-activating factor-1 (Apaf-1), ATP/dATP, and procaspase-9.18 Following its activation within the apoptosome, caspase-9 activates the downstream effector caspase cascade.19 Initiation of the extrinsic apoptosis pathway involves ligand-induced aggregation of death receptors and activation of procaspase-8 or procaspase-10 within the death-inducing signaling complex (DISC).20,21 Activated procaspase-8 or -10 is released into the cytoplasm, where it induces activation of downstream effector caspases. The intrinsic and extrinsic apoptotic pathways converge at the level of caspase-3 activation.
Leukemia subtypes with a relatively unfavorable prognosis have been associated with in vitro drug resistance.8-10 Moreover, cellular drug resistance is associated with decreased ability to induce apoptosis in pediatric ALL.22 Therefore, one of the factors that may contribute to the different treatment response of genetic leukemia subtypes may be a differential propensity to undergo apoptosis. Apoptosis is controlled by various positive and negative regulators, responding to stimuli from inside and outside the cell.23,24 Most papers to date addressing causes of cellular drug resistance, however, only focus on a limited number of apoptosis molecules. In the present study we analyzed the expression patterns of 70 key apoptosis genes in leukemic cells of 190 children at initial diagnosis of ALL. The expression of these genes was tested for association with (1) lineage and genetic subtype, (2) in vitro drug resistance to 4 widely used drugs in treatment of ALL (ie, prednisolone, vincristine, L-asparaginase, and daunorubicin), and (3) clinical outcome. Last, we analyzed the relation between the expression of active Apaf-1 isoforms and cellular drug resistance.
Patients, materials, and methods
Patient samples
Bone marrow (BM) and peripheral blood (PB) were obtained after informed consent from 190 children with newly diagnosed ALL who were enrolled on treatment protocols 92 and 97 at the hospitals participating in the German Cooperative Study Group for Acute Lymphoblastic Leukemia (COALL) study or the ALL-9 Dutch Childhood Oncology Group (DCOG) protocol at the Erasmus MC-Sophia Children's Hospital in Rotterdam (study cohort); and of 92 children enrolled as part of the Total Therapy protocols 13A25 and 13B26 of St Jude Children's Research Hospital (SJCRH) in Memphis, TN (validation cohort).27 Approval was obtained from the Erasmus MC-Sophia Children's Hospital and SJCRH institutional review board for these studies. Clinical characteristics of these patients are provided in Table 1.
Isolation of leukemia cells
Mononuclear cells were isolated by sucrose density gradient centrifugation (Lymphoprep, density 1.077 g/mL; Nycomed Pharma, Oslo, Norway) within 24 hours after sampling. Cells were resuspended in culture medium consisting of RPMI 1640 (Dutch modification without L-glutamine; Gibco BRL, Life Technologies, Breda, the Netherlands) supplemented with 20% fetal calf serum (FCS; Integro, Zaandam, the Netherlands), 2 mM L-glutamine, 200 μg/mL gentamycin (Gibco BRL), 100 IU/mL penicillin, 100 μg/mL streptomycin, 0.125 μg/mL fungizone (Gibco BRL), and 5 μg/mL insulin, 5 μg/mL transferrin, and 5 ng/mL sodium selenite (ITS media supplement; Sigma-Aldrich Chemie, Zwijndrecht, the Netherlands). If necessary, leukemic samples were further enriched to more than 90% leukemic blasts by removing nonmalignant cells with immunomagnetic beads (DynaBeads; Dynal, Oslo, Norway).28
In vitro drug-resistance assay
Responsiveness of leukemia cells to prednisolone (PRED; Bufa Pharmaceutical Products, Uitgeest, the Netherlands), vincristine (VCR; TEVA Pharma, Mijdrecht, the Netherlands), L-asparaginase (ASP; Paronal, Christiaens, Breda, the Netherlands), and daunorubicin (DNR; Cerubidine, Rhône-Poulenc Rorer, Amstelveen, the Netherlands) was determined by the 4-day in vitro MTT drug resistance assay.3 The concentration ranges tested for these drugs were PRED, 0.008-250 μg/mL; VCR, 0.05-50 μg/mL; ASP, 0.003-10 IU/mL; and DNR, 0.002-2.0 μg/mL. The drug concentration lethal to 50% of the ALL cells (LC50 value) was used as the measure of cellular drug resistance. The cut-off LC50 values, used to assign cases as sensitive or resistant to each agent, were those previously shown to be associated with a good or poor treatment outcome in children with ALL.3,4
