Abstract
Humans and rodents exhibit a peculiar type of placentation in which zygote-derived trophoblast cells, rather than endothelial cells, line the terminal maternal vascular space. This peculiar aspect of the placental vasculature raises important questions about the relative contribution of fetal and maternal factors in the local control of hemostasis in the placenta and how these might determine the phenotypic expression of thrombophilia-associated complications of pregnancy. Using genomewide expression analysis, we identify a panel of genes that determine the ability of fetal trophoblast cells to regulate hemostasis at the fetomaternal interface. We show that spontaneous differentiation of trophoblast stem cells is associated with the acquisition of an endothelial cell–like thromboregulatory gene expression program. This program is developmentally regulated and conserved between mice and humans. We further show that trophoblast cells sense, via the expression of protease activated receptors, the presence of activated coagulation factors. Engagement of these receptors results in cell-type specific changes in gene expression. Our observations define candidate fetal genes that are potential risk modifiers of maternal thrombophilia-associated pregnancy complications and provide evidence that coagulation activation at the fetomaternal interface can affect trophoblast physiology altering placental function in the absence of frank thrombosis.
Introduction
In the hemochorial type of placentation observed in humans and mice, fetal nutrition involves the direct uptake of nutrients by zygote-derived trophoblast cells from circulating maternal blood. The required placental morphology is achieved through a highly regulated process of trophoblast differentiation coupled with remodeling of maternal and fetal vasculature. As a consequence, in contrast to all other vascular beds in which the blood vessel endothelium is the principal gatekeeper between tissue and blood, the terminal vascular space of the placenta is lined by trophoblast cells.1,2 Trophoblast cells are genetically distinct from the maternal vascular endothelium and are derived from a different developmental lineage than endothelial cells.3 In all nonplacental vascular beds, normal endothelium proactively suppresses the activity of the coagulation system, thereby maintaining a nonthrombogenic surface. A survey of existing data suggests that trophoblast cells also produce endothelial regulators of hemostasis, such as thrombomodulin (TM), endothelial protein C receptor (EPCR), and tissue factor pathway inhibitor (TFPI).4-8 Such findings indicate that trophoblast cells might exhibit an endothelial cell-like ability to partake in the regulation of hemostasis at the fetomaternal interface. Indeed, the term “endothelial mimicry” has been coined to describe a process of remodeling of the maternal arteries, during which so-called “endovascular” trophoblast cells replace the maternal endothelium in these blood vessels and switch their expression from epithelial to endothelial adhesion receptor repertoire.9-11 It is unknown whether trophoblast cells acquire anticoagulant gene expression in a temporally and spatially controlled manner similar to that described for the subset of endovascular trophoblast cells or whether the acquisition of an endothelial cell-like anticoagulant phenotype is a cell type-defining feature of trophoblast cells in general.
The placenta also is a rich source of the initiator of coagulation, tissue factor (TF). TF antigen and procoagulant activity are detected in mouse giant and labrynthine trophoblasts and on human syncytiotrophoblast membranes.12-15 With the exception of angiogenic endothelium, and in endothelium subjected to inflammatory and thrombotic stimuli, TF expression is strictly excluded from endothelial cells. Proinflammatory cytokines, ligands for Toll-receptors, and the principal coagulation protease, thrombin, induce TF expression in cultured endothelial cells, evoke increased production of endothelial-leukocyte adhesion receptors, and simultaneously suppress the expression of anticoagulant gene products. This transition from a noncoagulant and antiadhesive phenotype to a state of enhanced coagulation and leukocyte interactions has been termed “endothelial activation” and appears to reflect a principal switch in a concerted gene-expression program.16 In contrast, trophoblast cells constitutively express TF, thus exhibiting, even under normal conditions, a hallmark of activated endothelium. At least in mice, constitutive expression of tissue factor by placental trophoblast cells is essential for normal placental function.14
Constitutive TF expression at the blood-tissue interface sets the vascular bed of the placenta aside from the circulatory system of other organs. This procoagulant feature of trophoblast cells could, unless tightly controlled by anticoagulant mechanisms, predispose the placental vascular bed to organ-specific thrombosis. Indeed, inherited and acquired thrombophilia of the mother, such as that caused by factor V (fV) and prothrombin gene mutations, correlate with an increased incidence of fetal loss at various stages of gestation, and with other obstetric complications, such as pre-eclampsia, intrauterine growth restriction (IUGR), placental abruption, and stillbirth.17-21 Yet, the strength of the association between maternal thrombophilia and adverse pregnancy outcome is highly variable between studies, indicating the existence of as-yet-uncharacterized risk modifiers. In theory, prothrombotic alterations in fetal trophoblast function could dramatically affect the local haemostatic balance in the placenta, since this would be precisely the locale where such risk factors would compound the systemic thrombophilia of the mother.
Tissue factor activity produced by trophoblast cells also may affect the interplay between coagulation activation and cellular signaling processes mediated by receptors for activated coagulation factors. Contact of TF-bearing trophoblast cells with blood-borne fVII and fX would yield signaling-competent TF-VIIa and TF-VIIa-Xa complexes that can activate protease activated receptor (PAR) 1 and PAR 2 directly or indirectly via the TF-initiated production of the coagulation proteases thrombin and fXa. Such ligand-receptor interactions may couple coagulation activation to trophoblast physiology and thereby affect the development and function of the placenta. For example, thrombin inhibits the proliferation and regulates the invasiveness of trophoblast cells, and these effects are mediated at least in part through the engagement of PARs.13,22-24 The role of such receptor-mediated, cellular effects of activated coagulation proteases on the pathogenesis of placental dysfunction caused by maternal thrombophilia is unclear.
