Several hematopoietic growth factors, including interleukin-10 (IL-10) and transforming growth factor-β1 (TGF-β1), promote the differentiation of tolerogenic dendritic cells (DCs). Hepatocyte growth factor (HGF) is a pleiotropic cytokine whose effects on human DC differentiation and function have not been investigated. Monocytes cultured with HGF (HGFMo) differentiated into accessory cells with DC-like morphology, released low amounts of IL-12p70 and up-regulated IL-10 both at the mRNA and at the protein level. Upon activation with HGFMo, allogeneic CD4+CD25- T cells expressed the T regulatory (Treg)-associated transcription factor FoxP3, proliferated poorly, and released high levels of IL-10. Interestingly, blockade of surface immunoglobulin-like transcript 3 (ILT3) on HGFMo or neutralization of secreted IL-10 translated into partial restoration of T-cell proliferation. Secondary stimulation of HGFMo-primed CD4+ T cells with immunogenic DCs differentiated with granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-4 from monocytes of the same donor resulted in measurable T-cell proliferation. HGFMo-primed CD4+ T cells significantly inhibited the proliferation of naive CD4+CD25- T cells in a cell-contact-dependent manner. Finally, DNA microarray analysis revealed a unique gene-expression profile of HGF-activated monocytes. Collectively, our findings point to a novel role for HGF in the regulation of monocyte/DC functions that might be exploited therapeutically. (Blood. 2006;108:218-227)
Introduction
Dendritic cells (DCs) are highly specialized antigen-presenting cells (APCs) with unique capacity to fully activate and induce clonal expansion of naive and memory T cells.1 Precursors for DCs circulate in human peripheral blood and comprise myeloid DC precursors (CD14+ monocytes) and plasmacytoid DC precursors (pDCs).2 Both myeloid and pDCs possess a remarkable functional plasticity, and the net effect of antigen (Ag) dose and DC maturational status, together with DC stimulation by pathogenderived products, determines whether a T-helper 1 (Th1)- or Th2-cell response develops.3,4 Importantly, pDCs express immunoglobulin-like inhibitory receptors, namely, immunoglobulin-like transcript 3 (ILT3) and ILT4.5 Recently, DCs have been implicated in the maintenance of Ag-specific unresponsiveness or tolerance in central lymphoid organs and in the periphery through the induction of regulatory T (Treg) cells.6,7 Both immature myeloid DCs and CD40 ligand (CD40L)-activated pDCs can promote Treg-cell differentiation either in vitro or in vivo.8 Among CD4+ T cells with inherent regulatory activity, interest has been focused on naturally occurring CD4+CD25+ Treg cells and on adaptive Treg cells, such as interleukin (IL)-10-secreting CD4+ type 1 Treg cells (Tr1). Expression of the forkhead/winged helix transcription factor, FoxP3, has been demonstrated in CD4+CD25+ Treg cells and correlates with the acquisition of suppressive activity.9 Conversion of CD4+CD25- T cells into CD4+CD25+ Treg cells might occur both in vitro and in vivo.10,11 Whereas CD4+CD25+ Treg cells induce cell-contact-dependent suppression, Tr1 cells can be differentiated in the periphery in the presence of IL-10 and, ultimately, regulate T-cell responses via secreted IL-10 and/or transforming growth factor-β (TGF-β).