Gene expression profiling: purification, labeling, and hybridization of RNA
Total cellular RNA was extracted from leukemic cells of 190 patients with acute lymphoblastic leukemia and hybridized to the U133A GeneChip oligonucleotide microarray containing 22 283 probe sets (∼12 700 genes) according to manufacturer's protocols (Affymetrix, Santa Clara, CA). Gene expression values were scaled to the target intensity of 2500 using Affymetrix Microarray Analysis Suite (MAS) 5.0 software.29,30 Probe sets expressed in fewer than 5 patients were omitted, leaving 14 550 probe sets in the filtered dataset for subsequent analyses. Gene expression analysis of 173 out of these 190 patients was published previously;29 in this paper, we focus solely on genes involved in apoptosis. All analyses were carried out on log2-transformed gene-expression values.
Real-time quantitative PCR
Total cellular mRNA was extracted using Trizol reagent (Gibco BRL) and cDNA was synthesized using random hexamers and oligo dT. mRNA expression levels of total Apaf-1, “active” Apaf-1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a reference were quantified using real-time quantitative (RTQ) polymerase chain reaction (PCR) analysis on a ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA) as previously described.31,32 The comparative cycle time (Ct) value of the target PCR was normalized by subtracting the Ct value of GAPDH (ΔCt). The ΔCt value was used to calculate the relative expression level to GAPDH for each target PCR using the following formula: relative mRNA expression = 2-ΔCt × 100%.33 Primer sequences used were as follows: upper, 5′-ACCAGC CGCATACTCTT-3; lower, 5′-CAGGGCCTACAAGTTCTG-3′ (total Apaf-1); upper, 5′-GGACCCTCAAGAGGATATG-3′; lower, 5′-GTGGGGAGAAGT CACAGTAC-3′ (“active” Apaf-1); and upper, 5′-GTCGGAGTCAACGGATT-3′; lower, 5′-AAGCTTCCCGTTCTCAG-3′ (GAPDH). Probe sequences were 5′-CACATGGCCAGTGCCAAGAT-3′ (total Apaf-1); 5′-AAGTGTTGTTCGTGGTCTGCTGAT-3′ (“active” Apaf-1); and 5′-TCAACTACATGGTTTACATGT TCCAA-3′ (GAPDH).
Statistical analysis
A selection of genes with known involvement in apoptosis was made by a search using GenMaPP version 2.0 and in literature (Gladstone Institutes, University of California, San Francisco, CA). Corresponding probe sets were retrieved using Affymetrix NetAffx (http://www.affymetrix.com/analysis/index.affx). From the total of 179 selected probe sets, 118 were present in the filtered dataset corresponding to 70 apoptosis genes (intrinsic pathway, 40; extrinsic pathway, 30) for subsequent analysis.
We applied the global test34 to identify those probe sets that are simultaneously differentially expressed between different subgroups defined by lineage, genetic subtype, in vitro drug resistance, and clinical outcome. Briefly, the global test compares 2 or more groups taking into account the association between probe sets as well as their individual effects.34 The advantage of the global test is that it is applied to the entire set of probe sets under study at the same time, yielding a single overall P value, rather than on individual probe sets consecutively. Thus, there are no multiple testing issues associated with the global test. In addition, the global test can be applied to multiple probe sets encoding one gene, since this test investigates the influence of each single probe set on the discrimination between the 2 studied groups. One of the outputs of this test is a so-called gene plot, which displays the individual influences of the probe sets on the test result. The gene plot was used to select those probe sets that were most strongly explaining the difference between 2 subgroups.
