A treatment strategy that combines arsenic trioxide (ATO) with the tyrosine kinase inhibitor imatinib mesylate (STI571, Gleevec) appears to induce markedly more cell apoptosis than imatinib mesylate alone in chronic myeloid leukemia (CML). To understand the mechanisms underlying the synergistic/additive action of these agents, we applied cDNA microarrays, component plane presentation integrated self-organizing map (CPP-SOM), and methods of protein biochemistry to study cell apoptosis induced by imatinib mesylate, ATO, and the combination of the 2 agents in the CML cell line K562. Numerous features with temporospatial relationships were revealed, indicating the coordinated regulation of molecular networks from various aspects of proapoptotic and apoptotic activities in CML. Imatinib mesylate appears to induce mainly the intrinsic pathway of cell apoptosis, whereas ATO induces the endoplasmic reticulum (ER) stress-mediated pathway of cell apoptosis, and the combination of the 2 agents seems to more effectively induce the intrinsic, extrinsic, and ER stress-mediated pathways of cell apoptosis, which results in a more effective and efficient induction of programmed cell death in K562 cells. This finding appears to be supported also by data derived from bone marrow cells of 2 patients with CML, one in chronic phase and the other in blast-crisis phase of the disease.
Introduction
Advances in molecular pathogenesis have facilitated the development of therapeutic strategies targeted to molecular events critical for human malignancies. This is represented by the treatment of chronic myeloid leukemia (CML) with imatinib mesylate (STI571), a specifically designed inhibitor that targets the tyrosine kinase activity of the BCR-ABL protein and consequently induces apoptosis in vitro as well as in vivo in CML cells.1-6 Recent clinical trials in the chronic phase of CML have also demonstrated the remarkable efficacy of this molecularly targeted agent to patients with CML.7 However, a significant proportion of the treated patients with previously failed experiences of interferon therapy remained predominantly BCR-ABL+, suggesting a risk of later relapse.8 Furthermore, patients in the accelerated and blast-crisis phase revealed a high frequency of relapse or resistance to imatinib mesylate.9-11 As a result, much interest is now focused on the development of combination therapies to improve response rates and prevent resistance or relapse.12 Arsenic, the oldest and also the newest form of antileukemia drug, may promote apoptosis and exert anti-CML effects.13 A treatment strategy that combines arsenic compounds that lower BCR-ABL levels, with imatinib mesylate that inhibits BCR-ABL tyrosine kinase activity, has indeed shown promising potential in inducing more apoptosis in BCR-ABL+ cells.14-16 Clinical applications of similar strategies may potentially strengthen the curative effects of imatinib mesylate. To better evaluate additive or synergistic effects of the combination of ATO with imatinib mesylate in CML cells, and to develop more sophisticated clinical protocols, we treated the CML cell line K562 with ATO, imatinib mesylate, and a combination of the 2 agents in a time series. We analyzed samples of each treatment series using a systems approach integrating methods of cell biology, cDNA microarrays, computational biology, and protein biochemistry, which allowed us to depict dynamic changes underlying proapoptotic and apoptotic activities occurring on drug exposure of K562 cells.
Materials and methods
Cell culture, sample treatments, growth, and apoptosis assessments
The CML-derived cell line K562 was maintained in RPMI 1640 supplemented with fetal bovine serum and antibiotics. Imatinib mesylate was kindly provided by Novartis Pharma (Basel, Switzerland) and ATO was purchased from Sigma (St Louis, MO). Cells were separately treated with 0.25 μM imatinib mesylate, 1.0 μM ATO, and the combination of the 2 agents, and analyzed at 0, 3, 8, 12, 24, 48, and 72 hours of treatment. Fresh bone marrow cells were obtained with informed consent according to the Declaration of Helsinki from 2 patients with CML with no previous exposure to drug treatment. One patient was in chronic phase and the other in blast-crisis phase of the disease based on diagnosis criteria of clinical, pathologic, and molecular examinations. Approval was obtained from the School of Medicine of Shanghai Jiao Tong University Institutional Review Board for these studies. Apoptotic assays were performed using ApoAlert Annexin V kit (Clontech, Mountain View, CA) and followed by flow cytometry analysis. Mitochondrial transmembrane potential (ΔΨm) was evaluated through rhodamine 123 and propidium iodide (Sigma) staining and followed by flow cytometry analysis.
Microarray and data mining
Array fabrication, reverse transcriptional labeling, and hybridization were performed as described previously.17 To reduce potential biases generated from tissue culture, untreated K562 cells were collected at 0, 3, 8, 12, 24, 48, and 72 hours, and the collections were pooled, before they were used as the reference control. A conservative 2-fold change threshold (ie, treated sample versus untreated reference) was used to determine regulated genes. Quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR) was conducted for 30 randomly selected genes with the Bio-Rad iCycler iQ detection system (Richmond, CA) using SYBR Green I (Applied Biosystems, Foster City, CA). For data mining and gene clustering, a software package of self-organizing map (SOM), implemented with the Matlab 6.5 environment was used for SOM training with 400 (20 × 20) neurons. Illustration of the SOM outputs by component plane presentations (CPPs) was conducted in the Matlab 6.5 environment as described previously.17,18 SOM outputs and functionally important genes were tabulated (Tables S1 and S2, available on the Blood website; see on the Supplemental Tables link at the top of the online article).
Methods of protein biochemistry
Western blot analyses were performed with caspase-8, -9, and -3, cleaved caspase-8, PARP, HSPA5 and DDIT3 antibodies, commercially obtained from BD Pharmingen (San Diego, CA), Oncogene (Boston, MA), Calbiochem (San Diego, CA), Cell Signaling Technology (Beverly, MA), Santa Cruz Biotechnology (Santa Cruz, CA), and Abcam (Cambridge, MA), using previously described methods.16 The activity assay of caspase-8 was performed according to the manufacturer's instructions (R&D Systems, Minneapolis, MN).
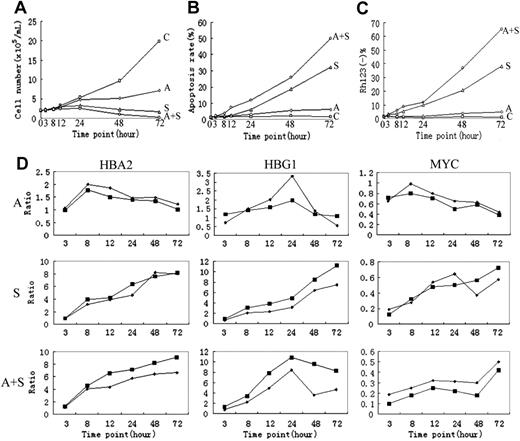
Cellular and molecular assays. (A) Cell viability as evaluated by cell counting through trypan blue staining. (B) Apoptotic induction, as determined through annexin V-specific antibody and propidium iodide (PI) staining, and followed by flow cytometry analysis. (C) Loss of mitochondria ΔΨm, as evaluated by rhodamine 123 and PI double staining, and followed by flow cytometry analysis. (D) Comparison between microarray (▪) and real-time RT-PCR data (♦). C indicates control; A, ATO; S, imatinib mesylate; A+S, ATO plus imatinib mesylate.