Factors governing the development of Treg-inducing, regulatory DCs are not fully elucidated. DCs can be rendered tolerogenic by a variety of means, including exposure to individual cytokines or cytokine combinations (eg, IL-10 and interferon-α [IFN-α]) by treatment with immunosuppressive agents and by exposure to microbial products.7,12 Previously, we showed that monocytederived DCs differentiated in vitro after granulocyte colonystimulating factor (G-CSF) administration to healthy donors and cancer patients acquire the functional properties of regulatory DCs and induce the differentiation of CD4+ Treg cells that release high quantities of immunosuppressive TGF-β1 and IL-10.13,14
Tumor cells might stimulate the production of immature DCs and/or favor abnormal DC differentiation through soluble factors, such as vascular endothelial growth factor, IL-10, monocyte CSF (M-CSF), or granulocyte-macrophage CSF (GM-CSF) and IL-6.15,16 In this respect, hepatocyte growth factor (HGF), originally identified and cloned as a potent mitogen for hepatocytes,17 is greatly elevated in immune suppressive conditions (eg, tumors) and in graft rejection.18 High serum levels of HGF confer unfavorable prognosis to solid tumors by promoting cell motility, invasiveness, angiogenesis, and tumor-cell proliferation.19-21
Conflicting results have been published regarding the ability of HGF to modulate the Ag-presenting function of mouse DCs.22,23 In humans, HGF up-regulates the expression of c-met/HGF receptor, induces HGF secretion, and stimulates monocyte motility and invasiveness.24,25 So far, the effects of HGF on human monocyte differentiation and function have not been investigated. It is demonstrated here that HGF promotes monocyte differentiation toward the generation of IL-10-producing, IL-12low/neg accessory cells with DC features (HGFMo). Such regulatory HGFMo have reduced T-cell allostimulatory capacity, favor the differentiation of cell-contact-dependent CD4+CD25+FoxP3+ Treg cells, and possess a unique gene signature.
Materials and methods
Generation of DCs from peripheral blood monocytes
Peripheral blood mononuclear cells (PBMNCs) were separated by Ficoll-Hypaque density gradient, as already reported.13 CD14+ monocytes were purified by positive selection (Monocyte Isolation Kit; Miltenyi Biotec, Bergisch Gladbach, Germany). Monocytes were cultured for 6 days at 37°C under serum-free (SF) conditions (10% BIT HCC-9500; StemCell Technologies, Vancouver, BC, Canada). The following cytokine combinations were used to promote DC differentiation: 800 IU/mL recombinant human GM-CSF and 500 IU/mL IL-4 (immunogenic DCs); 800 IU/mL GM-CSF and 20 ng/mL HGF; or 20 ng/mL HGF alone (all from R&D Systems, Oxon, Cambridge, United Kingdom). Immature DCs (iDCs) were incubated for an additional 48 hours with 500 IU/mL tumor necrosis factor-α (TNF-α; R&D Systems) to promote maturation.
Evaluation of endocytosis by FACS analysis
Cytokine-treated monocytes were suspended in culture medium supplemented with 10% fetal calf serum (FCS) in the presence of 100 μg/mL FITC-dextran (Sigma Chemical, St Louis, MO) for 1 hour at 37°C.26 Control DC cultures were pulsed with FITC-dextran at 4°C. After extensive washings with ice-cold phosphate-buffered saline (PBS) containing 1% FCS and 0.01% NaN3, cells were analyzed on a FACSCanto flow cytometer (Becton Dickinson, Mountain View, CA).
T-cell isolation, primary and secondary MLR
CD25+ cells were purified from umbilical cord blood samples by positive selection using directly conjugated anti-CD25 magnetic microbeads (4 μL/107 cells; Miltenyi Biotec). After the double-column procedure, CD4+CD25+ cells were routinely more than 94% pure by fluorescence-activated cell-sorting (FACS) analysis (data not shown). The remaining non-CD25+ fraction was used for the isolation of CD4+CD25- cells by positive selection with anti-CD4 monoclonal antibody (mAb)-coated microbeads (Miltenyi Biotec).
CD4+CD25- T cells were activated with the mixed leukocyte reaction (MLR). Briefly, graded doses of irradiated (25 Gy) DCs were cultured with 5 × 104 allogeneic CD4+CD25- T cells for 7 days. For secondary Ag-specific stimulation, CD4+ T cells recovered from the primary MLR were rechallenged with the original stimulator cells that were cryopreserved at the start of the experiment or with third-party GM-CSF/IL-4-induced DCs (immunogenic DCs). In selected experiments, neutralizing anti-TGF-β1 (20 ng/mL) and/or anti-IL-10 antibodies (10 μg/mL; both from R&D Systems) were used during the MLR, as previously reported.13 For secondary polyclonal stimulation, T cells recovered from the primary MLR were rested for 24 hours and then rechallenged for 72 hours with super-paramagnetic polystyrene beads coated with anti-CD3 and anti-CD28 mAbs (spvT3b clone and L293 clone, respectively; Dynabeads CD3/CD28 T Cell Expander; Dynal Biotech, Oslo, Norway) following the manufacturer's instructions, in the presence of 10 IU/mL recombinant human IL-2 (R&D Systems).