In addition, we applied the Wilcoxon rank-sum test to each probe set to identify those probe sets that were individually associated with the subgroups. P values were corrected for multiple testing using the false discovery rate (FDR) step-up procedure proposed by Benjamini and Hochberg.35 The global test has more power to detect differential expression when dealing with multiple probe sets with small effects, compared with tests applied probe set-wise, such as the Wilcoxon rank-sum test. The output of the global test and the FDR-corrected Wilcoxon rank-sum test is summarized in Figures 1 and 2 and Table 2.
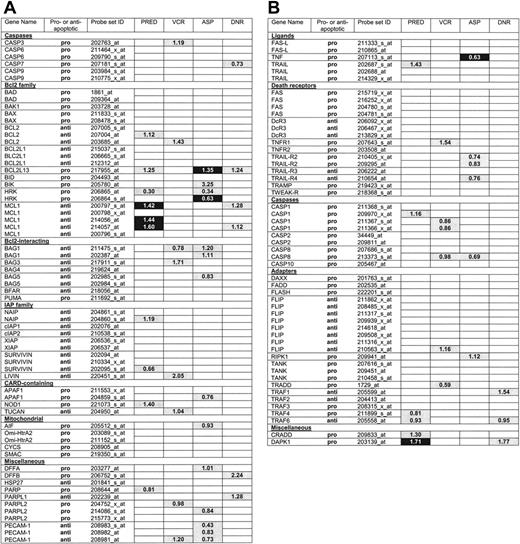
Differential expression of apoptosis genes between ALL subtypes. The global test and Wilcoxon rank-sum test were performed to identify which of the indicated 118 apoptosis probe sets were differentially expressed in various ALL subgroups defined by T-versus B-lineage ALL (lineage), or genetic subtype (ie, TEL-AML1 status [TA], ploidy, and E2A status [E2A]). For each subgroup, probe sets selected only by the global test (P <.001) are light gray, probe sets selected only by Wilcoxon rank-sum test with false discovery rate (FDR) less than 5% are dark gray, probe sets selected by the global test (P <.001) and Wilcoxon rank-sum test with FDR are black, and probe sets selected by none of these tests are white. The numbers indicated in colored boxes are the ratio per significant gene between T-lineage ALL and B-lineage ALL samples (Lineage), hyperdiploid and nonhyperdiploid B-lineage ALL samples (Ploidy), TEL-AML1-positive and TEL-AML1-negative B-lineage ALL samples (TA), and E2A-rearranged and E2A-germline B-lineage ALL samples (E2A).
Differential expression of apoptosis genes between ALL subtypes. The global test and Wilcoxon rank-sum test were performed to identify which of the indicated 118 apoptosis probe sets were differentially expressed in various ALL subgroups defined by T-versus B-lineage ALL (lineage), or genetic subtype (ie, TEL-AML1 status [TA], ploidy, and E2A status [E2A]). For each subgroup, probe sets selected only by the global test (P <.001) are light gray, probe sets selected only by Wilcoxon rank-sum test with false discovery rate (FDR) less than 5% are dark gray, probe sets selected by the global test (P <.001) and Wilcoxon rank-sum test with FDR are black, and probe sets selected by none of these tests are white. The numbers indicated in colored boxes are the ratio per significant gene between T-lineage ALL and B-lineage ALL samples (Lineage), hyperdiploid and nonhyperdiploid B-lineage ALL samples (Ploidy), TEL-AML1-positive and TEL-AML1-negative B-lineage ALL samples (TA), and E2A-rearranged and E2A-germline B-lineage ALL samples (E2A).
Apoptosis genes associated with resistance to four individual chemotherapeutic agents in B-lineage ALL. The global test and Wilcoxon rank-sum test were performed to identify which of the indicated 118 apoptosis probe sets were differentially expressed in B-lineage ALL cells sensitive and resistant to prednisolone (PRED), vincristine (VCR), L-asparaginase (ASP), and daunorubicin (DNR). For each drug, probe sets selected only by the global test (P <.001) are light gray, probe sets selected by the global test (P < .001) and Wilcoxon rank-sum test with FDR < 5% are black, and probe sets not selected by none of the tests are white. The numbers indicated in colored boxes are the ratio per significant gene between PRED-resistant and PRED-sensitive ALL samples (PRED), VCR-resistant and VCR-sensitive ALL samples (VCR), ASP-resistant and ASP-sensitive ALL samples (ASP), and DNR-resistant and DNR-sensitive ALL samples (DNR).
