Abstract
Recent studies suggest plasmacytoid predendritic cells (pDCs) and myeloid dendritic cells (mDCs) have the functional plasticity to produce similar amounts of type 1 interferons (IFNs) and interleukin-12 (IL-12), challenging the concept and existence of DC subsets with distinct function. In this study, we demonstrate that previous studies showed human pDCs produce large amounts of IL-12 because of contaminating mDCs. Using highly purified human DC subsets, we found that although pDCs make 300 times more IFN-α than mDCs and mDCs make 13 times more IL-12 p70 than pDCs in response to all the toll-like receptor ligands and CD40 ligands, pDCs rapidly make large amounts of IFN-α within the first 12 hours of activation and become refractory to further stimulation. pDCs preferentially expressed the transcriptional factors critical for type 1 IFN, but not for IL-12 transcription, and they dedicated 60% of new transcriptional activity to make 19 type 1 IFN subtypes. This study provides formal proof that the plasticity of DC subsets is limited and that different DC subsets evolve to perform distinct functions in linking innate and adaptive immunity. (Blood. 2006;107:2423-2431)
Introduction
Although all nucleated mammalian cell types have the ability to produce type 1 interferons (IFNs) in response to infection by an appropriated virus, enabling neighboring cells to resist viral infection, human plasmacytoid predendritic cells (pDCs), or type 1 IFN-producing cells (IPCs), have the ability to produce hundreds to thousands times more type 1 IFNs than other cell types after encounters with DNA and RNA viruses.1-6 This phenomenon, together with the selective expression of toll-like receptor 7 (TLR7) and TLR97-9 —which, respectively, recognize viral RNA and DNA10,11 —by human pDCs/IPCs, suggests that pDCs/IPCs are specialized in the viral recognition and production of type 1 IFNs. By contrast, human myeloid dendritic cells (mDCs) selectively express TLR2, TLR3, TLR4, TLR5, TLR6, and TLR8 and have the ability to produce high levels of interleukin-12 (IL-12), but only low levels of type 1 IFNs, on bacterial and viral infection.7,8 These findings have led to an evolution model suggesting that there are 2 unique antigen-presenting cell (APC) systems in humans, with pDCs specialized in making type 1 IFNs and mDCs specialized in making IL-12. This hypothesis is challenged, however, by studies showing that, in response to lipopolysaccharide (LPS) or CpG-oligodeoxynucleotide (ODN) plus CD40 ligand (CD40L), pDCs produce as much IL-12 as do mDCs2,9 and that, in response to electroporated poly(I:C), mDCs produce as much IFN-α as pDCs.12 An alternative instruction model, therefore, has been proposed, suggesting that pDCs and mDCs have the functional plasticity to produce similar amounts of IFN-α and IL-12, depending on the stimulus.
The question is whether the evolution model and the instruction model of DC development and maturation are mutually exclusive. Current literature on DC subsets is confusing. The confusion appears to be caused by species differences between human and mouse DC subsets13 and by diverse experimental conditions from various laboratories. For example, the expression patterns of TLRs by DC subsets in mice and humans are different.13 In mice, pDCs clearly have the ability to produce IL-12 and IFN-α in response to viral stimulation.1,14 However, in humans, pDCs appear to have a limited ability to produce IL-12. In humans, pDCs express high levels of IL-3 receptor and are highly responsive to IL-3 stimulation.15 Mouse pDCs, however, do not express IL-3 receptor and do not respond to IL-3.16 Another important factor that contributes to the contradictory data on the function of DC subsets has been the different degree of purity of DC subsets used by different laboratories.
One particular focus of the debates has been whether human pDCs and mDCs have equal ability to produce IL-12. By using highly purified human DC subsets and all the known TLR-ligands (TLRLs) and their combination with CD40L, we found that though pDCs preferentially produce type 1 IFNs, mDCs preferentially produce IL-12 regardless of the type of stimulation. We discovered several novel functional features of pDCs/IPCs that further support the concept that pDCs are professional type 1 IFN-producing cells.
We suggest that the functional plasticity of DCs is not unlimited. Experiments using DC subsets with low purity may generate confusing data about the functional plasticity of DCs. Although pDCs preferentially produce type 1 IFNs, mDCs preferentially produce IL-12 during antimicrobial innate immune responses.
Materials and methods
Media and reagents
Peptidoglycan (PGN from Bacillus subtilis, 5 μg/mL), zymosan (10 μg/mL), poly(I:C) (25 μg/mL), LPS (from Escherichia coli 0111:B4, 1 μg/mL), flagellin (from Salmonella typhimurium, 5 μg/mL), loxoribine (100 μM), and R848 (1 μg/mL) were purchased from InvivoGen (San Diego, CA). CpG type A (2216, 5 μM), type B (2006, 5 μM), type C (C274, 5 μM), γ-irradiated herpes simplex virus (HSV)-1 (KOS strain; 10 PFU/cell), influenza virus (PR8 strain; 10 PFU/cell), Sendai virus (Cantell strain; 5 PFU/cell), anti-CD40 monoclonal antibody (mAb) (G28.5, 3 μg/mL) and IL-3 (10 ng/mL; R&D Systems, Minneapolis, MN) were used for cell stimulation.
Cell isolation and culture
pDCs and CD11c+ mDCs were isolated from the buffy coat of blood from healthy volunteers and were obtained with informed consent, as described.7,17 CD123high/CD11c-/lineage-/CD4+ pDCs and CD11c+/lineage-/CD4+ CD11c+ mDCs were isolated by fluorescence-activated cell sorting (FACS) Aria (BD Biosciences, Sunnyvale, CA) using anti-CD123-PE; anti-CD11c-APC or anti-CD11c-PE; a mixture of FITC-labeled mAbs against CD3, CD14, CD15, CD16, CD19, and CD56; and anti-CD4-APC-Cy7 to reach greater than 99% purity according to BDCA2 expression. Blood monocytes or B cells were obtained using CD14 microbeads or CD19 microbeads (Miltenyi Biotec, Auburn, CA), followed by cell sorting by staining with anti-CD14-PE or anti-CD19-PE, respectively. The purity of these cells was greater than 99%. Immature monocyte-derived DCs were obtained by 5-day culture of CD14+ monocytes with granulocyte macrophate colony-stimulating factor (GM-CSF) (100 ng/mL) and IL-4 (200 ng/mL). BDCA4+ cells were positively isolated directly from PBMCs using a BDCA4+ Isolation Kit (Miltenyi Biotec) and were stained with anti-CD123-PE, anti-CD3-PE, anti-CD14-PE, or anti-CD56-PE; anti-BDCA2-FITC; anti-CD11c-APC; and anti-HLA-DR-PerCP. Cells were seeded at a density of 105 cells/200 μL medium in flat-bottomed, 96-well plates with each virus, each TLRL alone, TLRL plus irradiated CD40L-transfected L cells (at a 1:4 ratio), or medium alone. In some experiments, 5 × 104 CD11c+ mDCs were cocultured with different numbers of pDCs for 24 hours with R848 alone or with CpG plus CD40L-transfected L cells. IFN-α 2a (1000 U/mL), a mixture of neutralizing rabbit anti-human IFN-α Ab (2000 neutralization U/mL), neutralizing rabbit anti-human IFN-β Ab (1000 neutralization U/mL), and mouse anti-human IFN-α/β receptor mAb (10 μg/mL) (all from PBL Biomedical Laboratories, Piscataway, NJ) or a mixture of rabbit immunoglobulin (IgG) and mouse IgG (both from R&D Systems) was added to the mDC-pDC cocultures.