Immunologic markers and flow cytometry
Cells were incubated for 20 min at 4°C with FITC, PE, or PerCP fluorochromes conjugated with cyanine-5 (TRICOLOR [TC]; Caltag Laboratories, Burlingame, CA): CD1a, CD4, CD11c, CD14, CD25, CD62L, CD80, CD86, CD83 (Caltag Laboratories, Burlingame, CA), CD123 (IL-3 receptor α-chain), human leukocyte antigen (HLA)-DR, CC chemokine receptor 6 (CCR6), CD11c and CD123 (Becton Dickinson), blood DC antigen-4 (BDCA-4) and BDCA-2 (Miltenyi Biotec), ILT3, DC-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN; CD209; Immunotech, Marseille, France), or with the appropriate fluorochrome-conjugated, isotype-matched irrelevant mAbs to establish background fluorescence. For the detection of intracellular IL-10, cells were activated with PMA (5 ng/mL) and ionomycin (500 ng/mL; both from Sigma Chemical) in the presence of GolgiStop solution (Becton Dickinson) for 4 hours. Following fixation and permeabilization with Cytofix/Cytoperm solution (Becton Dickinson), cells were stained with FITC-conjugated rat anti-human IL-10 mAb (Caltag Laboratories). Expression of FoxP3 was detected in fixed/permeabilized T cells after labeling with PE-conjugated rat anti-human FoxP3 mAb (PCH101 clone; Human Regulatory T Cell Staining Kit; eBioscience, San Diego, CA). Samples were run through a FACSCanto flow cytometer (Becton Dickinson) with standard equipment.
Analysis of cytokine production
IL-2, IL-4, IL-5, IL-10, IL-12p70, IFN-γ, and TGF-β1 levels were quantified by enzyme-linked immunosorbent assays (ELISAs) using commercially available kits (R&D Systems). The limits of detection were as follows: 1 pg/mL IL-10 (Ultra Quantikine HS); 7 pg/mL TGF-β1, 0.5 pg/mL IL-12p70 (Quantikine HS), 10 pg/mL IL-2, 0.11 pg/mL IL-4, 3 pg/mL IL-5, and 15.6 pg/mL IFN-γ.
Split-well cultures and assays for Treg function
Coculture experiments for the assessment of regulatory properties of CD4+ T cells were performed either in the presence or in the absence of Transwell systems (MilliCell inserts, 0.4 μM; Millipore, Watford, United Kingdom), as previously detailed.13 Briefly, the proliferation of nonregulatory CD4+CD25- T cells plated in the lower chamber of the Transwell was monitored in the absence of direct contact with 5 × 105 DC-primed CD4+ T cells, which were placed in the upper compartment (primary cocultures).
Cell proliferation tracking
Freshly isolated CD4+ T cells were resuspended in PBS containing carboxyfluorescein-diacetate succinimidyl-ester (CFSE, 2.5 μM; Molecular Probes, Eugene, OR) for 10 minutes at room temperature. To quench the labeling process, an equal volume of FCS was added; after washings in PBS supplemented with 3% FCS, cells were used for MLR experiments. The analysis of CFSE fluorescence was performed with the proliferation wizard tool in ModFit LT 2.0 software (Verity Software House, Topsham, ME). Replication data were expressed in terms of “proliferation index” (PI), which was calculated as previously reported.27
RNA isolation and RT-PCR
Total RNA was isolated with the RNeasy minikit (Qiagen, Hilden, Germany) and was reverse-transcribed with 25 units of MMLV reverse transcriptase (RT; Perkin Elmer Cetus, San Diego, CA) at 42°C for 30 minutes in the presence of random hexamers. Polymerase chain reaction (PCR) amplification was performed using primer pairs specific for the molecules of interest (Table 1) and for human aldolase-A as a control. The PCR products were separated on 1% agarose gel and visualized by ethidium bromide staining. Band intensity was quantified with Photoretix 1D (Photoretix International, Newcastle-Upon-Tyne, United Kingdom) and was expressed as relative absorbance units.