Cellular and molecular assays. (A) Cell viability as evaluated by cell counting through trypan blue staining. (B) Apoptotic induction, as determined through annexin V-specific antibody and propidium iodide (PI) staining, and followed by flow cytometry analysis. (C) Loss of mitochondria ΔΨm, as evaluated by rhodamine 123 and PI double staining, and followed by flow cytometry analysis. (D) Comparison between microarray (▪) and real-time RT-PCR data (♦). C indicates control; A, ATO; S, imatinib mesylate; A+S, ATO plus imatinib mesylate.
Results
Cellular and molecular assays and microarray data validation
Cell samples were collected at 6 time points from each of the 3 drug treatment series and first subjected to different assays to evaluate cellular and molecular responses of K562 cells. As demonstrated in Figure 1A, cell viability is markedly reduced in cells treated with ATO, imatinib mesylate, or imatinib mesylate plus ATO, and the strongest reduction of cell viability is revealed in the cotreatment series. When cell apoptosis was evaluated, a synergistic induction of apoptosis was observed in cells cotreated with imatinib mesylate and ATO (Figure 1B-C). Although treatment with ATO alone results in a considerable reduction of cell viability, its impact on apoptosis seems to be limited within the observation time (ie, 72 hours). Time series samples of K562 cells were then used for RNA extraction and microarray hybridization. A real-time RT-PCR assay was performed on a number of genes, as an independent validation of the array data, typically represented by HBA2, HBG1, and MYC (Figure 1D).
Data mining and visualization by CPP-SOM
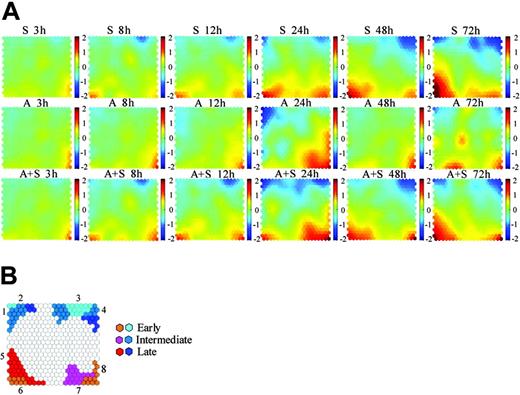
CPP-SOM has shown promising potential in both gene clustering and its visualization.17,18 As shown in Figure 2A, each presentation illustrates a treatment- or sample-specific, global transcriptional map, allowing us not only to directly correlate the functional significance of genes in each unit with respect to the sample or treatment condition, but also to compare global changes within and between the different treatment series. For instance, presentations of the imatinib mesylate (S) series share some similar regulatory patterns with those of the ATO (A) series, implicating the presence of some commonly/similarly regulated genes in these 2 treatment series. Prominently, dramatic changes observed after 24 hours of ATO treatment (A24h) highly overlap with those of the cotreatment at the same time point (A+S 24h), suggesting that these changes may contribute significantly to the synergistic induction of apoptosis in the cotreated cells. Obviously, the cotreatment series shares many commonly/similarly regulated patterns with the imatinib mesylate treatment series throughout the time course, indicating that most genes modulated in the imatinib mesylate series are also modulated in the cotreatment series. It is clearly evident that the apoptosis induced by imatinib mesylate, ATO, or the combination of the 2 agents is a complex process involving multiple gene activations and inactivations. In particular, comparison of transcriptome patterns between the imatinib mesylate and the cotreatment series permits the partition of the process into the early (3-12h), intermediate (24h), and late (48-72h) 3 major stages. Similarly, the ATO treatment series seems to be also separable using the same criteria. Gene clusters modulated in these stages are highlighted in Figure 2B. Based on a conservative 2-fold change threshold, the number of regulated genes in cells treated with either imatinib mesylate (1537) or imatinib mesylate plus ATO (1661) is much larger than that (915) in ATO-treated cells. Moreover, down-regulation in the imatinib mesylate or imatinib mesylate plus ATO treatment series appears to be more prominent than up-regulation (ie, 884 versus 653 and 870 versus 791), highlighting its functional importance to triggering growth arrest and probably apoptosis as well.
CPP-SOM of microarray data. (A) Illustration of self-organizing map outputs of microarray data by component plane presentations. Each presentation illustrates a sample specific global map in which all up-regulated units (red), down-regulated units (blue), and moderately regulated units (yellow and green) are well delineated, permitting direct comparison of regulatory status/biologic significance of clustered genes within and between treatment series (that is, imatinib mesylate treatment series (S series), ATO treatment series (A series), and imatinib mesylate plus ATO treatment series (A+S series), respectively, shown in rows 1, 2 and 3). Color index stands for log ratio with base 2. The brighter the color, the higher the ratio. (B) An enlarged grid ideogram summarizing modulated gene clusters shown on the component plane presentations. Seven major regulatory categories are recognizable. Each category appears to be associated with characteristic features: 1, regulators/players of cell cycle and survival; 2, members of BCR-ABL signaling and mitochondria respiration complexes; 3, regulators/players of cell cycle and negative regulators of the intrinsic apoptotic pathway; 4, members of BCR-ABL signaling and RNA splicing; 5, regulators/players of the intrinsic apoptotic pathway and erythroid differentiation; 6, transcription factors associated with cell apoptosis and erythroid differentiation; 7, molecules associated with oxidative stress; and 8, regulators/players of the extrinsic apoptotic pathway and negative regulators of cell survival/antiapoptosis.
CPP-SOM of microarray data. (A) Illustration of self-organizing map outputs of microarray data by component plane presentations. Each presentation illustrates a sample specific global map in which all up-regulated units (red), down-regulated units (blue), and moderately regulated units (yellow and green) are well delineated, permitting direct comparison of regulatory status/biologic significance of clustered genes within and between treatment series (that is, imatinib mesylate treatment series (S series), ATO treatment series (A series), and imatinib mesylate plus ATO treatment series (A+S series), respectively, shown in rows 1, 2 and 3). Color index stands for log ratio with base 2. The brighter the color, the higher the ratio. (B) An enlarged grid ideogram summarizing modulated gene clusters shown on the component plane presentations. Seven major regulatory categories are recognizable. Each category appears to be associated with characteristic features: 1, regulators/players of cell cycle and survival; 2, members of BCR-ABL signaling and mitochondria respiration complexes; 3, regulators/players of cell cycle and negative regulators of the intrinsic apoptotic pathway; 4, members of BCR-ABL signaling and RNA splicing; 5, regulators/players of the intrinsic apoptotic pathway and erythroid differentiation; 6, transcription factors associated with cell apoptosis and erythroid differentiation; 7, molecules associated with oxidative stress; and 8, regulators/players of the extrinsic apoptotic pathway and negative regulators of cell survival/antiapoptosis.