Apoptosis genes associated with resistance to four individual chemotherapeutic agents in B-lineage ALL. The global test and Wilcoxon rank-sum test were performed to identify which of the indicated 118 apoptosis probe sets were differentially expressed in B-lineage ALL cells sensitive and resistant to prednisolone (PRED), vincristine (VCR), L-asparaginase (ASP), and daunorubicin (DNR). For each drug, probe sets selected only by the global test (P <.001) are light gray, probe sets selected by the global test (P < .001) and Wilcoxon rank-sum test with FDR < 5% are black, and probe sets not selected by none of the tests are white. The numbers indicated in colored boxes are the ratio per significant gene between PRED-resistant and PRED-sensitive ALL samples (PRED), VCR-resistant and VCR-sensitive ALL samples (VCR), ASP-resistant and ASP-sensitive ALL samples (ASP), and DNR-resistant and DNR-sensitive ALL samples (DNR).
The duration of disease-free survival (DFS) was defined as the time from diagnosis until the date of leukemia relapse (event) or the last follow-up (censored). Univariate analysis using Cox proportional hazard regression models estimated the relative risk of an event. Significant probe sets from the univariate analysis were entered in a multivariate analysis using Cox proportional hazards regression model, which included known risk factors white blood cell count (WBC), age, lineage, and genetic subtype. DFS curves were calculated by reversing the cumulative incidence curve.37 Presence of competing events were accounted for in comparisons of DFS curves,36,38 and in multivariate analysis.37
The Wilcoxon rank-sum test was applied to compare Apaf-1 isoform mRNA expression in sensitive and resistant patients for each individual drug.
Results
Apoptosis-related genes and immunophenotypic and genetic subtypes of pediatric ALL
The expression of 118 probe sets corresponding to 70 apoptosis-associated genes was compared between various leukemic subgroups (ie, T-lineage and B-lineage ALL [lineage]); hyperdiploid (ie, more than 50 chromosomes present at cytogenetic analysis) and non-hyperdiploid B-lineage ALL patients (ploidy); TEL-AML1-positive and -negative B-lineage ALL patients (TA); and E2A-rearranged and E2A germ-line B-lineage ALL patients (E2A). The global test applied to all 118 probe sets generated P values less than .001 for lineage, ploidy, TA, and E2A. Gene plots that visualize influences for individual probe sets on the global test P value are indicated in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Probe sets were selected that had an influence on the global test P value of larger than 2 standard deviations above the expected value under the null hypothesis of no association (Figure 1). Figure 1 also includes the probe sets that were selected by Wilcoxon rank-sum test after correction for multiple testing (FDR controlled at 5%). Sixty-six probe sets (44 different genes) were differentially expressed in T-lineage versus B-lineage ALL and selected by both tests (indicated in black in Figure 1), 31 probe sets (22 different genes), genes in hyperdiploid versus nonhyperdiploid B-lineage ALL, 20 probe sets (16 different genes) in TEL-AML1-positive versus-negative B-lineageALL, and 15 probe sets (13 different genes) in E2A-rearranged versus E2A germ-line B-lineage ALL. The probe-set identification, gene names, and median expression of all probe sets are listed in Table S1.