Probe preparation for microarray analysis and data bioinformatics
RNA extraction. Total cellular RNA was isolated from cultured cells as described. Disposable RNA chips (Agilent RNA 6000 Nano LabChip kit; Agilent Technologies, Palo Alto, CA) were used to determine the concentration and purity/integrity of RNA samples using an Agilent 2100 bioanalyzer.
Target synthesis, GeneChip hybridization, and data acquisition. Biotin-labeled target synthesis was performed starting from 2 μg total cellular RNA, according to the protocol supplied by the manufacturer (Affymetrix, Santa Clara, CA). Labeled cRNA was purified using RNeasy spin columns (Qiagen, Valencia, CA) and fragmented (15 μg) as described in the Affymetrix GeneChip protocol. Disposable RNA chips (Agilent RNA 6000 Nano LabChip kit) and an Agilent 2100 bioanalyzer were used to determine the concentration and quality of cRNAs as well as to optimize the fragmentation. The fragmented cRNAs were then hybridized to Affymetrix HG-U133A GeneChip arrays for 16 hours. GeneChips were washed and stained using the instrument's standard protocol, using antibody-mediated signal amplification. GeneChips were finally scanned using the Affymetrix GeneChip scanner.
Normalization and intensity calculation. The amount of a transcript mRNA (signal) was determined, using the Gene Chip Operating Software (GCOS) 1.2 absolute analysis algorithm as already described.28 All expression values for the genes in the GCOS 1.2 absolute analyses were determined using the global scaling option that allows a number of experiments to be normalized to 1 target intensity. Alternatively, probe level data have been converted to expression values using Gene Spring's (Silicon Genetics, Redwood City, CA) robust multiarray average (RMA) preprocessing procedure.29
Statistical filtering and analysis. The GCOS as well as the RMA preprocessed data were additionally normalized using Gene Spring software version 7.2 (Silicon Genetics, Redwood City, CA). Each signal was divided by the 50th percentile of all measurements in that sample. A “per gene” normalization was achieved dividing each signal by the median of its values in all samples. In order to remove genes not expressed or always expressed at low levels, expression data were filtered using GeneSpring to select genes detected as “present” in at least 10% of samples. Then genes whose normalized expression levels were always between 0.66 and 1.5 across all of the samples were filtered out.
ANOVA and hierarchic agglomerative clustering. Analysis of variance (ANOVA) was performed with Gene Spring, using parametric tests and multiple testing corrections, to find genes expressed at different levels between the compared sample classes. Tukey post-hoc test was used to determine which pairs among the groups under study had expression means that differed in a statistically significant manner.
Hierarchic agglomerative clustering of the selected probe lists was performed in Gene Spring using the Pearson correlation coefficient and average-linkage as distance and linkage methods. EASE software (Science Applications International-Frederick, Frederick, MD) was used to examine selected lists of genes in order to identify overrepresentation of functional classes according to gene-ontology classification.30
Immunoblotting and immunocytochemistry
Cytokine-differentiated monocytes were lysed with a buffer solution containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100, 50 mM protease, and 50 mM phosphatase inhibitor cocktail. Protein extraction and measurements of protein concentration were performed as previously reported.31 Equivalent amounts of proteins (20 μg) were fractionated on a 10% Tris-glycine gel and electrotransferred to a nitrocellulose membrane (Novex, San Diego, CA). Nonspecific binding was blocked by incubation with 5% milk in 0.1% Tween 20-TBS (Fisher Scientific, Hanover Park, IL), followed by overnight incubation at 4°C with the primary mouse anti-human indoleamine 2,3-dioxygenase (IDO) antibody (MAB5412 clone, IgG3; Chemicon International, Temecula, CA). Membranes were washed 6 times with 0.1% Tween 20-TBS and were incubated first with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and then with the chemiluminescent substrate (Super Signal; Pierce, Rockford, IL) for 5 minutes prior to exposure to the film (Amersham, Arlington, Heights, IL).