Apoptosis induced by imatinib mesylate
Transcriptional remodeling. The specific targeting of the tyrosine kinase activity of the BCR-ABL oncoprotein by imatinib mesylate may exert blocking effects on the BCR-ABL signaling pathway and hence inactivate downstream gene transcription in leukemic cells. This appears to be indeed the case for genes modulated during the process of imatinib mesylate-induced apoptosis. For instance, genes modulated at the early stage are largely represented by those encoding transcription factors/cofactors (TFs/CoFs), of which down-regulated ones are mostly downstream of the BCR-ABL signaling (eg, MYC and ETV5) or related to transcriptional activities of cell growth (eg, TBPL1, HSGT1, and PC4; Figure 3). Accordingly, considerably reduced gene-expression levels suggest repressed activities mediated by these TFs/CoFs. In contrast, TF/CoF genes up-regulated at the same time period, such as KLF1, LMO2, NFE2, and TADA3L, are typically associated with transcriptional activities involved in erythroid differentiation and cell apoptosis. Changes in transcriptional regulation at the early stage appear to be extensive, not only involving genes of TFs/CoFs but also genes of chromatin remodeling (eg, HDAC2, CHD1, HTATIP, and HAT1), which regulate the accessibility of chromatin DNA to the transcriptional machinery through noncovalent modifications of chromatin structures. It is therefore logical to speculate the occurrence of a global remodeling at the level of transcriptional regulation on treatment with imatinib mesylate, which may in turn exert versatile impacts on various cellular activities.
Growth arrest. Genes encoding several cell-cycle regulators such as CCND2, GSPT1, YWHAG, and CTNNB1 are markedly down-regulated at the early stage (Figure 3). These regulators are known to be involved in the G1/S and G2/M transition, and their decreased gene-expression levels and thus reduced activities are probably essential for cell-cycle arrest at the early stage. Along with the progression of the apoptotic process, many more cell-cycle-related genes are down-regulated. These include genes of key regulators for the G1/S transition (ie, CCNH, CDK2, and CDK4), the G2/M transition (ie, CDC2, CCNB1, and STK6), S phase progression (ie, ASK, CDC7L1, CCNA2, CDK2, PCNA, and CDC45L) and S or M phase checkpoint control (ie, CHEK1 and MAD2L1; Figure 3). Obviously, down-regulation/inactivation of such a large number of cell-cycle-related genes may abolish the cell-cycle machinery, probably a prerequisite for apoptosis in imatinib mesylate-treated CML cells. On the other hand, down-regulation of these genes can be largely the result of chain reactions of transcriptional inactivation. For instance, CCND2 is known to be directly regulated by MYC at transcriptional sites19,20 and its down-regulation is logically linked to down-regulated MYC at the early stage.
Suppression of oncogenic signals. Although biochemical blocking of the BCR-ABL signaling pathway by imatinib mesylate is a primary event, transcriptional changes of the BCR-ABL-activated genes appear to occur mainly at the intermediate and late stages (Figures 2B and 3). This is probably due to feedback mechanisms operating at multiple levels. For instance, inactivation of the BCR-ABL fusion protein by imatinib mesylate blocks the transduction of oncogenic signals, which in turn results in inactivation of downstream transcription factors, and thus represses expression of target genes such as those encoding signal transduction molecules, as highlighted by members of the RAS/MAPK pathway (eg, HRAS, RAB18, RAB31, RGL2, MAPK6, MAP2K1IP1, and MAP2K4) and those of the PI3K/AKT pathway (eg, PIK3CB and HSPCB; Figure 3). Oncogenic signals of the RAS/MAPK and PI3K/AKT pathways are essential for both cell growth and antiapoptotic/survival activities in CML.21 Notably, suppression/inactivation of the PI3K/AKT pathway can be of particular value for understanding proapoptotic activities such as the restoration of apoptotic potential and the elimination of cell-survival competence in CML cells treated with imatinib mesylate.
Suppression of antiapoptotic/survival activities mediated by PI3K/AKT. AKT, a serine/threonine kinase downstream of PI3K, mediates many antiapoptotic activities through phosphorylation of a wide variety of target proteins including proapoptotic and cell-survival factors.22-24 On one hand, it phosphorylates proapoptotic proteins such as CASP9 and BAD to prevent proapoptotic activities and, on the other hand, it phosphorylates cell-survival molecules such as IKK of the NF-κB pathway to promote cell-survival signals. Importantly, 2 cofactors appear to be critical for AKT-mediated antiapoptotic/survival activities. One is HSP90 that binds to phospho-AKT to prevent AKT from undergoing dephosphorylation by PP2A, and the other is the 14-3-3 protein that interacts with phospho-BAD to prevent the formation of proapoptotic BAX homodimers. Accordingly, transcriptional down-regulation of HSP90- (ie, HSPCB), 14-3-3- (ie, YWHAG), AKT kinase PI3K- (ie, PIK3CB), and AKT-stimulated genes, such as members of the RAS family, strongly suggests suppression/inactivation of antiapoptotic/survival activities mediated by PI3K/AKT, which may correlate with the restoration of apoptotic potential in imatinib mesylate-treated K562 cells. Additional evidence supporting this notion is provided by reduced expression of genes encoding NF-κB-associated molecules (eg, VAPA, NDFIP2, NPM1, and ECT2), and increased expression of those encoding AKT phosphatase PP2A (eg, PPP2R5A and PPP2R5C) as well as other negative regulators of the BCR-ABL signaling (eg, RASA4 and TMOD1).
Functional classification of representative genes modulated by imatinib mesylate, ATO, and imatinib mesylate plus ATO. Functional feature of each gene is determined based on detailed literature reading (see references in Table S2). All the relevant genes are grouped by hierarchical clustering based on expression values (log2 ratios) across all the samples (Table S2). Log2 ratios are color coded as indicated. Detailed descriptions of important genes mentioned in the text are listed in the “Appendix.”