Apoptosis-related genes and cellular drug resistance in pediatric B-lineage ALL
Due to the large difference in the expression of apoptosis genes between T- and B-lineage ALL (P < .001, global test) and the limited number of T-lineage ALL patients, differences in expression of apoptosis genes between drug-sensitive and -resistant patients was only addressed in the B-lineage ALL group. The global test generated significant P values for prednisolone (16 probe sets corresponding to 14 different genes, P = .007) vincristine (14 probe sets corresponding to 13 different genes, P = .002), and L-asparaginase (20 probe sets corresponding to 15 different genes, P < .001), but not for daunorubicin (Figure 2). The probe sets most strongly associated with resistance to individual drugs in the global test and the Wilcoxon rank-sum test (FDR controlled at 5%) are indicated in black in Figure 2. While no probe sets were associated with resistance to vincristine or daunorubicin, 4 probe sets (corresponding to 2 genes, MCL1 and DAPK1) and 3 probe sets (BCL2L13, HRK, and TNF) were significantly associated with resistance in both tests toward prednisolone or L-asparaginase, respectively. Gene plots for each drug are shown in Figure S2 and probe-set identification, gene names, and median expression are shown for each drug in Table S2.
The expression of apoptosis-related genes and clinical outcome in pediatric ALL
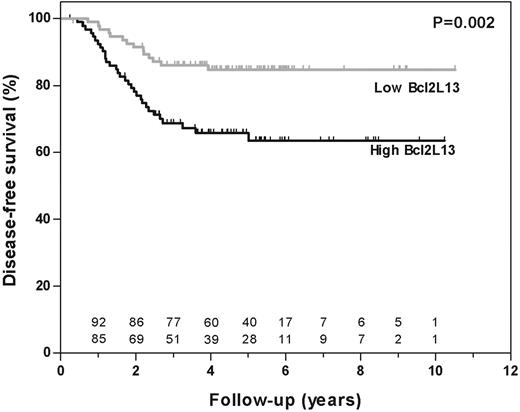
From the 190 patients included in this study (median follow-up at risk of event, 4.8 years; range, 0.3-10.5 years), 45 had disease-related events and 2 had a competing event, which was censored at the time of occurrence. Apoptosis gene expression profiles measured at initial diagnosis were compared between patients who entered and remained in continuous complete remission (CCR) and those who relapsed during follow-up. Gene plots for each drug are shown in Figure S3 and probe-set identification, gene names and median expression are shown for each drug in Table S3. Seventeen probe sets (13 genes) influenced the global test P value by more than 2 standard deviations. Out of these 17 probe sets, 4 probe sets (3 genes) were also selected by the univariate Cox regression analysis with FDR controlled at 5% (marked with an asterisk in Table 2). Subsequently, each of these 4 significant probe sets was analyzed in a multivariate Cox regression analysis with inclusion of conventional risk criteria (ie, age, white blood cell count, lineage, and genetic subtype [Tables 3-4]). BCL2L13 was the only gene that was independently and significantly associated with treatment outcome (P = .011; Table 3). BCL2L13 expression was significantly associated with treatment outcome when used as continuous variable (P < .001) and when divided into 2 equally sized groups (P = .002; Figure 3). The 5-year probability of disease-free survival (pDFS) plus or minus SE was 85% ± 5.2% for patients with low (below median) and 66% ± 7.3% for patients with high (above median) expression of BCL2L13.
Disease-free survival according to BCL2L13 expression in pediatric ALL. The disease-free survival (DFS) of patients was estimated according to a Kaplan-Meier plot among 190 patients of the COALL/DCOG study cohort. Patients were grouped according to their expression of BCL2L13 (ie, expression higher than (black line) or lower than (gray line) the median).
Disease-free survival according to BCL2L13 expression in pediatric ALL. The disease-free survival (DFS) of patients was estimated according to a Kaplan-Meier plot among 190 patients of the COALL/DCOG study cohort. Patients were grouped according to their expression of BCL2L13 (ie, expression higher than (black line) or lower than (gray line) the median).
For 92 patients enrolled at St Jude the median follow-up was 6.2 years. Of these patients, 15 had disease-related events and 8 had a competing event, which was censored at the time of occurrence. In this independent cohort treated with the same chemotherapeutic agents but on a different protocol at the St Jude Children's Research Hospital the association between BCL2L13 expression and outcome was significant in a univariate analysis when treated as a continuous variable (P = .025), but not significant when patients below and above the median were compared (P = .28). In a multivariate Cox analysis including the above-mentioned known risk factors BCL2L13 expression showed the same trend for an association with outcome in patients treated according to St Jude protocols (P = .051; Table 4).