Measurement of cytokine production
Supernatants of each APC subset were collected 4, 8, 12, 24, 36, and 48 hours after stimulation. After supernatant removal and extensive cell wash, the adjusted number of viable cells (105 cells/200 μL medium) were recultured for another 12 or 24 hours. Production of IL-12 p70, IL-10, tumor necrosis factor-α (TNF-α), IL-6 (kits from R&D Systems), and IFN-α (kits from PBL) in the supernatants was determined by enzyme-linked immunosorbent assay (ELISA).
Intracellular cytokine analyses
Intracellular cytokine staining in DC subsets was performed after 8, 10, or 12 hours of culture with different TLRLs plus CD40L-transfected L cells. Brefeldin A (10 μg/mL; Sigma, St Louis, MO) was added during the last 2 hours. After stimulation, cells were fixed and permeabilized using the Fix and Perm kit (Caltag, Burlingame, CA) and were stained with FITC-labeled anti-IFN-α2 mAb (Chromaprobe, Maryland Heights, MO) and APC-labeled anti-IL-12 p40+p70 mAb (BD Biosciences).
Microarray analysis
Total RNA from pDCs and CD11c+ mDCs, freshly isolated or activated with viruses or TLRLs for 20 hours, were immediately isolated with the RNeasy kit (Qiagen, Valencia, CA) and were used to generate cDNA according to the Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA). cRNA samples were generated with the Bioarray High-Yield RNA Transcript Labeling kit (ENZO, Farmingdale, NY) and the human genome U133 plus 2.0 array according to the manufacturer's protocol (Affymetrix). Scanned images were aligned and analyzed using the Affymetrix GeneChip software Microarray Suite 5.0 according to the manufacturer's instructions. Signal intensities were normalized to the mean intensity of all the genes represented on the array, and global scaling (scaling to all probe sets) was applied before comparison analysis. For genes represented by multiple probe sets, results for only one representative probe set are shown.
Real-time PCR and reverse transcriptase PCR
Total RNA was extracted with the Qiagen RNeasy mini protocol and was converted to cDNA using oligo-dT, random hexamers, and SuperScript II RT (Invitrogen, Carlsbad, CA). Real-time polymerase chain reaction (PCR) was performed using a sequence detector (model ABI PRISM 9600; PerkinElmer, Wellesley, MA) and target mixes (Applied Biosystems, Foster City, CA): IFN-α1 (Hs00256882_s1), IFN-α2 (Hs00265051_s1), IFN-β (Hs00277188_s1), IFN-ω (Hs00357857_s1), IFN-λ1 (Hs00601677_g1), IFN-γ (Hs00174143_m1), IL-12 p35 (Hs00168405_m1), IRF1 (Hs00233698_m1), IRF3 (Hs00155574_m1), IRF4 (Hs00180031_m1), IRF5 (Hs00158114_m1), IRF7 (Hs00242190_g1), IRF8 (Hs00175238_m1), C/EBPα (Hs00269972_s1), C/EBPβ (Hs00270923_s1), C/EBPγ (Hs00156454_m1), c-REL (Hs00231279_m1), ETS2 (Hs00232009_m1), PU.1 (Hs00231368_m1), and 18s (Hs99999901_s1). Threshold cycle (CT) values for each gene were normalized to 18s using the equation 2.0(18s-GENE) (105), where 18s was the CT obtained for 18s, GENE was the CT of the gene of interest, and 105 was an arbitrarily chosen factor to bring all values above 0. Reverse transcriptase (RT-PCR) was performed using primers of IFN-α and S14, as previously described.5
Suppression subtraction hybridization
Suppression subtraction hybridization was performed with total RNA prepared after 6- to 8-hour incubation of purified pDCs with IL-3 (10 ng/mL) and HSV-1 (10 PFU/cell) or with IL-3 alone. Material from several donors was pooled for each condition. The cDNAs were amplified according to the SMARTtechnology protocol (BD Clontech, Palo Alto, CA). Subsequent hybridization was performed using IL-3 and HSV-1 cDNA as tester and IL-3 cDNA as driver with the PCR-Select kit protocol (BD Clontech), generating a library enriched in HSV-1-induced pDC transcripts. Clones from this library were automatically sequenced (Applied Biosystems), and sequences were compared against GenBank and dbEST databases using BLAST.
Ethical principles
This study was approved by the institutional review board at the University of Texas M.D. Anderson Cancer Center and was conducted in accordance with the tenets of the Declaration of Helsinki.