Functional classification of representative genes modulated by imatinib mesylate, ATO, and imatinib mesylate plus ATO. Functional feature of each gene is determined based on detailed literature reading (see references in Table S2). All the relevant genes are grouped by hierarchical clustering based on expression values (log2 ratios) across all the samples (Table S2). Log2 ratios are color coded as indicated. Detailed descriptions of important genes mentioned in the text are listed in the “Appendix.”
Other aspects of activities potentially affected by suppression of BCR-ABL signaling. Splice variants appear to be abundant in tumors, probably due to variation in activities of spliceosomes and other mRNA-processing complexes.25 It is known that the mTOR pathway, downstream of PI3K/AKT, coordinates mRNA splicing and translation, which is largely mediated through CDC42 and RPS6KB1 (S6K1). Interestingly, these 2 molecules are also found to be frequently associated with oncogenesis.26 Accordingly, down-regulation of the CDC42 and RPS6KB1 genes, as observed in this study, can be of particular value in the context of antileukemic treatments. Equally intriguing is the down-regulation of genes involved in RNA processing machinery, including those encoding small nuclear ribonucleoproteins (snRNPs) such as SNRPD3, SNRPF, SNRPE, SNRPD2, SNRPG, and LSM10 and non-snRNP proteins such as SFRS1, SFRS2, SFRS5, SFRS10, and FUSIP1 (Figure 3), as well as factors involved in protein translation such as EIF4B, EEF1E1, and IF2. Moreover, genes encoding cytoskeletal proteins or cytoskeleton-associated proteins appear to be markedly modulated at the intermediate and late stages as well, implicating the occurrence of cytoskeleton reorganization, which is probably also coupled with inactivation of the RAS/MAPK and PI3K/AKT pathways.
Coordinated regulation of proapoptotic and apoptotic activities toward mitochondria-mediated apoptosis pathways. In addition to suppression of cell growth and survival/antiapoptotic activities, stimulation of proapoptotic and apoptotic cascades is probably essential to ensure cells entering into programmed cell death. This appears to be supported by modulation of specific genes during imatinib mesylate-induced apoptosis. For instance, up-regulated genes are prominently represented by those encoding potential regulators/players of proapoptotic and apoptotic cascades, and increased activities of these regulators/players can be correlated with cell apoptosis. At the early stage, genes typically belonging to this category are represented by those of an essential CoF of p53-mediated gene transcription (ie, TADA3L), a p53-interacting TF (ARID3A), a programmed cell death protein (PDCD7), caspase-7 (CASP7), caspase-9 (CASP9), TNF-α receptor 1 (TNFR1), a TNF-α receptor 1-interacting protein (MADD), and calpain-1 (CAPN1), an upstream element essential for BAX and caspase-3 activation. Accordingly, markedly increased expression levels of these genes at the early stage suggest their functional relevance to proapoptotic activities. Along with the progression of apoptosis, some more proapoptotic/apoptotic genes are up-regulated, such as SH3GLB, RTN4, BCL2L14, SLC25A6, and PRSS25, typical regulators/players of mitochondria-mediated pathways of apoptosis. In accordance, genes encoding a number of negative regulators are down-regulated, such as BIRC6, NCL, HMGB1, and CFLAR (Figure 3). Mitochondria-mediated apoptosis is characterized by the collapse of the mitochondrial transmembrane potential (ΔΨm), resulting in the release of mitochondrial proteins such as cytochrome c, thus triggering the activation of caspases typically represented by caspase-9, -7, and -3. This essential process is characteristically activated by proapoptotic BAX and inhibited by antiapoptotic BCL2 and BCL-XL. Accordingly, transcriptionally increased expression of positive regulators (eg, CAPN1, SH3GLB1, and RTN4) and decreased expression of negative regulators (eg, NCL and HMGB1) of BAX, and transcriptionally increased expression of inhibitors (eg, BCL2L14 and SLC25A6) of BCL2/BCL-XL, strongly suggest the collapse of the mitochondria potential and thus release of mitochondria proteins at the late stage of imatinib mesylate-induced apoptosis. Additional evidence for mitochondria-mediated apoptosis is provided by increased expression of caspase-9 and caspase-7, and decreased expression of caspase inhibitors such as BIRC6. Although some members of the endoplasmic reticulum (ER)-mediated and extrinsic pathways of apoptosis are transcriptionally modulated as well, involvement of these apoptotic mechanisms in imatinib mesylate-treated CML cells is probably minor or less prominent than that of the intrinsic pathway.
Regulation of erythroid-specific genes. Expression of erythroid-specific genes seems to be a prominent phenomenon at the early stage of imatinib mesylate-induced apoptosis in K562 cells, including those encoding erythroid-specific transcription factors (ie, KLF1, LMO2, and NFE2), erythrocyte membrane antigenic determinants (ie, GYPB and GYPE), a terminal enzyme in the heme biosynthetic pathway (ie, FECH), and hemoglobins (ie, HBA2 and HBG2; Figure 3). Early induction of erythroid-specific genes is probably due to the fact that K562 is derived from an erythroid blast crisis.27,28 It has been recently reported that hemin-mediated erythroid differentiation may sensitize CML cells to TRAIL-induced apoptosis.29 In imatinib mesylate-treated K562 cells, however, the potential role of erythroid differentiation in cell apoptosis remains to be determined, because mechanisms of apoptosis induced by imatinib mesylate can be different from those induced by TRAIL.