Expression of Apaf-1 isoforms and cellular drug resistance in pediatric ALL
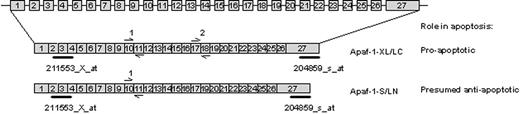
The expression of Apaf-1 splice variants has been linked to functional apoptosis in tumor cell lines.39-41 The presence of an additional C-terminal WD-40 repeat encoded by exon 18 appears to be required for in vitro activation of procaspase-9 and -3. The Affymetrix probe sets are unable to distinguish between the individual isoforms of Apaf-1 (Figure 4). To investigate whether the relative expression of proapoptotic (active) Apaf-1 isoforms (ie, the isoforms containing exon 18) is linked to sensitivity to antileukemic agents, real-time quantitative PCR was carried out in 36 children with ALL at initial diagnosis. Two primer pairs were used: one pair recognizes both pro- and antiapoptotic Apaf-1 isoforms, and one pair hybridizes to exon 18 and is thus specific for the proapoptotic isoform of Apaf-1 (Figure 4). The relative expression of proapoptotic Apaf-1 isoform ranged between 2% and 69% of total Apaf-1 and did not differ significantly in patients sensitive and resistant to prednisolone (P = .74), vincristine (P = .33), L-asparaginase (P = .79), or daunorubicin (P = .95). In addition, the absolute expression of the proapoptotic isoform did not differ significantly in patients sensitive and resistant to prednisolone (P = .96), vincristine (P = .20), L-asparaginase (P = .25), or daunorubicin (P = .67).
Quantification of the expression of Apaf-1 isoforms in pediatric ALL. The structure of the Apaf-1 gene and 2 Apaf-1 transcript variants. Indicated is the location of the Affymetrix probe sets (211553_x_at and 204859_s_at) and the Taqman primer pairs (1 and 2).
Quantification of the expression of Apaf-1 isoforms in pediatric ALL. The structure of the Apaf-1 gene and 2 Apaf-1 transcript variants. Indicated is the location of the Affymetrix probe sets (211553_x_at and 204859_s_at) and the Taqman primer pairs (1 and 2).
Discussion
Leukemic subtypes with an unfavorable prognosis may have a decreased tendency to undergo apoptosis compared with subtypes with a favorable prognosis. Gene expression signatures discriminative for lineage,27,42 genetic subtype,27,42 in vitro,29 and in vivo29,42 drug response were previously reported. Among the discriminative genes identified in these studies were virtually no apoptosis genes. This does not rule out a role for apoptosis genes in these leukemic subtypes per se, because these genes may be significant at a lower level than the cut-off P values used for the construction of these signature models. Therefore, we analyzed the expression patterns of 70 selected key apoptotic genes in leukemic cells of 190 children at initial diagnosis of ALL and correlated the expression of these genes to lineage, genetic subtype, in vitro drug resistance, and clinical outcome.
Children with T-lineage ALL have an increased risk of treatment failures compared with children with B-lineage ALL,43 which can be attributed to the presence of numerous adverse presenting features, such as older age, high white blood cell count, and in vitro resistance to a variety of drugs.8,44 However, intensification of treatment regimens has resulted in remarkably improved outcomes for children with T-ALL.45 Although T-ALL has been associated with aberrant expression of some apoptosis genes,46,47 the underlying causes of in vitro drug resistance have not yet been fully determined. Global test analysis indicated that the expression of apoptosis genes differs between T-lineage and B-lineage ALL (P < .001). A large number of apoptosis genes (44 of 70 examined genes) were most discriminative between T-lineage and B-lineage ALL (as defined by ≥ 2 SD influence on the global test P value; “Patients, materials, and methods”).