In the current study, we conduct a genomewide analysis of the thromboregulatory expression program that is acquired by trophoblast cells and delineate the in vivo expression pattern of thromboregulatory molecules in the placenta of mice and humans. Second, we investigate how the engagement of coagulation receptors alters the intrinsic phenotype of trophoblast cells, by characterizing the changes in gene expression induced by engagement of PAR1, 2, and 4 receptors on trophoblast cells.
Materials and methods
Cell culture
During placental organogenesis a small population of self-renewing and lineage-restricted fetal precursor cells (trophoblast stem cells; TS cells) give rise to these various subtypes of differentiated trophoblast cells (DT cells), which includes trophoblast giant cells (TGCs), spongiotrophoblast cells, and labyrinthine trophoblasts. TS cell cultures can be established from blastocyst outgrowths, and withdrawal of specific growth factors results in the spontaneous differentiation into DT cells.3 Mouse TS cells (kindly provided by J. Cross, University of Calgary, Calgary, Canada) were maintained as previously described.3 To obtain DT cells, TS cells were cultured without FGF4 and heparin for 96 hours. Jeg3 and BeWo human choriocarcinoma cell lines (ATCC, Manassas, VA) were maintained following the supplier's recommendations.
Microdissection of mouse placental tissues
Trophoblasts and embryos were microdissected from E8.5 fetuses obtained from timed CD1 mouse matings at 8.5 days postcoitum (dpc). The trophoblast shell containing the embryo was peeled from the surrounding decidual tissue and subsequently separated from Reichert's membrane, underlying visceral yolk sac, and the embryo proper. The staging was confirmed by microscopic observation. Tissue was stored in RNAlater (Ambion, Austin, TX) until analysis.
Microarray hybridization and data analysis
MGU74Av2 chips (Affymetrix, Santa Clara, CA) were used for all array experiments. Probe synthesis, hybridizations, washing, and scanning were performed according to Affymetrix protocols. Data were analyzed using MICROARRAY SUITE and DATA MINING TOOLS software (Affymetrix, Santa Clara, CA). “Present,” “absent,” “marginal” calls, and “difference” calls were assigned by applying the DATA MINING TOOLS algorithm to data from 4 independent profiling experiments. Final score reflecting aggregate results from all 4 experiments was determined using the following criteria. Probe sets that scored “present” in at least 2 of 4 expression profiles by the DATA MINING TOOLS algorithm were assigned a “present” call. The remaining probe sets that scored a signal of more than 60 (which equaled average background plus average noise in these experiments) in at least 2 of 4 expression profiles were assigned a “marginal” call. All the rest were assigned an “absent” call. A gene was assigned “increased” in DT only if (1) it scored as “increased” in all 4 comparisons, and (2) it scored “present” in DT cells. The average fold increase was calculated from all 4 comparisons. Cluster analyses were carried out using GeneSpring 7.2 software (Silicon Genetics, Redwood City, CA). Standard correlation was used as the similarity measure for these analyses.
PCR analysis
Total RNA was reverse-transcribed using superscript reverse transcriptase (Invitrogen, Carlsbad, CA) and polymerase chain reaction (PCR)–amplified (primer sequences available upon request). Input c-DNA in PCR reaction was equalized by quantification with trace amounts of α[32P]-dCTP added to the RT reaction. Quantitative PCR was performed in iCycler iQ Real-Time Detection system (Biorad Laboratories, Hercules, CA) using iQ supermix (Biorad Laboratories, Hercules, CA), 200 nM primers, and the intercalating dye SYBR Green (Molecular Probes, Eugene, OR). Transcript abundance was calculated relative to GAPDH.
Calcium mobilization studies
Intracellular Ca+2 measurements were performed as previously described.25 Briefly, cells were grown on cover slips, loaded with Fura-2-AM (Molecular Probes, Eugene, OR), and real-time changes in the ratio of fluorescence levels upon excitation of clusters of 6 to 12 cells at 340 and 380 nm were measured in response to 10 nM thrombin or 100 μM peptide agonists for Par1 (TFLLRNPND-NH2), Par2 (SLIGRL-NH2), Par4 (AYPGKF-NH2), and corresponding control peptides (FSLLRN-NH2, LSIGRL-NH2, and GAPGKF-NH2).
In situ hybridization
In situ hybridizations were performed as previously described13 on frozen (mouse) or paraffin (human) placental sections with 300 to 400 ng/mL antisense and sense dioxygenin-labeled riboprobes (Roche Diagnostics, Indianapolis, IN). Bound probe was visualized with alkaline phosphate–conjugated antidioxygenin antibody, developed with nitro blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Roche Diagnostics, Indianapolis, IN). Slides were mounted in Permount (Fischer Scientific) and examined under a Nikon Eclipse TE200 microscope (Nikon, Melville, NY). Pictures were taken using a 20 ×/0.45 objective lens and an RT color spot camera (Diagnostic Instruments, Sterling Heights, MI). Spotsoftware 3.5.9.1 and Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA) software programs were used for image acquisition and processing.