Apoptosis induced by ATO
ATO can effectively induce cell apoptosis in CML at high dose (eg, 10 μM).30 At subtoxic or clinically achievable concentrations (0.5-2 μM), however, the effectiveness of ATO in apoptotic induction appears to be significantly reduced, resulting in prolonged proapoptotic periods (eg, 7-10 days).31 Accordingly, transcriptome changes depicted by CPP-SOM of the ATO-treated series may mainly reflect proapoptotic activities induced by a low dose of ATO. By comparing presentations of the ATO and imatinib mesylate treatment series (Figure 2), overlaps of regulatory patterns are visible, particularly at the early stage. This is probably due to ATO-targeted down-regulation of the BCR-ABL protein level,32 with a proportional decrease of the tyrosine kinase activity,13 which might provide feedback to transcriptional regulation. Because ATO is also a toxic and oxidative agent, it is logical to expect complex cellular and molecular responses to the drug to prevent toxicity and promote self-defense. This seems to be supported by the occurrence of dramatic transcriptional changes after 24 hours of treatment. Some of these changes are reduced in later stages and others are sustained (Figure 2). Of note, transcriptional changes underlying responses to oxidative stress appear to be particularly prominent and can be relevant to cell apoptosis. These changes typically affect genes encoding proteins of the metallothionein family (eg, MT1G, MT2A, MT1X, MT1L, MT1B, MT1H, and MT1E), disulfide isomerase-like proteins (eg, ERP70 [ERP72], GRP58 [ERP57], C12orf8, P4HB, TXNDC7, and DNAJB11), mitochondria respiratory complex proteins (eg, CYC1, COX5B, NDUFB8, NDUFA2, and NDUFA8), and protein-folding and chaperone proteins (eg, TRA1, DNAJC10, HYOU1, HSPA5, EIF2AK3, ATF6, and DDIT3; Figure 3). Metallothioneins are metal-binding proteins that maintain metal ion homoeostasis and redox balance, and may help to scavenge superoxide and hydroxyl radicals.33 Similarly, disulfide isomerase-like proteins, located in the ER, may also be able to maintain the redox balance by catalyzing thiol-disulfide exchange reactions. Moreover, these proteins are probably involved in the oxidative folding of newly synthesized proteins.34-36 Equally significant is the modulation of genes encoding proteins of the respiration complexes. Among those up-regulated are mostly members of the respiration complexes I, III, and IV families. Under physiologic conditions, reactive oxygen species (ROSs) are produced through the NADH respiratory chain, which is composed of complex I, III, and IV (Figure 3). It appears therefore that ATO may inhibit some of the mitochondria respiration cascades, probably those composed of complexes II, III, and IV, and enhance others such as the cascades composed of complexes I and III, resulting in significantly high levels of ROSs as reported before.37 High levels of ROSs can be an effective inducer of cell apoptosis, probably through the ER stress-mediated pathway. This appears to be implicated by modulation of a large number of genes encoding ER proteins such as those encoding disulfide isomerase-like proteins, protein-folding proteins, and particularly the ER stress markers including HSPA5 (GRP78), EIF2AK3, ATF6, and DDIT3 (GADD153).38 Although the impact of ER stress on the induction of apoptosis in ATO alone-treated series seems to be limited, it can be substantially significant to the synergistic induction of apoptosis in the cotreatment series.
Apoptosis induced by imatinib mesylate plus ATO
Coordinated regulation of the intrinsic, extrinsic, and ER stress-mediated pathways. The combination of imatinib mesylate with ATO induces markedly more apoptosis in K562 cells than either agent alone. At the transcriptome level, dynamic changes induced by the combined treatment appear to overlap strongly with those induced by imatinib mesylate (Figure 2A), but with some distinct features that may consequently contribute to the apoptotic synergy as observed at the cellular level. For instance, modulation of many important genes in the combined treatment series appears to be either more profound or to occur earlier than in the imatinib mesylate treatment series (Figure 3). These genes encode key regulators/players of cell growth, the BCR-ABL signaling and cell-survival pathways, and proapoptotic/apoptotic cascades. Their synergistic/additive modulations may consequently result in enhanced proapoptotic/apoptotic activities both quantitatively and qualitatively. The intrinsic apoptotic pathway seems to be quantitatively enhanced, as highlighted by enhanced expression of the genes encoding proapoptotic/apoptotic factors (CASP7, CASP9, and BCL2L14), and further suppressed expression of those encoding negative regulators (NCL and SON; Figure 3). Enhanced proapoptotic/apoptotic activities are probably also suggested by addition of the extrinsic apoptotic pathway, as highlighted by synergistic/additive modulation of genes encoding proapoptotic factors (CASP8, TNFR1, MADD, and DEDD2), and a number of negative regulators (Figure 3). The extrinsic pathway of apoptosis is characterized by the engagement of TNF death receptors (TNFRs), which deliver proapoptotic signals through death domain-mediated recruitment of adaptor proteins. It has been recently reported that 2 types of TNF signaling complexes can be formed in cells.39 One type of complex binding to the plasma membrane may trigger NF-κB responses, and the other formed in the cytoplasm can activate caspases such as CASP8. Once the cytoplasmic complex functions, the NF-κB signal from the membrane-bound complex fails to induce antiapoptotic activities. Accordingly, transcriptionally synergistic/additive up-regulation of CASP8, TNFR1, MADD, and DEDD2, and down-regulation of negative regulators or NF-κB survival factors encoded by genes such as SLC20A1 and VAPA, may suggest the involvement of the extrinsic pathway of cell apoptosis in the imatinib mesylate plus ATO-treated CML cells. Moreover, ER stress-mediated apoptosis appears to be involved as well, as highlighted by modulation of a number of genes encoding various factors involved in ER stress cascades. Modulation of these genes is obviously largely contributed by ATO-induced oxidative stress as discussed in previous sections, though modulation of some members of the cascades appears to be potentiated by imatinib mesylate (Figure 3). Taken together, it can be reasoned that the in vitro synergistic activity of imatinib mesylate and ATO is probably related to the activation of multiple apoptotic pathways including the intrinsic, extrinsic, and ER stress pathways of cell apoptosis.
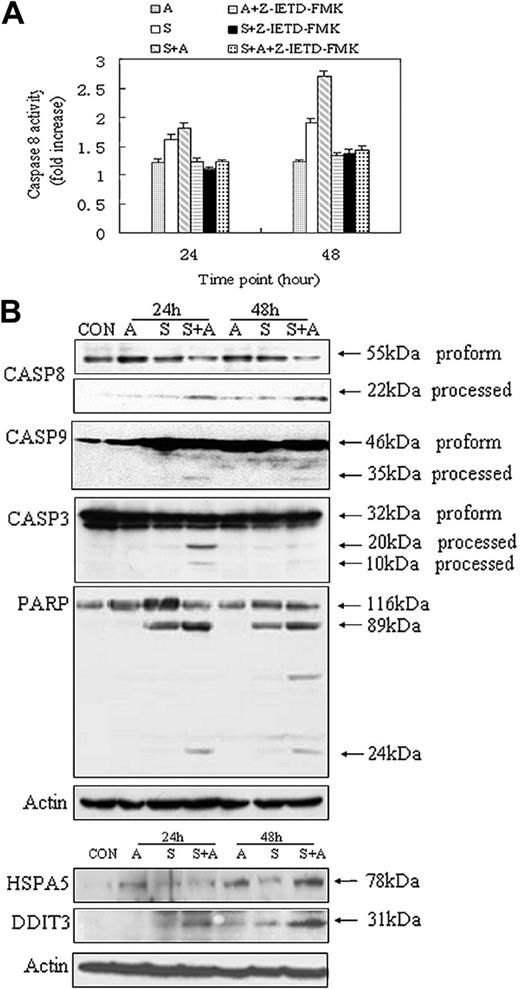
Evidence of protein biochemistry for the involvement of the intrinsic, extrinsic, and ER stress-mediated apoptosis. (A) Caspase-8 activity assays. Caspase-8 inhibitor Z-IETD-FMK was added into culture medium 30 minutes before drug treatment, as recommended by the manufacturer (R&D Systems). Data were obtained from 3 independent experiments. (B) Western blot analysis of caspases and marker molecules involved in the intrinsic, extrinsic, and ER stress-mediated apoptosis. Specific antibodies were obtained from commercial sources (see “Materials and methods”) and reacted with Western blots of cell lysates of the drug treatment series as indicated.