Results
Differential capacity of pDCs and mDCs to produce IL-12 and IFN-α in response to TLR signaling
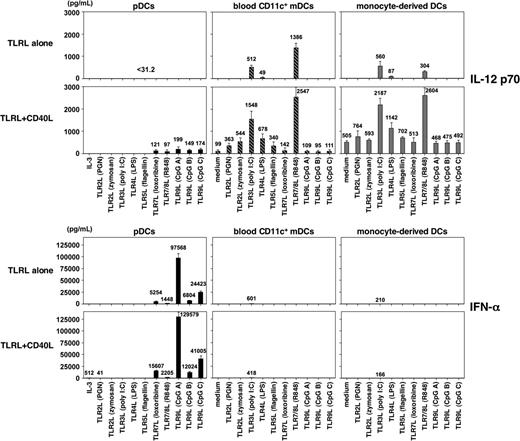
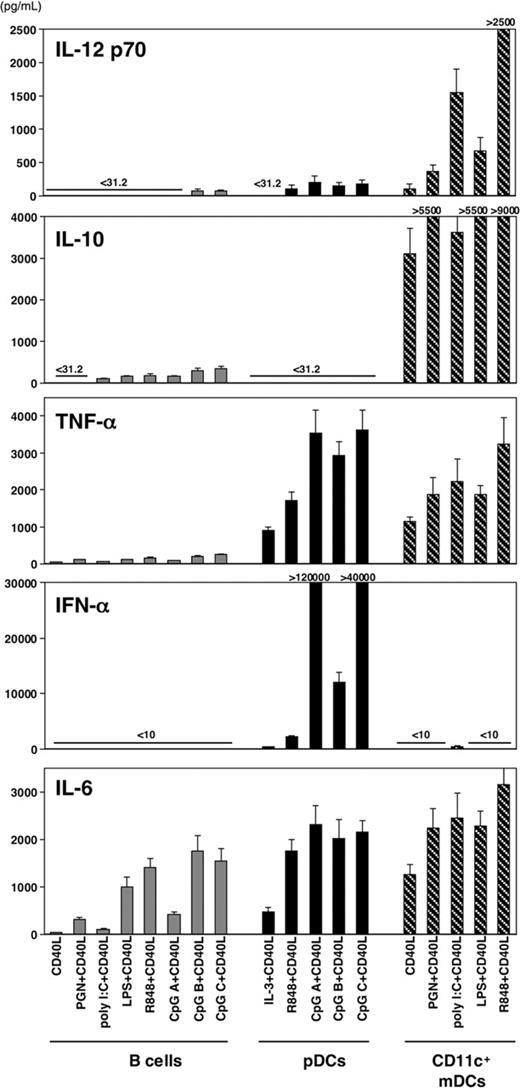
To clarify the previous conflicting data on the ability of pDCs to produce IL-12 and the ability of mDCs to produce IFN-α, it is important to carry out experiments using highly purified cell populations and the microbial ligand to each of the known TLRs. Human pDCs, CD11c+ mDCs, and CD14+ monocytes were isolated from peripheral blood by cell sorting to greater than 99% purity. Monocytes were cultured with GM-CSF plus IL-4 for 5 days to generate immature monocyte-derived DCs (mono-DCs). pDCs, CD11c+ mDCs, and mono-DCs were cultured with 10 TLRLs, and their ability to produce cytokines IFN-α and IL-12 p70 were evaluated by ELISA (Figure 1). Consistent with their TLR expression pattern, pDCs produced IFN-α, but not IL-12, in response to TLR7Ls (loxoribine and R848) and TLR9Ls (CpGs A, B, and C). CD11c+ mDCs and mono-DCs produced IL-12 p70 in response to TLR3L (poly(I:C)), TLR4L (LPS), and TLR8L (R848) but only a small amount of IFN-α in response to TLR3L. The maximal IFN-α production by pDCs in response to CpG A was at least 150 times more than the maximal IFN-α produced by mDCs in response to poly(I:C) (97 568 ± 9054 pg/mL compared with 601 ± 183 pg/mL). These results confirm and extend previous findings7,8 that human pDCs preferentially produce IFN-α and mDCs preferentially produce IL-12 in response to appropriate TLRLs.
Capacity of DC subsets to produce IL-12 and IFN-α in response to TLRL. Human pDCs, CD11c+ mDCs, and in vitro-generated mono-DCs from the same donor isolated to more than 99% purity were cultured with different TLRLs individually and TLRLs plus CD40L. IL-3 was added only in the pDC culture without TLRL. After 24 hours, the concentrations of IL-12 p70 and IFN-α in the culture supernatants were measured by ELISA. Data are shown as mean ± SEM of 5 independent experiments; mean values are indicated above the bars. Undetectable level for IL-12 p70 is less than 31.2 pg/mL; that for IFN-α is less than 10 pg/mL.
Capacity of DC subsets to produce IL-12 and IFN-α in response to TLRL. Human pDCs, CD11c+ mDCs, and in vitro-generated mono-DCs from the same donor isolated to more than 99% purity were cultured with different TLRLs individually and TLRLs plus CD40L. IL-3 was added only in the pDC culture without TLRL. After 24 hours, the concentrations of IL-12 p70 and IFN-α in the culture supernatants were measured by ELISA. Data are shown as mean ± SEM of 5 independent experiments; mean values are indicated above the bars. Undetectable level for IL-12 p70 is less than 31.2 pg/mL; that for IFN-α is less than 10 pg/mL.
Cytokine production by pDCs and mDCs in response to TLR and CD40 dual signaling
A recent study suggests that human pDCs/IPCs may require 2 signals mediated by TLRL and CD40L to produce IL-12.9 Therefore, we examined the ability of highly purified pDCs, mDCs, and mono-DCs to produce IL-12 and IFN-α in response to each of the known TLRLs in the presence of CD40L (Figure 1). CD40L strongly promoted IFN-α production by pDCs induced by the TLR7Ls and the TLR9Ls. In contrast, CD40L failed to induce mDCs to produce IFN-α in combination with TLR2L, TLR4L, TLR5L, or TLT7/8L and failed to promote IFN-α production by mDCs induced by TLR3L. Dual signaling by CD40L in combination with the TLR7Ls or TLR9Ls did induce pDCs to produce small amounts of IL-12 p70 (97-199 pg/mL); however, dual signaling by CD40L plus the TLR3L or TLR8L synergistically promoted mDC subsets to produce 10 times more IL-12 p70 (1548-2547 pg/mL from mDCs and 2187-2604 pg/mL from mono-DCs). Although individual signals by TLR2L and TLR5L failed to induce mDC subsets to produce bioactive IL-12 p70, dual signaling with these TLRLs and CD40L were capable of inducing IL-12 p70 production by mDCs, as previously demonstrated.18,19
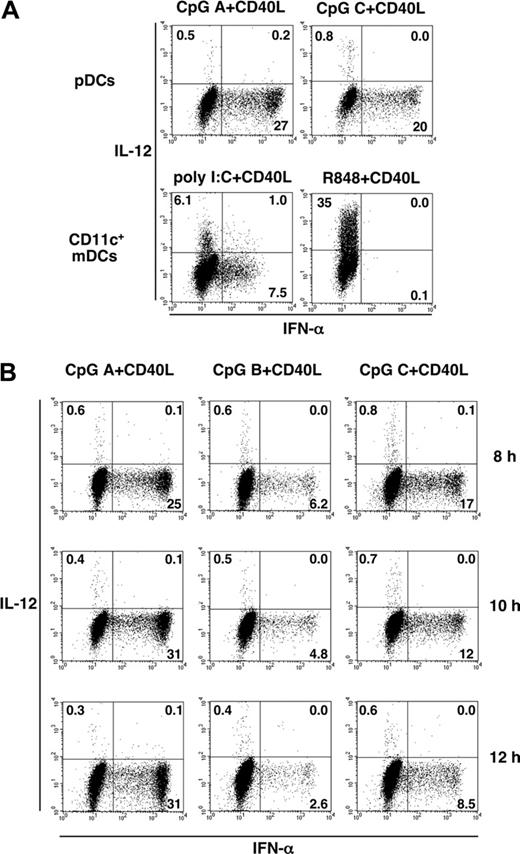
The ability of pDCs and mDCs to produce IFN-α and IL-12 was further examined by 2-color staining by intracellular anti-IFN-α and anti-IL-12 at the single-cell level (Figure 2). At 8 hours after activation by CD40L plus CpG A or CpG C, less than 1% of pDCs produced IL-12, but 20% to 27% of pDCs produced high levels of IFN-α (Figure 2A, upper panels). At 8 hours after activation by CD40L plus R848, less than 1% of mDCs produced IFN-α, but 35% of mDCs produced IL-12. At 8 hours after activation by CD40L plus poly(I:C), some mDCs produced IL-12 (6.1%) and others produced IFN-α (7.5%) (Figure 2A, lower panels). Few cells among the mDCs and pDCs produced both IL-12 and IFN-α after activation by CD40L plus TLRL, consistent with the findings of Duramad et al.20 Although 7.5% of the mDCs were found to produce IFN-α in response to poly(I:C) plus CD40L, their level of intracellular IFN-α expression was 1 log lower than that of activated pDCs.