The death-receptor Fas has been linked to apoptosis and nuclear factor-κB (NF-κB)-related inflammatory response pathways.48 Activation of NF-κB inhibits drug-induced apoptosis in various cell-line studies and was shown to be linked to drug resistance in childhood ALL.49-51 Interestingly, we observed simultaneous upregulation of Fas and its downstream effectors (ie, Fas-associated death domain [FADD], caspase-8, and caspase-10) in T-compared with B-lineage ALL and up-regulation of several NF-κB target genes, such as cIAP1, cIAP2, survivin, and FLIP. The relative high expression of NF-κB-associated genes may point to enhanced NF-κB activity in T-lineage compared with B-lineage ALL.
Children with hyperdiploid and TEL-AML1-positive ALL have a favorable prognosis, which is associated with a relatively high in vitro sensitivity to various drugs, including L-asparaginase.11,12 Interestingly, the TNF receptor ligand (TNF) is expressed higher in hyperdiploid and TEL-AML1-positive B-lineage ALL patients (Figure 1). Moreover, TNF is 0.6-fold less expressed in L-asparaginase-sensitive cases (Figure 2). Since both hyperdiploid and TEL-AML1-positive B-lineage ALL are in vitro sensitive to L-asparaginase,11,12 these data point to novel insights in the apoptotic changes underlying L-asparaginase cytotoxicity. Another notable feature of hyperdiploid B-lineage ALL cells is the simultaneous overexpression of TNF-R1, TRAIL-R2, and TRAIL-R4. The overexpression of these cytokine receptors was not previously observed in hyperdiploid B-lineage ALL but may contribute to their marked apoptotic propensity in allogeneic bone marrow-derived stromal layers that contain the microenvironment to trigger these receptors.52
The relation between the expression of apoptosis genes and in vitro53-55 or in vivo response56-59 has been extensively studied in ALL. However, these studies each focused on the expression of only a few genes out of the large family of apoptosis-related genes. In this study, analysis of 70 key apoptotic genes revealed that only 2 and 3 genes were significantly associated with resistance toward prednisolone and L-asparaginase, respectively. Bcl-2 family members are thought of as the central regulators of apoptosis by regulating cytochrome c release upstream of the mitochondria.60 We observed increased expression of the antiapoptotic Bcl-2 family member MCL1 in prednisolone-resistant B-lineage ALL cells and decreased expression of the proapoptotic Bcl-2 family member HRK in L-asparaginase-resistant B-lineage ALL cells. The differential expression of these Bcl-2 family members may contribute to the apoptotic blockage we previously observed upstream of the mitochondria in prednisolone- and L-asparaginase-resistant ALL cells.22 The fact that we observed decreased apoptosis in vincristine- and daunorubicin-resistant ALL cells in the former study,22 and that no apoptosis gene was associated with vincristine and daunorubicin resistance in the present study, suggests that resistance to these drugs is caused by mechanisms that do not appear transcriptionally. Alternatively, VCR and DNR resistance in childhood ALL may be caused by a defect further upstream of the mitochondria. Aberrant expression and function of cytoskeleton-associated genes29,61 and lack of ceramide generation62 are examples of upstream defects that were previously observed in leukemic samples resistant to VCR and DNR, respectively.