Evidence of protein biochemistry for the involvement of the intrinsic, extrinsic, and ER stress-mediated apoptosis. (A) Caspase-8 activity assays. Caspase-8 inhibitor Z-IETD-FMK was added into culture medium 30 minutes before drug treatment, as recommended by the manufacturer (R&D Systems). Data were obtained from 3 independent experiments. (B) Western blot analysis of caspases and marker molecules involved in the intrinsic, extrinsic, and ER stress-mediated apoptosis. Specific antibodies were obtained from commercial sources (see “Materials and methods”) and reacted with Western blots of cell lysates of the drug treatment series as indicated.
Lines of evidence from protein biochemistry studies. CASP8 represents a key factor in the extrinsic pathway of cell apoptosis. To evaluate the potential role of CASP8 in apoptosis induced by the combined treatment, a specific enzymatic assay as well as an inhibitory test was performed on cells treated, respectively, with imatinib mesylate, ATO, and imatinib mesylate plus ATO (Figure 4A). As expected, the activity level of CASP8 in the combined treatment series is considerably higher than that in the imatinib mesylate treatment or ATO treatment series, particularly after 48 hours of treatment. Additional lines of evidence for the involvement of the intrinsic and the extrinsic pathways of cell apoptosis are provided by Western blot analysis of CASP8, cleaved CASP8, CASP9, CASP3, and PARP (Figure 4B). The proapoptotic form of CASP8 is markedly reduced, whereas its active (processed) form is markedly increased in the cotreatment samples, implicating the activation of CASP8 in the cotreatment series. Processed CASP8 may, in turn, activate other caspases such as CASP3 and CASP9.40 Indeed, activation of CASP3 appears to be more obvious in the cotreatment series, particularly at 24 hours of treatment, than that in the imatinib mesylate treatment series. Its apoptotic role, however, is probably transitional, because the amount of activated CASP3 seems to be relatively reduced in the later stage of the apoptosis. Of note, the basal level of the proapoptotic form of CASP3 is high across all the samples, including the untreated sample. This not only implies a functional importance of this form of CASP3 in K562 cells, but also explains why gene-expression levels of CASP3 are unchanged in all the samples. Conversely, the proapoptotic form of CASP9 is considerably increased in imatinib mesylate or imatinib mesylate plus ATO-treated samples, which is in accordance with the gene-expression data, and processed CASP9 is obviously enhanced in the cotreatment samples after 24 and 48 hours of treatment. PARP is a downstream target of CASP3 and some other caspases, and its gene-expression levels appear to be increased in samples treated with imatinib mesylate and imatinib mesylate plus ATO. In accordance, strongly activated forms of PARP protein are also observed in these 2 treatment series, particularly with respect to samples cotreated with imatinib mesylate and ATO (Figure 4B). At the transcriptome level, a large number of ER stress-related genes are significantly regulated in both ATO and ATO plus imatinib mesylate treatment series, and many of them appear to be synergistically/additively enhanced in the latter setting (Figure 3). To validate the involvement of ER stress-mediated apoptosis, Western blot analysis with specific antibodies of ER stress markers HSPA5 and DDIT3 was performed (Figure 4B). Obviously, the protein level of HSPA5 is markedly higher in both ATO- and ATO plus imatinib mesylate-treated samples than that in either untreated or imatinib mesylate-treated samples, particularly at 48 hours of treatment. Of note, the cotreatment sample at this time point reveals the most prominent expression of the HSPA5 protein. This appears to be also the case in the Western blot detected by the DDIT3 specific antibody.
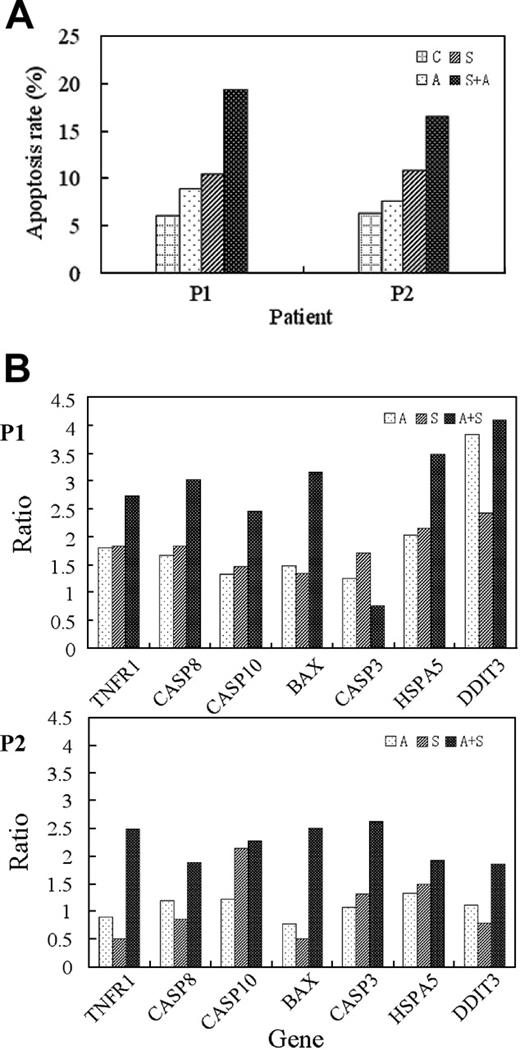
Evidence from bone marrow cells of patients with CML. (A) Apoptotic induction, as evaluated through annexin V-specific antibody and propidium iodide (PI) staining, and followed by flow cytometry analysis. (B) Real-time RT-PCR analysis of specific marker genes. Total RNA was prepared from each of the patient samples and quantitatively analyzed by iCycler iQ real-time PCR detection system (Bio-Rad) using SYBR Green I (Applied Biosystems). P1: the patient in blast crisis; P2: the patient in chronic phase. Treatment conditions are indicated in the top right corner of each panel. Ratio stands for value of treated sample versus that of untreated sample of the same patient.
Evidence from bone marrow cells of patients with CML. (A) Apoptotic induction, as evaluated through annexin V-specific antibody and propidium iodide (PI) staining, and followed by flow cytometry analysis. (B) Real-time RT-PCR analysis of specific marker genes. Total RNA was prepared from each of the patient samples and quantitatively analyzed by iCycler iQ real-time PCR detection system (Bio-Rad) using SYBR Green I (Applied Biosystems). P1: the patient in blast crisis; P2: the patient in chronic phase. Treatment conditions are indicated in the top right corner of each panel. Ratio stands for value of treated sample versus that of untreated sample of the same patient.