We next investigated the kinetics of intracellular expression of IFN-α and IL-12 in pDCs at 8, 10, and 12 hours after stimulation. The proportion of IL-12-producing cells did not exceed 1% at any time point examined (Figure 2B), suggesting that pDCs have limited ability to produce IL-12.
BDCA4+ pDCs/IPCs contain a fraction of CD11c+ mDCs that contribute to IL-12 production
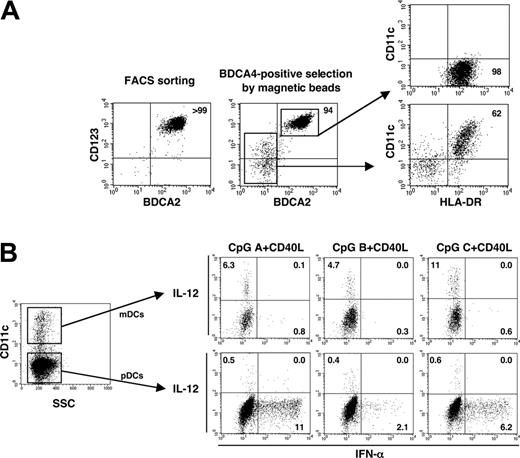
The conclusion that pDCs/IPCs have the capacity to produce as much IL-12 as mDCs was based on a study using human pDCs isolated by anti-BDCA4 magnetic beads and activation using CD40L plus CpG-ODN.9 We had 3 major concerns about this study: (1) it is difficult to obtain a highly purified population of pDCs by magnetic bead separation; (2) BDCA4, also known as neuropilin-1, is expressed not only by pDCs but also by a small population of mDCs, CD3+CD57+ T cells, and B cells21 ; (3) using CD40L and CpG-ODN as stimuli to test the capacity of pDCs and mDCs to produce IL-12 is not appropriate because, though pDCs are activated by CD40L and CpG-ODN, human mDCs are activated by CD40L only, given that they do not express TLR9 and are unable to respond to CpG-ODN.7
To investigate whether BDCA4+ cells isolated by magnetic bead sorting contained mDCs that might have contributed to IL-12 production, we first examined the composition of the cell population. In contrast to the population purified by cell sorting using a combination of several markers (fraction of lineage-/CD11c-/CD123high/CD4+), the population positively selected by BDCA4-conjugated magnetic beads contained approximately 5% to 6% BDCA2- cells (Figure 3A). We found that more than 60% of these BDCA2- cells expressed CD11c and HLA-DR (Figure 3A), but they did not express CD14 CD3, or CD56 (data not shown), indicating that BDCA4+ cells isolated by magnetic bead selection contained CD11c+ mDCs. These BDCA4+ cells were activated by CD40L plus each of the 3 types of CpG-ODNs, and intracellular IFN-α and IL-12 and surface CD11c expression were analyzed by 3-color immunofluorescence flow cytometry. Although IL-12-producing cells were found exclusively within the CD11c+-contaminating mDCs, IFN-α-producing cells were found within the CD11c- pDC population (Figure 3B). Few cells within either the CD11c+ mDC population or the CD11c- pDC population produced both IL-12 and IFN-α. These results suggested that IL-12 production by BDCA4+ cells isolated by magnetic bead sorting was mainly derived from the contaminating CD11c+ mDCs rather than from CD11c- pDCs/IPCs in response to CpG plus CD40L. In addition, few mDCs or pDCs were found to produce both IL-12 and IFN-α.
Intracellular cytokine analyses in pDCs and CD11c+ mDCs in response to TLRL plus CD40L. pDCs and CD11c+ mDCs purified by cell sorter to more than 99% purity were cultured with TLRLs plus CD40L. (A) After 8 hours, intracellular cytokine production by activated pDCs and CD11c+ mDCs was analyzed by flow cytometry. (B) After 8, 10, and 12 hours of culture, cytokine production by activated pDCs was analyzed by flow cytometry. Percentages of the cytokine-producing DCs are indicated in each dot blot profile. Data are from 1 of 3 independent experiments.
Intracellular cytokine analyses in pDCs and CD11c+ mDCs in response to TLRL plus CD40L. pDCs and CD11c+ mDCs purified by cell sorter to more than 99% purity were cultured with TLRLs plus CD40L. (A) After 8 hours, intracellular cytokine production by activated pDCs and CD11c+ mDCs was analyzed by flow cytometry. (B) After 8, 10, and 12 hours of culture, cytokine production by activated pDCs was analyzed by flow cytometry. Percentages of the cytokine-producing DCs are indicated in each dot blot profile. Data are from 1 of 3 independent experiments.