TF, TM, and TFPI functional assays
TF activity of mouse cells was determined by a 2-stage fXa generation assay, with human fX, and plasma from fibrinogen-knockout mice as a source of murine fVII.26 Cells were washed and resuspended in Tris-buffered saline (0.05 M Tris-HCl, 0.2 M NaCl, pH 8.4) at 106 cells/mL and incubated for 15 minutes with 12.5 mM CaCl2 and 125 nM FX (Enzyme Research Laboratories, South Bend, IN) and pooled citrated plasma from fibrinogen-null mice containing 4 units/mL of human hirudin (Sigma, St Louis, MO). The reaction was quenched in 0.25M EDTA, and FXa generation was measured using the chromogenic substrate S2222 (Chromogenix, Milano, Italy). TM cofactor activity was measured by a 2-step assay for PC activation.27 Functional TFPI was determined by treating cells with phosphatidylinositol-specific phospholipase C [1 U/mL; 1 hour, RT] to release cell-associated TFPI. TF-inhibitory activity in the cell-free supernatant was measured as previously described.28
Results
Differentiation-dependent acquisition of an anticoagulant gene expression program by cultured trophoblast cells
The global gene expression profiles of TS and DT cells were analyzed using oligonucleotide arrays containing 12 488 probe sets corresponding to 9218 unigene entries. 6233 probe sets were expressed in TS or DT cells. Expression of approximately 19% (1177) of probe sets was altered (586 increased, 591 decreased) upon differentiation of TS into DT cells. Differentially expressed genes included those previously documented to be regulated during trophoblast differentiation in vivo.3,29-31 For example, DT cells, but not TS cells, expressed TGC markers, placental lactogen 1 and 2, prolactin-like proteins A and B, proliferin, proliferin 2, the spongiotrophoblast marker Tpbpa (trophoblast specific protein/4311), and genes that are in vivo expressed by labyrinthine trophoblasts, such as Hsd11b2, the gap junction protein Gjb2/Cx26, PDGF receptor α, and the transcription factors Pparg, Dlx3, and Tcfeb. In contrast, genes characteristic of undifferentiated cells, such as Fgfr2, Eomes, Cdx2, Idb1, and Idb2 were highly transcribed in TS cells but not in DT cells. The identification of TS- and DT-specific genes in these experiments provides an internal control for consistency of the in vitro differentiation protocol and validates the notion that in vitro–derived DT cells represent a heterogeneous cell population that includes TGC, labyrinthine, and spongiotrophoblast derivatives of TS cells.
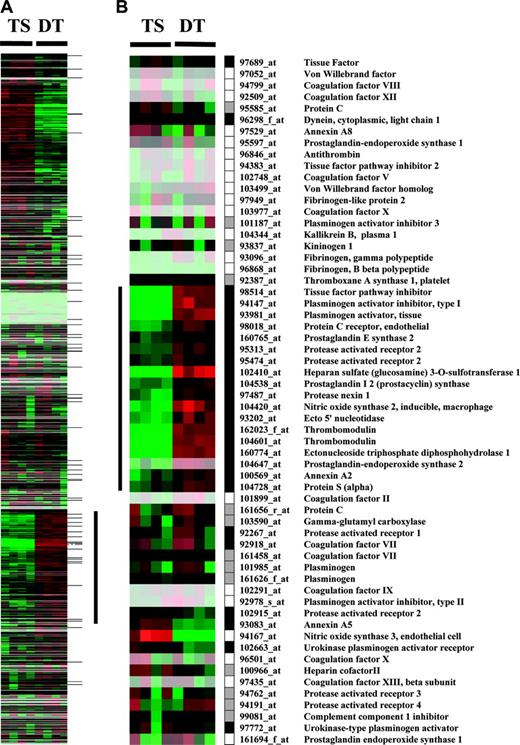
Cluster display of expression data from TS and DT cells. Entire data set from MGU74Av2 chips (A) or hemostasis related probe sets (B) is represented. Rows represent data for each probe set; columns represent experiments. Expression values are normalized to the median of all measurements for a probe set. Below median value are colored green (down-regulated); above median value are colored red (up-regulated); median values (unchanged) are colored black. Lighter colors reflect probe sets with lower median expression values. Black, white, and gray boxes represent present, absent, and marginal expression status of probe sets in DT cells. The vertical bar in panel A indicates a cluster of genes up-regulated during trophoblast differentiation. This cluster contains 16 hemostasis-related genes highlighted by the vertical bar in panel B. Horizontal marks to the right of panel A indicate the position of hemostasis-related genes.
Cluster display of expression data from TS and DT cells. Entire data set from MGU74Av2 chips (A) or hemostasis related probe sets (B) is represented. Rows represent data for each probe set; columns represent experiments. Expression values are normalized to the median of all measurements for a probe set. Below median value are colored green (down-regulated); above median value are colored red (up-regulated); median values (unchanged) are colored black. Lighter colors reflect probe sets with lower median expression values. Black, white, and gray boxes represent present, absent, and marginal expression status of probe sets in DT cells. The vertical bar in panel A indicates a cluster of genes up-regulated during trophoblast differentiation. This cluster contains 16 hemostasis-related genes highlighted by the vertical bar in panel B. Horizontal marks to the right of panel A indicate the position of hemostasis-related genes.