For intracellular detection of IDO protein, 106 cells were cytospun and then stained with the anti-IDO mAb for immunocytochemistry analysis. Mouse IgGs were used as isotype-matched, control antibodies.
Statistical analysis
The approximation of data distribution to normality was preliminarily tested with statistics for kurtosis and symmetry. Results were presented as means and SDs. All comparisons were performed with the Student t test for paired or unpaired determinations or with ANOVA, as appropriate. The criterion for statistical significance was defined as a P value of .05 or less.
Results
HGF modulates the expression of differentiation/maturation Ags on monocytes
Highly purified PB monocytes were cultured with established protocols either in the presence of GM-CSF and IL-4 (subsequently designated GM4-DCs)32 or in the presence of HGF (subsequently designated HGFMo). By day 6 of culture, cell recovery averaged 72.5% ± 7% and 62.5% ± 8% in cultures supplemented with GM-CSF + IL-4 and with HGF, respectively. The effects of HGF provision on monocyte phenotypic features and cytokine release were maximal after 6 days of culture (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) and this culture protocol was maintained in all subsequent experiments.
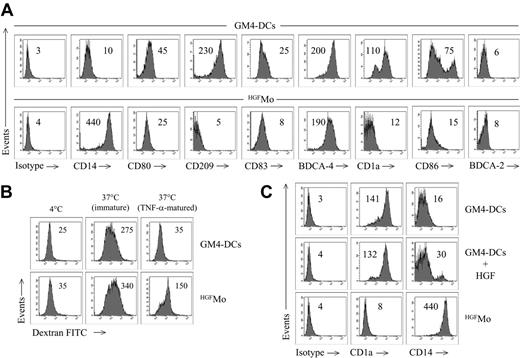
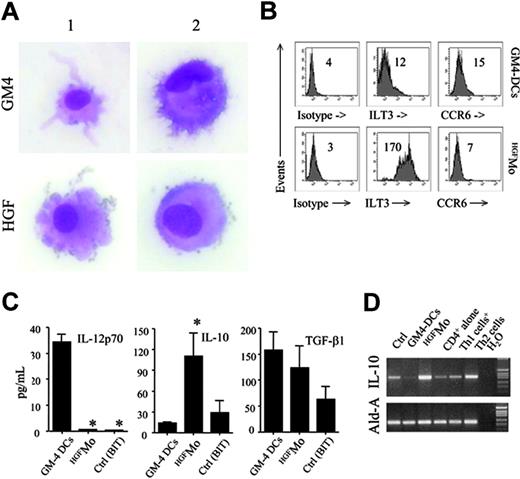
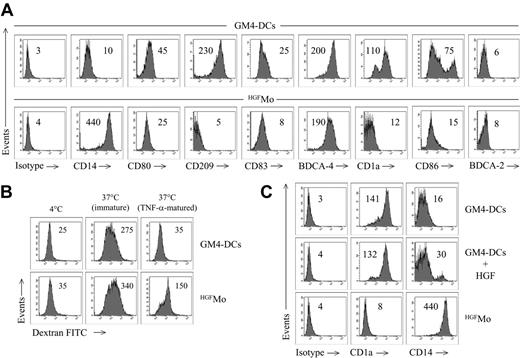
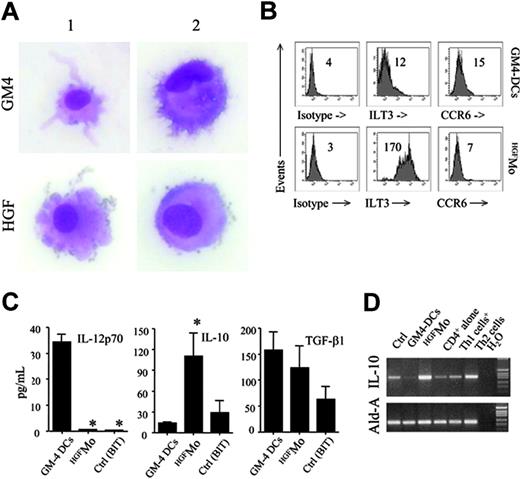
GM4-DCs were CD14-CD1a+, whereas HGFMo maintained a CD14+CD1a- phenotype (Figure 1A). At variance with immunogenic GM4-DCs, HGFMo were CD80low, CD86low, and CD83low, and stained negatively for CD209. In particular, the mean fluorescence intensities (MFIs) of CD80 and CD86 on GM4-DCs were 50 ± 8 and 78 ± 10, respectively, compared with 30 ± 5(P < .01) and 18 ± 5 (P < .001) on HGFMo. No differences in expression levels of other DC-associated Ags (HLA-DR, CD11c, CD40, BDCA-4, BDCA-2, CD11c, and CD123) were measured when comparing GM4-DCs and HGFMo (Figures 1A, S2). Interestingly, immature HGF-DCs accumulated FITC-dextran, a conventional marker of endocytic capacity (Figure 1B).26 When HGF was added to GM-CSF and IL-4, the phenotype of DCs was similar to that described for GM4-DCs (Figure 1C), indicating that IL-4 competed with HGF in the generation of DCs before the HGF-specific differentiation program initiated. HGF provision translated into the acquisition of a DC-like morphology with irregular cytoplasmic outline and tiny cytoplasmic projections (Figure 2A).