pDCs/IPCs promote IL-12 production by mDCs in response to TLRL or CD40L
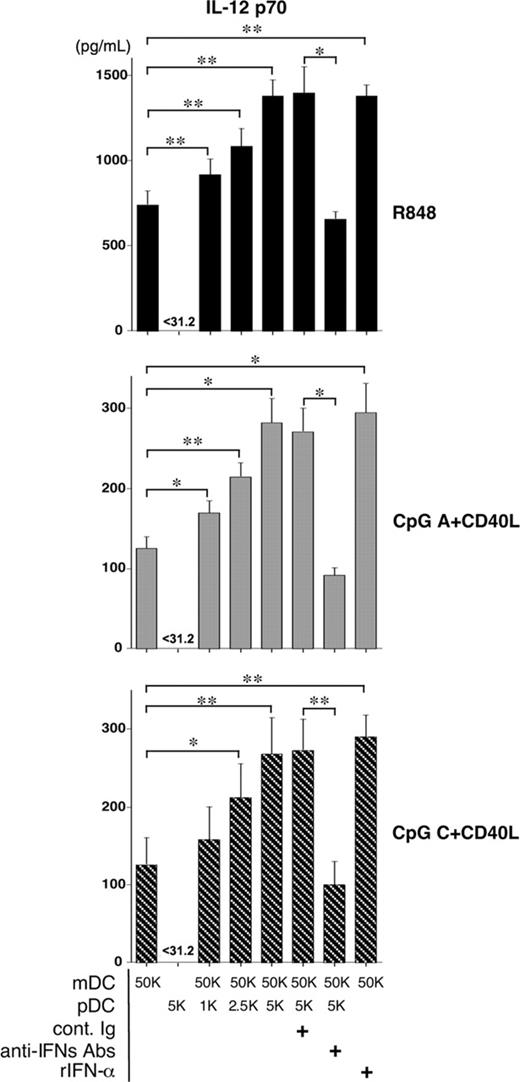
It has been reported that type 1 IFNs may positively or negatively regulate IL-12 production by mDCs, depending on their maturation stage.22,23 We therefore hypothesized that after activation by CD40L plus CpG or R848, pDCs would be able to regulate IL-12 production by mDCs through type 1 IFNs. CD11c+ mDCs were cultured with R848 or CpG plus CD40L for 24 hours, in the absence or presence of different numbers of pDCs at 50:1, 20:1, and 10:1 ratios. mDCs, but not pDCs, produced high levels of IL-12 p70 after stimulation (Figure 4). Adding pDCs to the mDC culture promoted IL-12 production by mDCs in a dose-dependent fashion. The ability of pDCs to promote IL-12 production by mDCs depended on type 1 IFNs because neutralizing antibodies to type 1 IFNs significantly blocked the enhancement of IL-12 production in the cocultures of pDCs and mDCs. Statistical analyses of the above data showing that the ability of type 1 IFNs produced by pDCs to enhance IL-12 production by mDCs are significant. These data suggest that pDCs activated by virus or TLRs have the ability to promote IL-12 production by mDCs activated by CD40L or TLR8L. They also suggest the possibility that the CD11c+ mDCs contaminating the BDCA4+ pDC preparations may be helped by pDCs to produce IL-12 in cultures with CpG and CD40L.
BDCA4+pDCs isolated by magnetic beads contain CD11c+mDCs that contribute to IL-12 production. (A) pDCs isolated by cell sorter or by BDCA4-conjugated magnetic beads were analyzed by staining with anti-CD123, anti-BDCA2, anti-CD11c, and anti-HLA-DR mAbs. Data shown are from 1 of 4 independent donors. (B) Using the cells isolated by BDCA4-conjugated magnetic beads, intracellular cytokine production and surface CD11c expression were analyzed by flow cytometry after 8 hours of stimulation with CpG plus CD40L. Numbers in each dot blot profile indicate the percentages of the indicated fractionated cells. Data are from 1 of 3 independent experiments.
BDCA4+pDCs isolated by magnetic beads contain CD11c+mDCs that contribute to IL-12 production. (A) pDCs isolated by cell sorter or by BDCA4-conjugated magnetic beads were analyzed by staining with anti-CD123, anti-BDCA2, anti-CD11c, and anti-HLA-DR mAbs. Data shown are from 1 of 4 independent donors. (B) Using the cells isolated by BDCA4-conjugated magnetic beads, intracellular cytokine production and surface CD11c expression were analyzed by flow cytometry after 8 hours of stimulation with CpG plus CD40L. Numbers in each dot blot profile indicate the percentages of the indicated fractionated cells. Data are from 1 of 3 independent experiments.
pDCs and B cells produce different cytokines in response to TLR7 and TLR9 signaling
Previous studies in humans and mice showed that pDCs express many genes related to B lymphocytes—including RAG1, RAG2, SPIB, and λ5—and IgH d-J gene rearrangements.24-26 In the human system, pDCs and B cells express TLR7 and TLR9.27,28 The question was whether pDCs and B cells make similar cytokine responses to signaling through TLR7 and TLR9. To address this, human pDCs, CD11c+ mDCs, and CD19+ B cells were isolated from the same donor and were cultured for 24 hours with each of the TLRLs plus CD40L, and then cytokine production was analyzed by ELISA (Figure 5). mDCs produced large amounts of IL-10, TNF-α, IL-6, and IL-12 but not IFN-α. pDCs produced large amounts of IFN-α, TNF-α, and IL-6 but small amounts of IL-12 and no IL-10. B cells produced large amounts of IL-6 but low levels of IL-12, TNF-α, and IL-10 and no IFN-α. pDCs did not produce detectable amounts of IL-10, and B cells did not produce detectable amounts of IFN-α. These data suggest that although pDCs and B cells express TLR7 and TLR9, they produced different cytokines in response to TLR7 and TLR9 signaling in the presence of CD40L.
pDCs promote IL-12 production by mDCs through type 1 IFNs. Purified CD11c+ mDCs were cocultured with or without different doses of autologous purified pDCs for 24 hours in the presence of the indicated stimuli. Numbers of mDCs and pDCs indicate 103 × the cell number per well. Concentrations of IL-12 p70 in the supernatant cultures were measured by ELISA. Mean ± SEM obtained from 3 independent experiments are shown. Statistical significance was determined using paired Student t test. *P < .05; **P < .01. rIFN-α; recombinant human IFN-α 2a, anti-IFNs Abs; mixture of neutralizing Abs to IFN-α, IFN-β Ab, and IFN-α/β receptor, cont. Ig; mixture of rabbit IgG and mouse IgG.
pDCs promote IL-12 production by mDCs through type 1 IFNs. Purified CD11c+ mDCs were cocultured with or without different doses of autologous purified pDCs for 24 hours in the presence of the indicated stimuli. Numbers of mDCs and pDCs indicate 103 × the cell number per well. Concentrations of IL-12 p70 in the supernatant cultures were measured by ELISA. Mean ± SEM obtained from 3 independent experiments are shown. Statistical significance was determined using paired Student t test. *P < .05; **P < .01. rIFN-α; recombinant human IFN-α 2a, anti-IFNs Abs; mixture of neutralizing Abs to IFN-α, IFN-β Ab, and IFN-α/β receptor, cont. Ig; mixture of rabbit IgG and mouse IgG.