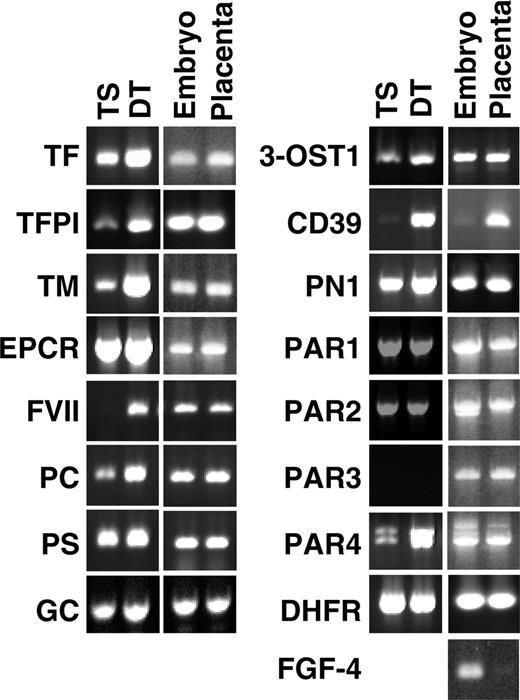
Expression of coagulation-related genes in TS and DT cells and in E8.5 microdissected placenta and embryos determined by semiquantitative RT-PCR. Placenta consisting of trophoblast shell were carefully peeled from the surrounding decidual tissue and separated from Reichert's membrane, visceral yolk sac, and the embryo proper. Dihydrofolate reductase (DHFR; housekeeping gene) and fibroblast growth factor 4 (FGF4; expressed only by the embryo) were included as controls.
Expression of coagulation-related genes in TS and DT cells and in E8.5 microdissected placenta and embryos determined by semiquantitative RT-PCR. Placenta consisting of trophoblast shell were carefully peeled from the surrounding decidual tissue and separated from Reichert's membrane, visceral yolk sac, and the embryo proper. Dihydrofolate reductase (DHFR; housekeeping gene) and fibroblast growth factor 4 (FGF4; expressed only by the embryo) were included as controls.
Probe sets involved in the control of hemostasis were short-listed from the array to generate a subset of 60 probe sets corresponding to 51 unique genes (Figure 1B and Table S1; see the Supplemental Materials link at the top of the online article, at the Blood website). Of the 51 coagulation-related genes, 23 were “present” in DT cells by the array analysis (Figure 1B), and 13 were detected as present in TS cells. All genes expressed at a detectable level in TS cells were also present in DT cells (Table S1). Cluster analysis of the entire array data showed that 16 (70%) of the 23 hemostasis-related genes expressed in DT cells co-segregate into a cluster of genes that is coordinately up-regulated during in vitro differentiation of TS cells (indicated by the vertical bar in Figure 1A). This cluster comprises genes for TFPI, TM, EPCR, protein S (PS), protease nexin 1 (PN1), heparansulfate-D-glucosaminyl-3-O-sulfotransferase-1 (3-OST1), PAR2, tissue plasminogen activator (tPA), plasminogen activator inhibitor (PAI-1), annexin A2, ectonucleoside triphosphate diphosphohydrolase (CD39), ecto5′nucleotidase, nitric oxide synthase 2 (NOS2), prostaglandin (PGE) synthase, prostacyclin (PGI2) synthase, and prostaglandin-endoperoxide synthase 2 (COX-2). Within this group, genes for TM, CD39, 3-OST-1, NOS2, and COX2 exhibited the largest differential in mRNA abundance, following essentially an off (TS cells)–on (DT cells) pattern. Transcripts encoding TFPI, TM, EPCR, annexinA2 and A5, PGI2-synthase, PAI-1, and tPA ranked among the 15% of genes with the highest absolute mRNA abundance in DT cells.
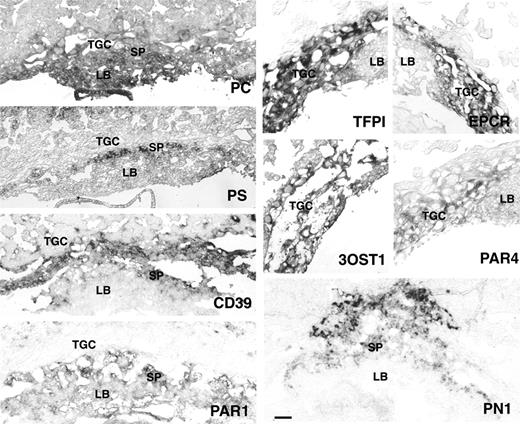
In situ hybridization analysis of PC, PS, CD39, PN1, TFPI, EPCR, 3-OST1, Par1, and Par4 in E9.5 mouse placenta. TGC indicates trophoblast giant cells; SP, spongiotrophoblast cell layer; and LB, labyrinth. TFPI, EPCR, 3-OST1, and PAR4 are expressed in TGC of E9.5 placenta. CD39 expression is observed in TGC and spongiotrophoblasts. PC is expressed in all 3 trophoblast layers. PS, PN1, and PAR1 are expressed in spongiotrophoblast cells. PAR1 expression extends to the labyrinth with staining in fetal blood cells. Bar represents 0.1 mm.
In situ hybridization analysis of PC, PS, CD39, PN1, TFPI, EPCR, 3-OST1, Par1, and Par4 in E9.5 mouse placenta. TGC indicates trophoblast giant cells; SP, spongiotrophoblast cell layer; and LB, labyrinth. TFPI, EPCR, 3-OST1, and PAR4 are expressed in TGC of E9.5 placenta. CD39 expression is observed in TGC and spongiotrophoblasts. PC is expressed in all 3 trophoblast layers. PS, PN1, and PAR1 are expressed in spongiotrophoblast cells. PAR1 expression extends to the labyrinth with staining in fetal blood cells. Bar represents 0.1 mm.
In vivo expression of hemostasis regulators in the mouse placenta
RT-PCR analysis of mRNA isolated from in vitro–cultured TS and DT cells and from micro-dissected placentas at embryonic day 8.5 (E8.5) confirmed the expression of all coagulation-related genes scored as “present” by the array analysis, and demonstrated expression of these genes in the mouse placenta in vivo (Figure 2). In addition, we detected mRNA encoding protein C (PC), coagulation factor VII, γ-glutamyl carboxylase (GC), and PAR4 in TS and DT cells cultured in vitro and in placental tissue in vivo. PAR3 mRNA remained undetectable in cultured trophoblast cells by this approach but was observed in placental tissue.