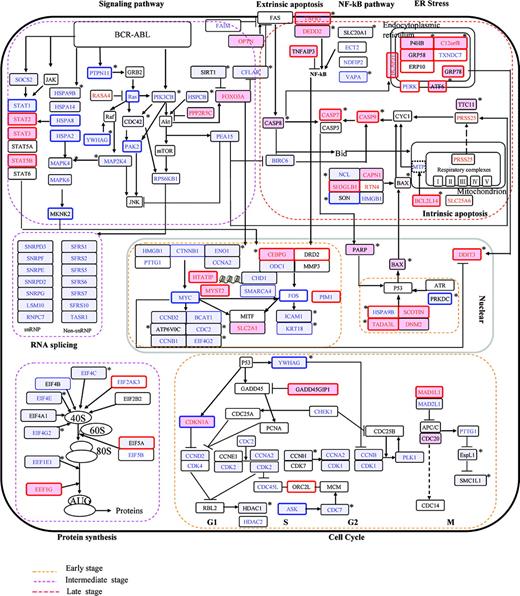
Ideogram illustration of dynamic changes underlying imatinib mesylate/ATO-induced apoptosis in CML. Genes modulated in the imatinib mesylate series are marked in red (up-regulation) and blue (down-regulation), whereas those modulated in the ATO series are framed by red (up-regulation) and blue (down-regulation), and those modulated in the cotreatment series are highlighted by pink (up-regulation) and light-blue (down-regulation) background. *Synergistically/additively affected genes in the cotreatment series. Unchanged genes are indicated in black-framed boxes. Molecular events at the early, intermediate, and late stages are rimmed, respectively, by yellow, pink, and red lines.
Ideogram illustration of dynamic changes underlying imatinib mesylate/ATO-induced apoptosis in CML. Genes modulated in the imatinib mesylate series are marked in red (up-regulation) and blue (down-regulation), whereas those modulated in the ATO series are framed by red (up-regulation) and blue (down-regulation), and those modulated in the cotreatment series are highlighted by pink (up-regulation) and light-blue (down-regulation) background. *Synergistically/additively affected genes in the cotreatment series. Unchanged genes are indicated in black-framed boxes. Molecular events at the early, intermediate, and late stages are rimmed, respectively, by yellow, pink, and red lines.
Evidence from bone marrow cells of patients with CML. Bone marrow cells from 2 patients with CML, one in the chronic phase and the other in the blast-crisis phase of the disease, were cultured in vitro, treated with imatinib mesylate, ATO, and imatinib mesylate plus ATO for 48 hours, and then subjected to different assays. At the cellular level, samples from both patients appear to respond to the apoptotic induction of the cotreatment in a synergistic/additive manner (Figure 5A). Based on RNA extracted from each treated sample, a quantitative RT-PCR analysis was conducted with a set of selected marker genes (Figure 5B). Specific markers of the extrinsic pathway of apoptosis (ie, TNFR1, CASP8, and CASP10) are prominently induced in the cotreatment samples of both patients, and those of the intrinsic pathway (ie, BAX) seem to be somewhat differentially expressed in the cotreatment samples between the 2 patients. Specific markers of the ER stress pathway (ie, HSPA5 and DDIT3) appear to be more prominently induced in the cotreatment samples of the blast crisis patient than those in the cotreatment samples of the chronic phase. Taken together, it appears that evidence derived from bone marrow cells of these patients supports the notion derived from the K562 cells that the combination of imatinib mesylate and ATO may more effectively induce the intrinsic, extrinsic, and ER stress-mediated pathways of apoptosis in CML.
Overview
Information obtained from this study permits recognition of a number of pathways with temporospatial relationships, which might contribute to the mechanisms underlying proapoptotic and apoptotic activities induced by imatinib mesylate, ATO, and the combination of the 2 agents (Figure 6). Blocking of the BCR-ABL signaling by imatinib mesylate at the biochemical level seems to primarily result in transcriptional remodeling at the global scale, which may, in turn, have a versatile impact on various aspects of cellular activities in imatinib mesylate-treated K562 cells. Probably through chain reactions and feedback mechanisms, genes involved in cell-cycle control are prominently suppressed at the early stage and those involved in some other leukemia activities, such as oncogenic signal transduction, cell survival/antiapoptosis, RNA processing, and protein synthesis, are mostly inactivated afterward. Activation of specific genes involved in proapoptotic and apoptotic cascades is probably essential to ensure cells entering into programmed cell death, as implicated by the observation of genes up-regulated throughout the apoptotic process. In particular, genes/proteins involved in the intrinsic pathway of cell apoptosis appear to be characteristically activated in K562 cells treated with imatinib mesylate. Although oxidative stress resulting from a subtoxic concentration (1 μM) of ATO alone seems to be insufficient to induce cell apoptosis in K562 cells within the observation time, its induced ER stress appears to contribute significantly to the synergistic induction of apoptosis in the cotreatment series. Genes/proteins modulated by imatinib mesylate plus ATO appear to be strongly overlapped with those modulated by imatinib mesylate, but with some distinct features that may consequently contribute to the apoptotic synergy as observed at the cellular level. The intrinsic apoptotic pathway seems to be quantitatively enhanced by synergistic/additive modulation of many members of the pathway. Enhanced proapoptotic/apoptotic activities are further suggested by addition of cascades of the ER stress-mediated pathway and probably the extrinsic apoptotic pathway as well. Coordinated regulation of the genes/proteins relevant to these pathways may consequently result in an effective and efficient induction of cell apoptosis in K562 cells treated by imatinib mesylate combined with ATO. Further evidence from bone marrow cells of patients with CML suggests that CML cells responding to cotreatment of imatinib mesylate and ATO may share similar/the same mechanisms as recognized from this setting. Accordingly, this study not only allows us to better understand the mechanisms underlying cell apoptosis induced by imatinib mesylate, ATO, and imatinib mesylate plus ATO in CML, but will also help us to eventually develop even more sophisticated protocols for the treatment of leukemia and probably other human malignancies as well in the future.