Capacity of B cells, pDCs, and CD11c+mDCs to produce cytokines. pDCs, CD11c+ mDCs, and CD19+ B cells isolated from the same donor were cultured with TLRL plus CD40L. After 24 hours, the concentrations of IL-12 p70, IL-10, IL-6, TNF-α, and IFN-α in the culture supernatants were measured by ELISA. Data are shown as mean ± SEM of 5 independent experiments.
Capacity of B cells, pDCs, and CD11c+mDCs to produce cytokines. pDCs, CD11c+ mDCs, and CD19+ B cells isolated from the same donor were cultured with TLRL plus CD40L. After 24 hours, the concentrations of IL-12 p70, IL-10, IL-6, TNF-α, and IFN-α in the culture supernatants were measured by ELISA. Data are shown as mean ± SEM of 5 independent experiments.
pDCs/IPCs do not make secondary type 1 IFN responses
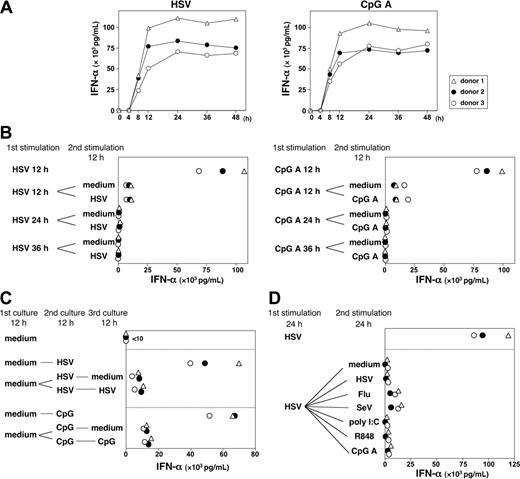
Although pDCs/IPCs promptly produced large amounts of type 1 IFNs after stimulation by virus or CpG-ODN,5,29 it has been unclear whether activated pDCs/IPCs can make secondary type 1 IFNs on restimulation. We first analyzed the kinetics of IFN-α production by pDCs in response to HSV and CpG A. pDCs secreted large amounts of IFN-α at 8 hours after stimulation by HSV and CpG A (Figure 6A). The IFN-α response plateaued at 12 hours after HSV or CpG activation and maintained that level for up to 48 hours. To determine whether pDCs could make a secondary IFN-α response after the initial IFN-α production on primary stimulation with HSV or CpG A, pDCs preactivated by HSV or CpG A or precultured with medium alone for 12 hours were collected and washed, and the same number of viable pDCs was recultured with medium or was restimulated with HSV or CpG A for another 12 hours. Figure 6B shows that pDCs preactivated with HSV or CpG A for 12, 24, or 36 hours lost the ability to produce large amounts of IFN-α on restimulation during the next 12 hours. By contrast, pDCs precultured with medium alone for 12 hours maintained the ability to produce large amounts of IFN-α on stimulation with HSV or CpG A, and they promptly lost the ability to produce IFN-α on next restimulation (Figure 6C). To further determine whether pDCs could make secondary IFN-α responses to an original stimulus or to different stimuli, pDCs were cultured with HSV for 24 hours and were subsequently restimulated with HSV or with different stimuli, including RNA viruses (influenza virus and Sendai virus) and TLRLs (poly(I:C), R848, and CpG A). pDCs produced much IFN-α during the first 24 hours of HSV stimulation and then produced less IFN-α whether without stimulation or with the original stimulus or with different stimuli (Figure 6D). These findings suggest that pDC/IPCs produced a large amount of IFN-α within the first 12 hours of activation by virus but then did not have the ability to mount a secondary IFN-α response to further stimulation by the same or by different viruses, which is a key feature of the innate immune response.
Kinetics of IFN-α antiviral response by pDCs. (A) pDCs were cultured with HSV-1 or CpG A. Concentrations of IFN-α in pDC culture supernatants at different time points (4, 8, 12, 24, 36, and 48 hours) after stimulation were measured by ELISA. (B) pDCs were first cultured for 12, 24, or 36 hours with HSV or CpG A. After extensive washing, the same number of viable pDCs was restimulated with the same stimulus or with medium alone for another 12 hours. (C). pDCs were first cultured for 12 hours with medium alone. After washing, the same number of viable pDCs was then stimulated with HSV or CpG A for another 12 hours. Cells were washed again and restimulated with the same stimulus or with medium alone for 12 more hours. (D) pDCs were first cultured for 24 hours with HSV. After extensive washing, the same number of viable pDCs was restimulated with the several stimuli for the next 24 hours. The concentration of IFN-α in each supernatant was measured in duplicate by ELISA. Three independent experiments are shown as ▵, ○, and •. Flu indicates influenza virus; SeV, Sendai virus.
Kinetics of IFN-α antiviral response by pDCs. (A) pDCs were cultured with HSV-1 or CpG A. Concentrations of IFN-α in pDC culture supernatants at different time points (4, 8, 12, 24, 36, and 48 hours) after stimulation were measured by ELISA. (B) pDCs were first cultured for 12, 24, or 36 hours with HSV or CpG A. After extensive washing, the same number of viable pDCs was restimulated with the same stimulus or with medium alone for another 12 hours. (C). pDCs were first cultured for 12 hours with medium alone. After washing, the same number of viable pDCs was then stimulated with HSV or CpG A for another 12 hours. Cells were washed again and restimulated with the same stimulus or with medium alone for 12 more hours. (D) pDCs were first cultured for 24 hours with HSV. After extensive washing, the same number of viable pDCs was restimulated with the several stimuli for the next 24 hours. The concentration of IFN-α in each supernatant was measured in duplicate by ELISA. Three independent experiments are shown as ▵, ○, and •. Flu indicates influenza virus; SeV, Sendai virus.