The anatomic site of expression of those gene products not previously associated with mouse placental trophoblasts (ie, TFPI, 3-OST-1, CD39, PN-1, factor VII, protein S, protein C, PAR1, PAR2, and PAR4) was examined by RNA in situ hybridizations on histologic sections of placentas at E8.5, 9.5, and 12.5 (Figure 3). Placental lactogen 1 (expressed by trophoblast giant cells) and Tpbpa (expressed by spongiotrophoblast) were used to localize subsets of trophoblast cells in adjacent sections.
TFPI mRNA was present in E8.5 and E9.5 TGC, overlapping with the expression of TF mRNA.13 Parietal endoderm cells produced TF but not TFPI mRNA (data not shown). At E12.5, TFPI expression was retained in the TGC population at the baso-lateral aspect of the placenta. Protein C mRNA was detected in TGC at E8.5 and 9.5. At E9.5, expression extended to the labyrinth and the parietal endoderm (not shown). At E12.5, placental expression of protein C was below the limit of detection. Protein S mRNA localized to the parietal endoderm, to patches of spongiotrophoblast cells and to maternal decidual cells in the mesometrial aspect of the decidua between E8.5 and 12.5. EPCR mRNA was present in TGC at E8.5 and 9.5, confirming published results obtained with EPCR-specific antibodies.7 CD39 mRNA was present in, and restricted to, the TGC at E8.5 and E9.5. At E12.5 expression was observed in the labyrinth, the spongiotrophoblast layers, and in a subpopulation of TGC at the baso-lateral aspect of the placenta. Protease nexin-1 mRNA was expressed in a small group of cells at the mesometrial pole of the ectoplacental cone at E8.5, colocalizing with the expression of spongiotrophoblast marker, Tpbpa. At E12.5 expression continued in spongiotrophoblast cells and extended to the labyrinth. 3-OST-1 expression was observed in TGC, and parietal endoderm at E8.5 and E9.5 and was restricted to cells apposing lacunar blood spaces at the base of the placenta (chorionic plate) at E12.5. PAR1 expression first appeared at E9.5 in the labyrinthine trophoblast cells and, as reported before,32 also was evident in fetal blood cells. At E12.5, PAR1 expression also appeared in the spongiotrophoblast layer and in decidual and fetal blood vessels. PAR4 was expressed in TGCs and parietal endoderm cells of the E8.5 and E9.5 placenta but was no longer detected at E12.5. Expression of factor VII- and PAR-2 mRNA was below the level of detection by in situ hybridization at all stages analyzed.
This survey revealed a dynamic temporal and spatial pattern of expression of anticoagulant genes in the placenta. Expression was not restricted to endovascular or interstitial trophoblasts but was high in trophoblast cells that remain resident in the placenta. TFPI, CD39, 3-OST1, and protein C, as well as the previously reported expression of TM and EPCR, colocalized to TGC at E9.5. The TGC layer forms chronologically and anatomically the first interface with maternal blood. It is developmentally derived from two different sources: mural trophectoderm, which gives rise to the primary giant cell layer, constituting the majority of trophoblast cells in the E8.5 mouse placenta; and the polar trophectoderm, which is the source of trophoblast stem cells, in turn giving rise to secondary giant cells.33 The combined in vitro and in vivo analysis therefore suggests that both primary and secondary trophoblast cell populations activate an endothelial cell-like thromboregulatory gene expression program. In contrast, in the E12.5 placenta, expression in TGC was diminished, and—at least in the case of EPCR and CD39—is observed in differentiated trophoblast cells other than giant cells. This transition in expression pattern coincides with the establishment of a functional definitive placenta in which the site of fetomaternal exchange moves to the labyrinth.
Functional activity of TF, TFPI, and TM expressed by mouse trophoblast cells
The analysis of mRNA expression suggested that both TS and DT cells constitutively produce the coagulation initiator, TF, whereas the expression of the coagulation inhibitors, TFPI and TM, occurs in a strictly differentiation-dependent manner only in DT cells. We confirmed these observations by quantifying TF, TFPI, and TM mRNA levels in TS and DT cells by real-time PCR relative to GAPDH. TF expression remained unchanged between TS and DT cells, while TFPI and TM expression increased 29 (± 3)– and 1530 (± 170)–fold, respectively (Figure S1). TF activity of DT cells, measured by fVII-dependent activation of fX, was significantly lower than that of TS cells (18 ± 2 versus 64 ± 2 nMoles of fXa generated per 103 cells). To examine whether the reduction in TF activity could be accounted for by a corresponding increase in TFPI expression, the presence of TF-inhibitory activity was determined in protein extracts prepared from TS and DT cells treated with phosphatidylinositol-specific phospholipase C, which releases GPI-anchored TFPI from the cell surface.28 An inhibitory activity equivalent to 2 ng TFPI per 106 cells was measured in DT cells, whereas TS cells did not exhibit TF-inhibitory activity in excess of controls (no cells). Likewise, TM cofactor activity in the augmentation of protein C activation by thrombin was below the level of detection in TS cells but was strongly induced in DT cells (0.25 ± 0.39 versus 48.97 ± 1.96 ng activated protein C generated per 103 TS or DT cells, respectively). These results demonstrate (1) that functional activities of TF, TFPI, and TM correlate with changes in the abundance of the respective mRNA species, and (2) that TS cells exhibit a pronounced procoagulant surface, whereas differentiated cells mitigate coagulation initiation and propagation by producing the direct TF inhibitor, TFPI, and a key component of the protein C–dependent anticoagulant pathway, TM.