Appendix
The fully expanded gene names corresponding to each of the gene symbols used in the paper are as follows: ARID3A: AT-rich interactive domain 3A; ASK: activator of S phase kinase; ATF6: activating transcription factor 6; BAX: BCL2-associated X protein; BCL2: B-cell CLL/lymphoma 2; BCL2L14: BCL2-like 14; BIRC6: baculoviral IAP repeat-containing 6; C12orf8: chromosome 12 open reading frame 8; CAPN1: calpain 1; CASP7: caspase-7; CASP8: caspase-8; CASP9: caspase-9; CCNA2: cyclin A2; CCNB1: cyclin B1; CCND2: cyclin D2; CCNH: cyclin H; CDC2: cell division cycle 2; CDC42: cell division cycle 42; CDC45L: CDC45 cell division cycle 45-like; CDC7L1: CDC7 cell division cycle 7; CDK2: cyclin-dependent kinase 2; CDK4: cyclin-dependent kinase 4; CFLAR: CASP8 and FADD-like apoptosis regulator; CHD1: chromodomain helicase DNA-binding protein 1; CHEK1: CHK1 checkpoint homolog; COX5B: cytochrome c oxidase subunit Vb; CTNNB1: catenin, β 1; CYC1: cytochrome c-1; DDIT3: DNA damage-inducible transcript 3; DEDD2: death effector domain containing 2; DNAJB11: DnaJ (Hsp40) homolog, subfamily B, member 11; DNAJC10: DnaJ (Hsp40) homolog, subfamily C, member 10; ECT2: epithelial cell-transforming sequence 2 oncogene; EEF1E1: eukaryotic translation elongation factor 1 ϵ 1; EIF2AK3: eukaryotic translation initiation factor 2-α kinase 3; EIF4B: eukaryotic translation initiation factor 4; ERP70: protein disulfide isomerase-associated 4; ETV5: Ets variant gene 5; FECH: ferrochelatase; FUSIP1: FUS-interacting protein 1; GRP58: protein disulfide isomerase-associated 3; GSPT1: G1-to-S phase transition 1; GYPB: glycophorin B; GYPE: glycophorin E; HAT1: histone acetyltransferase 1; HBA2: hemoglobin, α 1; HBG1: hemoglobin, γ A; HBG2: hemoglobin, γ G; HDAC2: histone deacetylase 2; HMGB1: high-mobility group box 1; HRAS: V-Ha-ras Harvey rat sarcoma viral oncogene homolog; HSGT1: suppressor of Saccharomyces cerevisiae gcr2; HSPA5: heat shock 70-kDa protein 5; HSPCB: heat shock 90-kDa protein 1, β; HTATIP: HIV-1 Tat-interacting protein, 60 kDa; HYOU1: hypoxia up-regulated 1; KLF1: Kruppel-like factor 1 (erythroid); IF2 (EIF5B): eukaryotic translation initiation factor 5B; LMO2: LIM domain only 2 (rhombotin-like 1); LSM10: LSM10, U7 small nuclear RNA associated; MAD2L1: MAD2 mitotic arrest deficient-like 1; MADD: MAP-kinase activating death domain; MAP2K1IP1: mitogen-activated protein kinase kinase 1 interacting protein 1; MAP2K4: mitogen-activated protein kinase kinase 4; MAPK6: mitogen-activated protein kinase 6; MCM7: MCM7 minichromosome maintenance deficient 7; MT1B: metallothionein 1B; MT1E: metallothionein 1E; MT1G: metallothionein 1G; MT1H: metallothionein 1H; MT1L: metallothionein 1L; MT1X: metallothionein 1X; MT2A: metallothionein 2A; MYC: V-myc myelocytomatosis viral oncogene homolog; NCL: nucleolin; NDFIP2: Nedd4 family interacting protein 2; NDUFA2: NADH dehydrogenase (ubiquinone) 1 α subcomplex, 2; NDUFA8: NADH dehydrogenase (ubiquinone) 1 α subcomplex, 8; NDUFB8: NADH dehydrogenase (ubiquinone) 1 β subcomplex, 8; NFE2: nuclear factor (erythroid-derived 2); NPM1: nucleophosmin (nucleolar phosphoprotein B23, numatrin); PARP: poly (ADP-ribose) polymerase family, member 1; P4HB: procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), β polypeptide (protein disulfide isomerase-associated 1); PC4: interferon-related developmental regulator 1; PCNA: proliferating cell nuclear antigen; PDCD7: programmed cell death 7; PIK3CB: phosphoinositide-3-kinase, catalytic, β polypeptide; PPP2R5A: protein phosphatase 2, regulatory subunit B (B56), α isoform; PPP2R5C: protein phosphatase 2, regulatory subunit B (B56), γ isoform; PRSS25: protease, serine, 25; RAB18: RAB18, member RAS oncogene family; RAB31: RAB31, member RAS oncogene family; RASA4: RAS p21 protein activator 4; RPS6KB1: ribosomal protein S6 kinase, polypeptide 1; RTN4: reticulon 4; SFRS1: splicing factor, arginine/serine-rich 1; SFRS10: splicing factor, arginine/serine-rich 10; SFRS2: splicing factor, arginine/serine-rich 2; SFRS5: splicing factor, arginine/serine-rich 5; SH3GLB1: SH3-domain GRB2-like endophilin B1; SLC20A1: solute carrier family 20 (phosphate transporter), member 1; SLC25A6: solute carrier family 25, member 6; SNRPD2: small nuclear ribonucleoprotein D2; SNRPD3: small nuclear ribonucleoprotein D3; SNRPE: small nuclear ribonucleoprotein polypeptide E; SNRPF: small nuclear ribonucleoprotein polypeptide F; SNRPG: small nuclear ribonucleoprotein polypeptide G; SON: SON DNA-binding protein; STK6: serine/threonine kinase 6; TADA3L: transcriptional adaptor 3-like; TBPL1: TATA box binding protein-like 1; TMOD1: tropomodulin 1; TNFR1: tumor necrosis factor receptor superfamily, member 1A; TRA1: tumor rejection antigen 1; TXNDC7: protein disulfide isomerase-associated 6; VAPA: VAMP-associated protein A; VDAC1: voltage-dependent anion channel 1; YWHAG: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, γ polypeptide.
Prepublished online as Blood First Edition Paper, October 25, 2005; DOI 10.1182/blood-2005-06-2318.
Supported in part by the Chinese National Key Program for Basic Research (973:2004CB518606), 100-Talent Program of Chinese Academy of Sciences (J.Z.), National Natural Science Foundation of China (30328028, 30300409, and 90209007), Shanghai Commission of Science and Technology (04DZ14004 and 04QMX1427), and China Postdoctoral Science Foundation (20040350503).
Y.K.D. and K.W. contributed equally to this work.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Special thanks are due to Prof Jiangye Chen, PhD, from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, for her support for this paper. Thanks to the anonymous reviewers as well. We thank all colleagues of the State Key Laboratory of Medical Genomics and the Sino-French Laboratory of Genomics and Life Sciences for their encouragement and support.