Dominant IFN-gene transcription in pDCs after viral activation
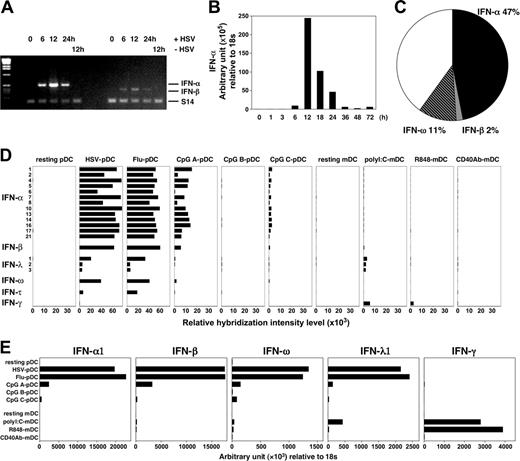
Immunoglobulin-secreting plasma cells contain abundant rearranged immunoglobulin mRNA transcript. Because pDCs are specialized in producing type 1 IFNs, we further investigated (1) whether pDCs contained preexisting mRNA transcripts for type 1 IFNs before viral stimulation, (2) the kinetics of type 1 IFN mRNA transcription, and (3) the quantity of type 1 IFN transcripts compared with other gene transcripts. We found that pDCs do not contain preexisting mRNA for type 1 IFNs before activation by viruses (Figure 7A-B). However, within a few hours of activation by HSV, IFN-α and IFN-β mRNAs were detected in pDCs. The levels of IFN-α and IFN-β mRNA peaked at 12 hours after HSV activation. To analyze gene expression by pDCs induced by HSV, a subtracted hybridization technique was applied. We constructed a cDNA library from pDCs activated by HSV and IL-3 (as tester) subtracted from pDCs activated by IL-3 only (as driver) for 6 to 8 hours. Within this cDNA library, we analyzed the numbers of transcripts for IFN-α, IFN-β, and IFN-ω in 258 random sequences. We found that IFN transcripts represented approximately 60% of all sequences (Figure 7C), including 47% IFN-α (121 sequences), 2% IFN-β (5 sequences), and 11% IFN-ω (29 sequences). These data suggest that in response to viral infection, pDCs may dedicate a large proportion of their transcription machinery to making type 1 IFNs.
Repertoire of type 1 IFN responses by pDCs/IPCs
In humans, the type 1 IFN family consists of at least 13 IFN-α members and single members of IFN-β, IFN-ω, and IFN-τ. The recently described IFN-λ1 (alternatively called IL-29), IFN-λ2 (IL-28A), and IFN-λ3 (IL-28B) are distantly related to type 1 IFNs based on their sequence similarities and antiviral activity.30,31 To determine the type 1 IFN repertoire expressed by pDCs/IPCs in an antiviral response, we performed microarray gene expression analyses in human pDCs resting or activated by DNA virus (HSV-1), RNA virus (influenza virus), or 3 types of CpGs for 20 hours. Relative expression levels of the genes encoding members of the type 1 family and type 2 IFN (IFN-γ) were analyzed. We showed again that pDCs did not express mRNA for type 1 IFNs before stimulation (Figure 7D). The 2 viruses and CpG A stimulated pDCs to express high levels of mRNA for type 1 IFNs, including 13 subtypes of IFN-α, IFN-β, IFN-ω, 3 subtypes of IFN-λ, and IFN-τ, but not IFN-γ. CpG C induced pDCs to express lower levels of type 1 IFN mRNA, in contrast to CpG A. CpG B did not induce pDCs to express marked levels of type 1 IFN mRNA. CD11c+ mDCs activated by poly(I:C) expressed a low level of mRNA for type 1 IFNs but a significant level of IFN-γ. Of these results of microarray analyses, mRNA expression of IFN-α1, IFN-β, IFN-ω, IFN-λ1, and IFN-γ were also quantitatively confirmed by real-time PCR (Figure 7E). These data suggest that pDCs can produce a wide repertoire of type 1 IFNs in response to viral infection.
pDCs/IPCs and mDCs express different sets of transcriptional factors critical for type 1 IFNs compared with IL-12 synthesis
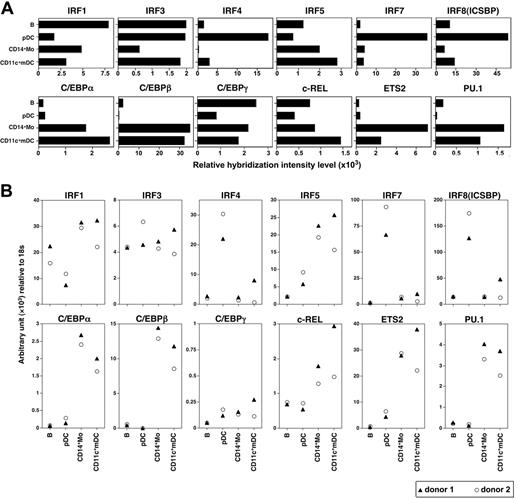
To further understand the molecular basis underlying the specialization of pDCs and mDCs in producing type 1 IFNs compared with IL-12, we analyzed the expression of 2 sets of transcriptional factors, IFN regulatory factors (IRFs) and the transcriptional factors critical for IL-12 production in pDCs and mDCs, together with B cells and monocytes by microarray gene expression analysis (Figure 8A) and real-time PCR (Figure 8B). pDCs selectively and constitutively expressed IRF4, IRF7, and IRF8. By contrast, monocytes and mDCs constitutively expressed IRF1 and IRF5 and selectively expressed CCAAT/enhancer-binding protein (C/EBP)-α, C/EBP-β, c-Rel, ETS2, and PU.1, transcriptional factors critical for IL-12 production.32
Dominant IFN gene transcription in pDCs after viral activation. (A-B) Purified pDCs were cultured with HSV-1, and cells were collected at different times after stimulation; mRNA expression of IFN-α and IFN-β were then examined by RT-PCR (A) and real-time quantitative PCR (B). (C) Suppression subtraction hybridization was performed with total RNA prepared from purified human pDCs activated with IL-3 and HSV-1, and from cells incubated with only IL-3 for 6 to 8 hours. Percentages of transcripts for IFN-α (121 sequences), IFN-β (5 sequences) and IFN-ω (29 sequences) within 258 random sequences are indicated. (D-E) Expression profiles of the type 1 and 2 IFN family genes in the DCs were analyzed by microarray (D) and real-time quantitative PCR (E). Each graph represents data from pDCs and CD11c+ mDCs, either resting or activated by virus or TLRL for 20 hours. Results of the gene expression profiles (D) are shown as relative hybridization intensity levels in the microarray analysis. Flu indicates influenza virus.