Functional activity of protease-activated receptors expressed by mouse trophoblasts
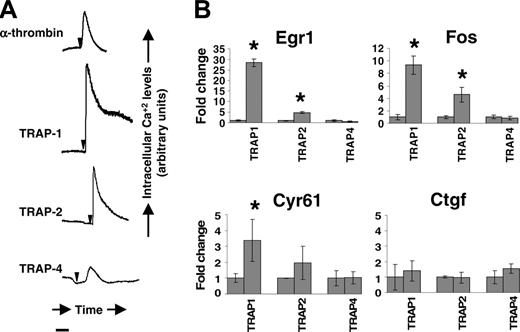
To assess the functional status of PARs expressed by trophoblast cells, we measured the ability of thrombin and receptor-specific agonist peptides to elicit changes in intracellular calcium levels in DT cells. Thrombin and agonist peptides for PAR1, PAR2, and PAR4, but not sequence-scrambled control peptides, transiently increased intracellular calcium levels (Figure 4A).
Egr1, Fos, Cyr61, and Ctgf have been previously identified as downstream transcriptional targets of PAR1 and PAR2 stimulation in the endothelium and other cell types.34-36 We measured their steady state levels in DT cells with and without PAR activation. Real-time PCR experiments showed that PAR1 engagement by agonist peptides resulted in augmented steady-state mRNA levels of Egr1, Fos, and Cyr61 in these cells. PAR2 agonist peptides augmented mRNA expression of Egr1 and Fos, whereas the augmentation of Cyr61 did not reach statistical significance (Figure 4B). PAR4 stimulation did not alter expression of these target genes.
To characterize the range of target genes regulated by individual PARs in trophoblast cells, TS and DT cells were treated with receptor-specific agonist peptides and subjected to array analysis (Table S2). This analysis showed the following: first, only PAR1 and PAR2, but not PAR4, stimulation elicited changes in gene expression detectable by the array-based assay; second, 112 (∼1%) genes of the total set of 9218 were sensitive to PAR engagement; 10 of these PAR-responsive genes also were contained within the subset of genes that were regulated in a differentiation-dependent manner; third, except for annexin A4, PAR1 or PAR2 stimulation did not alter the steady state mRNA level of hemostatic regulators queried on the array; fourth, there was very limited overlap in the set of genes induced by PAR1 or PAR2 engagement in a given cell type, or between TS and DT cells exposed to the same agonist. Only 4 genes each were induced to a similar extent by PAR1 or PAR2 engagement in TS (Klf4, Sgk, Rgs16, Lamna) and DT cells (Fos, FosB, Actn1, Zfbp574), respectively. Rather, Par stimulation affected the expression of receptor- and cell type–specific gene sets. Of note, a particular and unique set of 6 transcripts induced by PAR2 stimulation in DT cells followed essentially an “off-on” pattern of expression (Vav2, Gna12, Pdgfb, Src, Casp9, Adra2a). In contrast, all the other PAR-sensitive genes showed only comparably moderate changes in expression levels.
Protease-activated receptor activity in mouse trophoblast cells. Panel A shows real-time changes in intracellular calcium levels in response to thrombin (10 nM), peptide agonists for PAR1 (TRAP1, 100 μM), PAR2 (TRAP2, 100 μM), and PAR4 (TRAP4; 100 uM) in DT cells. Arrows indicate time of addition of the agonist. Bar represents 100 seconds. No change in intracellular calcium level was observed with scrambled peptides. Panel B shows Egr1, Fos, Cyr61, and Ctgf mRNA induction in mouse trophoblast cells determined by real-time PCR analysis of DT cells stimulated with TRAP1, TRAP2, or TRAP4 for 45 minutes. Transcript abundance was normalized to GAPDH and expressed as fold change over average abundance in unstimulated cells. *Statistically significant (P < .05) fold changes.
Protease-activated receptor activity in mouse trophoblast cells. Panel A shows real-time changes in intracellular calcium levels in response to thrombin (10 nM), peptide agonists for PAR1 (TRAP1, 100 μM), PAR2 (TRAP2, 100 μM), and PAR4 (TRAP4; 100 uM) in DT cells. Arrows indicate time of addition of the agonist. Bar represents 100 seconds. No change in intracellular calcium level was observed with scrambled peptides. Panel B shows Egr1, Fos, Cyr61, and Ctgf mRNA induction in mouse trophoblast cells determined by real-time PCR analysis of DT cells stimulated with TRAP1, TRAP2, or TRAP4 for 45 minutes. Transcript abundance was normalized to GAPDH and expressed as fold change over average abundance in unstimulated cells. *Statistically significant (P < .05) fold changes.
A comparison of PAR-responsive genes observed in trophoblast cells with published data on endothelial cells36-40 showed that only Jun, Cyr61, Pdgfb, and Gadd45b are regulated by PAR1-agonist peptide in both trophoblast and endothelial cells. The response of Gadd45b to PAR1 engagement in trophoblast cells is opposite to that observed in endothelial cells. A different set of genes is regulated by PAR1 engagement on cytokine-activated endothelial cells, of which Fos, Jun, Nr4a1, Sgk, and RNA messages for nuclear lamin filaments were also identified in our screen with trophoblast cells.41 The majority of PAR-responsive genes in trophoblast cells, therefore, appear to be different from those previously identified in endothelial cells.