BCL2L13 (Bcl-rambo) is a recently discovered member of the Bcl-2 family with proapoptotic activity.63,64 Remarkably, however, in this study we observed that a high mRNA expression of BCL2L13 was associated with in vitro L-asparaginase resistance and an unfavorable long-term clinical outcome in children with ALL. This finding suggests BCL2L13 may have a different apoptotic role in primary leukemic cells of children compared with the cell lines used to describe its apoptotic role. Alternative splicing is known to generate both anti- and proapoptotic variants of a single apoptosis gene (eg, Apaf-1; Figure 4).40,65,66 Therefore, an alternative explanation for our finding may be the existence of a previously unrecognized antiapoptotic splice variant. Probe sets designed by Affymetrix are (in general) not suitable to recognize differential expression of splice variants of a single gene. Most importantly, high expression of the BCL2L13 probe set was associated with resistance toward L-asparaginase and independently linked to an unfavorable prognosis compared with other known risk factors. Since BCL2L13 expression was also associated with an inferior outcome in a second (differently treated) validation cohort, this gene may represent a new risk factor in childhood ALL. The fact that only one of the 70 apoptosis genes was independently associated with treatment outcome in this study suggests that treatment outcome in childhood ALL is largely dependent on genes involved in other pathways than the apoptosis pathway. This notion is supported by the absence of apoptosis genes among the genes previously associated with treatment response in several studies in diagnostic childhood ALL samples.42,67,68
In conclusion, this study is the first to describe an association between the differential expression of key apoptosis genes and lineage, genetic subtype, and in vitro drug resistance in children with ALL. In addition, we identified a single gene, BCL2L13, which is related to both L-asparaginase resistance and treatment outcome independent from known prognostic factors in 2 independent cohorts of children with B-lineage ALL. To establish BCL2L13 expression as a true prognostic factor in childhood ALL, prospective validation is required. Also, the currently identified genes warrant further studies on expression and function at the protein level to further increase our insight in the causes of drug resistance and therapy failure in pediatric ALL. It was recently demonstrated that inhibition of Mcl-1 by the cyclin-dependent kinase (CDK) inhibitor seliciclib induced significant cytotoxicity in multiple myeloma cells sensitive and resistant to conventional therapy.69 In addition, depletion of Mcl-1 levels by antisense Mcl-1 oligonucleotides sensitized lung cancer cells to apoptosis induced by cytotoxic agents as well as by ionizing radiation.70 Likewise, it can be hypothesized that down-regulation of Bcl2l13 by antisense oligonucleotides or specific inhibitors may sensitize ALL cells to L-asparaginase and eventually other drugs.
Prepublished online as Blood First Edition Paper, September 27, 2005; DOI 10.1182/blood-2005-07-2930.
Supported by grants from the Pediatric Oncology Foundation Rotterdam, the Nijbakker-Morra Foundation, and the René Vogels Stipendium 2002, the Netherlands, and supported in part by grants from the National Institutes of Health and the American Lebanese Syrian Associated Charities.
W.E.E. and R.P. contributed equally to this study.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
We thank the medical staff and the patients participating in these studies at the centers of the Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia (COALL), the Erasmus MC-Sophia Children's Hospital, and St Jude Children's Research Hospital. The authors acknowledge the outstanding biostatistical assistance of Deiqing Pei.

![Figure 1. Differential expression of apoptosis genes between ALL subtypes. The global test and Wilcoxon rank-sum test were performed to identify which of the indicated 118 apoptosis probe sets were differentially expressed in various ALL subgroups defined by T-versus B-lineage ALL (lineage), or genetic subtype (ie, TEL-AML1 status [TA], ploidy, and E2A status [E2A]). For each subgroup, probe sets selected only by the global test (P <.001) are light gray, probe sets selected only by Wilcoxon rank-sum test with false discovery rate (FDR) less than 5% are dark gray, probe sets selected by the global test (P <.001) and Wilcoxon rank-sum test with FDR are black, and probe sets selected by none of these tests are white. The numbers indicated in colored boxes are the ratio per significant gene between T-lineage ALL and B-lineage ALL samples (Lineage), hyperdiploid and nonhyperdiploid B-lineage ALL samples (Ploidy), TEL-AML1-positive and TEL-AML1-negative B-lineage ALL samples (TA), and E2A-rearranged and E2A-germline B-lineage ALL samples (E2A).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-07-2930/4/m_zh80020689700001.jpeg?Expires=1770495169&Signature=Ny8o3ZBBBzEVFL6Fvn1huK0m420hTZQcyNSSC60oCxfLRXO1dSzVtTyW02qjnQr3YfL53~4yAtifx25xqgKsc24eVYKm6enu9QUEmGzLH9-1H6Ukoh0Xf3TEaLjGvHqxPIXX9DCauBBLlL3iEmHdYYn~H4~ASeQBQ-0kIN2TTnItseenrfgoceIQtP~tqnbiS15Qu-S9OrZ~fN-RZb8LodVzqiagRYV2nCF0PguUmf~2hJwEzfT8AZaaMPyBzuxW5-vj8DN447PelZoW--vXcFY5866zqfxc0LaOjC0W2h0T8v9rinuND2T9bWbPYXrIqkazWS7al8zULsdl-69brA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)