Dominant IFN gene transcription in pDCs after viral activation. (A-B) Purified pDCs were cultured with HSV-1, and cells were collected at different times after stimulation; mRNA expression of IFN-α and IFN-β were then examined by RT-PCR (A) and real-time quantitative PCR (B). (C) Suppression subtraction hybridization was performed with total RNA prepared from purified human pDCs activated with IL-3 and HSV-1, and from cells incubated with only IL-3 for 6 to 8 hours. Percentages of transcripts for IFN-α (121 sequences), IFN-β (5 sequences) and IFN-ω (29 sequences) within 258 random sequences are indicated. (D-E) Expression profiles of the type 1 and 2 IFN family genes in the DCs were analyzed by microarray (D) and real-time quantitative PCR (E). Each graph represents data from pDCs and CD11c+ mDCs, either resting or activated by virus or TLRL for 20 hours. Results of the gene expression profiles (D) are shown as relative hybridization intensity levels in the microarray analysis. Flu indicates influenza virus.
pDCs and mDCs express different sets of transcriptional factors. Expression profiles of the indicated transcriptional factor genes in the resting B cells, pDCs, monocytes, and mDCs were analyzed by microarray (A) and real-time quantitative PCR (B). Results of the gene expression profiles (A) are shown as relative hybridization intensity levels in the microarray analysis. IRF indicates IFN regulatory factor; C/EBP, CCAAT/enhancer-binding protein; ICSBP, IFN consensus sequence-binding protein.
pDCs and mDCs express different sets of transcriptional factors. Expression profiles of the indicated transcriptional factor genes in the resting B cells, pDCs, monocytes, and mDCs were analyzed by microarray (A) and real-time quantitative PCR (B). Results of the gene expression profiles (A) are shown as relative hybridization intensity levels in the microarray analysis. IRF indicates IFN regulatory factor; C/EBP, CCAAT/enhancer-binding protein; ICSBP, IFN consensus sequence-binding protein.
Discussion
By using highly purified human pDC and mDC subpopulations, we demonstrated that human pDCs preferentially produced IFN-α and that mDCs preferentially produced IL-12 in response to TLR signaling or TLR and CD40 dual signaling. Using 2-color intracellular anti-IL-12 and anti-IFN-α staining and flow cytometric analyses, we found that more than 20% of pDCs expressed high levels of intracellular IFN-α, that less than 1% of pDCs expressed intracellular IL-12, and that no cells expressed both IL-12 and IFN-α in response to CpG and CD40L. Using this technology, we directly demonstrated that previous findings that human pDCs/IPCs produced large amounts of IL-12 in response to CpG plus CD40L resulted from the contaminated CD11c+ mDCs within the pDCs/IPCs isolated by anti-BDCA4 magnetic beads. Thus, when these human pDCs isolated by anti-BDCA4 magnetic beads were activated by CpG plus CD40L, the contaminating CD11c+ mDCs contained intracellular IL-12 but not IFN-α, and the CD11c- pDCs contained intracellular IFN-α but not IL-12. We also showed that neither mDCs nor pDCs produced both IL-12 and IFN-α in response to TLRL and CD40L. These data further support the fact that human pDCs and mDCs have limited plasticity in producing IFN-α and IL-12.
Recent studies have further defined the molecular mechanisms underlying the unique function of pDCs as professional type 1 IFN-producing cells. pDC/IPCs, but not other hematopoietic cell types, constitutively express high levels of IRF7, a critical transcriptional factor required for type 1 IFN production.33 On TLR9 signaling, MyD88 recruits and directly interacts with IRF7 and the adaptor molecule TRAF6. This molecular supercomplex, together with IRAK4 and IRAK1, forms a cytoplasmic transductional-transcriptional processor for type 1 IFN production by pDCs.34,35 pDCs, but not other cell types, have a unique capacity to retain CpG-ODN in the endosome compartment for a prolonged period, allowing direct signaling through TLR9 within the endosome to trigger massive type 1 IFN production.36 Although mDCs have the ability to produce IFN-α in response to poly(I:C) or poly(I:C) plus CD40L, the maximum amount of IFN-α produced by mDCs in these conditions can be 1/300 that produced by pDCs activated by CpG plus CD40L. Recently, Coccia et al29 showed that TLR3 stimulation by poly(I:C) induces IFN-β and IFN-λ gene expression in mono-DCs. Indeed, mDCs activated by poly(I:C) expressed mRNA of type 1 IFNs, including IFN-β and IFN-λ in the present study, but their levels were very low. In response to R848, a small molecular compound that triggers TLR7 on pDCs and TLR8 on mDCs,37 pDCs preferentially produce IFN-α and mDCs preferentially produce IL-12.38 These results underscore the important fact that pDCs and mDCs not only express different sets of TLRs, they are equipped with different intracellular molecular machineries to activate different sets of cytokine genes. Although human pDCs and B cells express TLR7 and TLR9, only pDCs produce a large amount of IFN-α in response to signaling through TLR7 or TLR9 alone or in combination with CD40L. In contrast, B cells, but not pDCs, have the ability to produce IL-10. Thus, although pDCs and B cells express the same set of TLRs, they are equipped with different intracellular molecular machineries to activate different sets of cytokine genes. Indeed, we found that pDCs, but not B cells, monocytes, or mDCs, selectively and constitutively expressed IRF7 critical for type 1 IFN production.34,35 In addition, pDCs selectively and constitutively expressed IRF4 and IRF8, but their function in regulating type 1 IFN production remains to be determined. By contrast, monocytes and mDCs, but not pDCs and B cells, selectively and constitutively expressed C/EBP-α, C/EBP-β, c-Rel, ETS2, PU.1, and IRF1. Although IL-12 p35 and p40 gene expression are complicated, these transcriptional factors are most likely critical for IL-12 production.32,39-45
After establishing that pDCs are professional type 1 IFN-producing cells, we further characterized the type 1 IFN responses by pDCs, similar to the characterization of antibody responses by B cells. We found that type 1 IFN responses by pDCs have the following features: response reaches its peak at 12 hours after pDC encounter with virus or CpG-ODN, pDCs produce only a fraction of type 1 IFN during the next 48 hours and do not make secondary type 1 IFN responses when stimulated further, pDCs dedicate approximately 60% of their new transcriptional activity to make type 1 IFNs, and pDCs make all transcripts of 19 different type 1 IFN subtypes tested. We conclude that the plasticity of DC function is limited by its unique surface expression of distinct sets of pattern recognition receptor, transcriptional factors, and selective cytokine responses to different microbial ligands. pDCs are professional type 1 IFN-producing cells that have the ability to rapidly produce large amounts of almost every type 1 IFN subtype. Their selective expression of TLR7 and TLR9 and the dedication of 60% of their transcription activity to make type 1 IFN reflect the evolutionary memory of the innate antiviral immune system.
Prepublished online as Blood First Edition Paper, November 17, 2005; DOI 10.1182/blood-2005-07-2709.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Karen Ramirez, Zhiwei He, and Eric Wieder for cell sorting and support.