Cross-species conservation of trophoblast gene expression in the human placenta
RNA transcripts encoding TF, TM, TFPI, EPCR, PS, CD39, PN1, PAR1, and PAR2 could be readily detected in 2 human choriocarcinoma cell lines of trophoblastic origin, Jeg3 and BeWo (Figure S2). Corroborating earlier reports,42 both cell lines expressed high levels of TF activity (data not shown) and initiated intracellular calcium signals upon PAR1 and PAR2 activation (data not shown). Akin to murine trophoblast cells, PAR1 activation resulted in a 13-fold increase in EGR1 mRNA and a 5-fold increase in CYR61 mRNA expression in human Jeg3 cells (Figure S2).
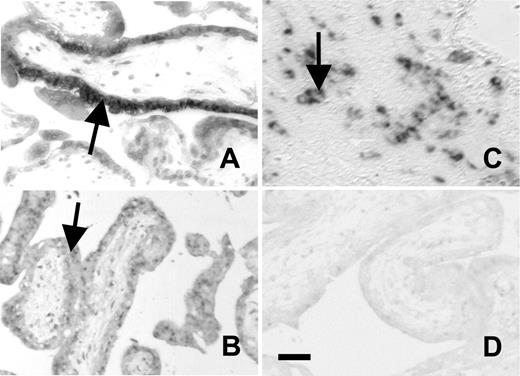
PAR1, PAR2, TM, and TFPI mRNA expression in syncytiotrophoblast cells of first trimester human placenta has been reported.6,8,22-24 We therefore limited the current analysis to the detection of EPCR, PS, PC, CD39, 3-OST-1, and PN1 transcripts by in situ RNA hybridization. Using probes generated with the same primer set employed to amplify these sequences by RT-PCR in Jeg3 and BeWo cells, we were unable to detect mRNA for protein C, protein S, and 3-OST-1 in first-trimester human placentas. EPCR mRNA was highly expressed in the cytotrophoblast-synctiotrophoblast bilayers of placental villi (Figure 5), corresponding to expression seen in TGC and labyrinthine trophoblasts in mice, and overlapping the previously reported sites of high TM and TFPI expression.6,8 The distribution of CD39 mRNA was identical to that of EPCR mRNA. PN1 mRNA was localized on cytotrophoblast columns, analogous to its expression in mouse spongiotrophoblasts.
In situ hybridizations on first trimester human placenta. In situ hybridizations on first trimester human placenta showing EPCR (A), CD39 (B), and PN1 (C) expression on trophoblast cells. Panel D shows negative stain with sense control. Bar represents 0.05 mm. EPCR and CD39 are expressed on cytotrophoblast-synctiotrophoblast bilayers of placental villi. PN1 is expressed at a different site on columns of cytotrophoblast cells.
In situ hybridizations on first trimester human placenta. In situ hybridizations on first trimester human placenta showing EPCR (A), CD39 (B), and PN1 (C) expression on trophoblast cells. Panel D shows negative stain with sense control. Bar represents 0.05 mm. EPCR and CD39 are expressed on cytotrophoblast-synctiotrophoblast bilayers of placental villi. PN1 is expressed at a different site on columns of cytotrophoblast cells.
Discussion
The goal of the current study was to delineate the role of trophoblast cells in placental hemostasis and to understand how activated coagulation factors may alter trophoblast cell function through the engagement of protease-activated receptors. We first show that cultured murine trophoblast cells activate in a differentiation-dependent manner a gene expression program that confers onto these cells a thromboresistant phenotype. Akin to the ability of endothelial cells to regulate various aspects of hemostatic system activity, the set of genes activated in fetal trophoblast cells comprises regulators of fibrinolysis, various aspects of platelet function, the initiation of coagulation, and of the thrombin-generating amplification of the coagulation reaction. The genome-wide expression analysis consolidates previous work addressing the expression of individual gene products in trophoblast cells and extends such findings to include genes, such as 3-OST-1, protein C, and protein S, not previously known to be associated with rodent or human trophoblasts, and others such as CD39 and PN-1, hitherto described only in term human placenta. Existing data on human placenta and results from the current study are consistent with the notion that the specific thromboregulatory properties described in mouse trophoblast cells also are a characteristic feature of human trophoblast cells. This shows that, although derived from two distinct developmental lineages, endothelial cells and trophoblast cells activate a similar gene expression program that enables both cell types to partake in the regulation of hemostasis at the blood-tissue interface. In spite of its cellular heterogeneity, the lining of the maternal blood space in the placenta is therefore homogeneous with respect to its thromboresistance. These observations extend the concept of “endothelial mimicry” by trophoblast cells beyond the subpopulation of endovascular trophoblast. In contrast to the temporally and spatially restricted phenotypic adaptation of endovascular trophoblast cells, the ability to regulate hemostasis in an endothelial cell-like fashion appears, at least in vitro, coupled to the spontaneous differentiation of trophoblast stem cells.
The group of thromboregulatory trophoblast gene products identified here represents a set of candidate genes that are potential modifiers of the risk for adverse pregnancy outcome experienced by mothers with a thrombophilic predisposition. Because trophoblast cells are of fetal origin, genetic polymorphisms of either parental genome have a 50% chance of germline transmission and thereby affecting trophoblast function. To determine the true risk for adverse pregnancy outcome associated with these modifiers, it might be necessary to conduct a retrospective analysis of placental/fetal tissue, or—if viable—the offspring. Cell surface–associated membrane receptors and ectoenzymes, but not soluble factors, are likely the most relevant candidates, because they directly affect hemostasis at the fetomaternal interface. In contrast, polymorphisms in fetal genes encoding circulating factors only will affect hemostasis within the fetal blood vessel compartment and therefore are unlikely to alter the risk conferred by maternal thrombophilia.
At present, direct proof for the critical role of trophoblast cells in the regulation of hemostasis at the fetomaternal interface derives exclusively from genetically modified mice: animals lacking TM or EPCR expression on trophoblast cells succumb to fatal defects in placental development.13,43-45 3-OST-1 knockout mice exhibit intrauterine growth restriction, but it is unknown whether this phenotype is caused by a trophoblast defect.46 Similarly, TFPI deficiency causes abnormalities, including large maternal blood pools and increased fibrin deposition in the placenta.47,48 In all these animal models, the defects are due solely to the complete absence of these factors from fetal trophoblast and are manifest in the absence of maternal thrombophilia. Whether or not partial deficiency or reduced function of the same components exacerbates in an epistatic manner the consequences of maternal thrombophilia is currently unclear. Pilot studies in mice conducted in our laboratory show that reduced, but not absent, expression of at least TM and EPCR by trophoblast cells leads to fetal loss in the first half of gestation in mothers that are homozygous carriers of the fV Leiden mutation, but not in mothers with normal or near-normal hemostatic system function (R.S., M. Zogg, and H.W.; unpublished observations, June 2005). Such preliminary findings strongly imply that epistasis between fetal and maternal risk factors indeed occurs, and may precipitate fetal loss. Our study defines a list of candidate fetal gene products predicted to represent risk modifiers of adverse pregnancy outcomes in thrombophilic mothers.
We demonstrate, second, that trophoblast cells are capable of sensing the presence of activated coagulation factors and that the latter elicit—via engagement of protease-activated receptors—alterations in trophoblast gene expression. The receptor-activated transcriptome is highly specific for both the receptor and for the differentiation state of the target trophoblast cells and differs substantially from corresponding transcriptomes identified in endothelial cells. Trophoblast stem cells exhibited a rather limited response, comprising 5 PAR1- and 9 PAR2-responsive genes. In contrast, differentiated trophoblast cells responded to PAR stimulation by altering the transcription of more than 100 genes. In particular, PAR2 engagement not just modulated, but essentially switched on a specific transcription program comprising 6 genes, whose expression cannot be detected in unstimulated cells. The exit from the stem cell compartment therefore appears associated with a substantial gain in the responsiveness to PAR agonists, although PAR1 and PAR2 are expressed at similar levels in differentiated and undifferentiated trophoblast cells. Since differentiated cells comprise various subsets of distinct trophoblast populations, the relatively large number of different genes responding to PAR agonists might therefore constitute the aggregate of gene subsets regulated in specific subpopulations, rather than reflecting the target transcriptome of any individual cell population. Pathway analysis using EASE (Expression analysis systematic explorer) software49 suggests that PAR1 agonists have the most relevant effect on the regulation of cell-cycle and protein biosynthesis. Of note, we show here that Cyr61 is a PAR1-responsive gene in trophoblast cells. Cyr61 deficiency results in defects in chorioallantoic fusion and undervascularization of the placenta.50 PAR1 activation of human extravillous trophoblasts is accompanied by increased invasiveness and cytoskeletal reorganization,24,51 properties that are likely regulated by Cyr61.52,53 PAR2 responses also target the regulation of cell cycle but predominantly affect genes contributing to intracellular signal transduction pathways, including GTPase regulator activity (P < .001), guanyl-nucleotide exchange factor activity (P < .001), and protein kinase cascades (P < .001). This indicates that PAR engagement may change the set point or responsiveness of trophoblast cells to various extracellular signals by altering the sensitivity of signal transduction pathways coupled to other cell-surface receptors. Such a cross talk has been previously described between PARs and the EGF receptor.54 In addition, signals transmitted through PARs modulate the expression levels of transcription factors, such as Klf4 or Nr4a1, which might further amplify the effect of PAR agonists on transcription. Having defined the receptor- and cell type–specific transcriptomes coupled to PAR engagement, we still need to confirm the array-based data through an independent approach in a similar manner as conducted for Egr1, Fos, Cyr61, and Ctgf; and the responses elicited by nonphysiologic agonist peptides must be compared to the responses elicited by physiologic ligands, including thrombin, signaling-competent TF complexes, and activated protein C.
Irrespective of these shortcomings, our results suggest a number of readily testable hypotheses, which address critical gaps in the knowledge about the role of coagulation and coagulation receptors in placental development. The precise pathogenesis of the placental abnormalities in thrombophilic women is still unclear and cannot fully be explained by frank thrombosis.55,56 In view of the findings reviewed in the discussion that coagulation factor signaling can have physiologically relevant effects on trophoblast function, the deregulated engagement of coagulation receptors is an attractive candidate mechanism that may contribute to placental abnormalities. However, to our knowledge, no in vivo data have been generated in support of this hypothesis. Our analysis of PAR responses defines a molecular fingerprint or signature that can be traced in vivo and should allow detecting the activation state of these receptors in trophoblast cells of the placenta. Mice lacking individual components of this coagulation receptor network (ie, PARs, TF, EPCR, TFPI, and TM) are available and amenable to such investigations.
Prepublished online as Blood First Edition Paper, December 27, 2005; DOI 10.1182/blood-2005-10-4111.
Supported by research grant HL60655 from the National Heart, Lung, and Blood Institute.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Trudy Holyst for synthesis of peptides, Sarah Alliman for technical assistance with TFPI functional assays, and Barbara Fleming for sectioning of tissue for immunohistologic analysis.