We next determined whether molecules previously assigned to DCs with tolerogenic function were preferentially expressed by HGFMo. Whereas CCR6 was detected on very low percentages of GM4-DCs and HGFMo (Figure 2B), ILT3 expression levels were significantly up-regulated on HGFMo (the MFI was 75 ± 6 compared with 10 ± 2 for GM4-DCs, P < .001; Figure 2B). Collectively, HGF promoted the differentiation of monocytes into accessory cells with DC features that were costimulationlowCD83lowCD209neg, stained negatively for Ags assigned to pDCs such as CD123 and BDCA-2, and acquired surface Ags potentially associated with regulatory features.
Phenotypic features of cytokine-differentiated monocytes. DCs were generated as detailed in “Materials and methods.” (A) Phenotypic profile of monocytes cultured with GM-CSF + IL-4 (GM4-DCs) or with HGF (HGFMo). Numbers indicate MFI of mAb staining measured in a representative experiment of 9 with similar results. (B) Endocytic capacity of HGFMo. DCs were either left immature or matured with TNF-α for 48 hours and were then incubated for 1 hour with FITC-dextran at 37°C. Control cultures were established at 4°C to measure background fluorescence.26 Numbers indicate MFI of mAb staining measured in a representative experiment of 4 with similar results. (C) Expression of informative DC Ags on monocytes cultured with GM-CSF, IL-4, and HGF, with GM-CSF and IL-4, or with HGF alone. Numbers indicate MFI of mAb staining measured in a representative experiment of 4 with similar results.
Phenotypic features of cytokine-differentiated monocytes. DCs were generated as detailed in “Materials and methods.” (A) Phenotypic profile of monocytes cultured with GM-CSF + IL-4 (GM4-DCs) or with HGF (HGFMo). Numbers indicate MFI of mAb staining measured in a representative experiment of 9 with similar results. (B) Endocytic capacity of HGFMo. DCs were either left immature or matured with TNF-α for 48 hours and were then incubated for 1 hour with FITC-dextran at 37°C. Control cultures were established at 4°C to measure background fluorescence.26 Numbers indicate MFI of mAb staining measured in a representative experiment of 4 with similar results. (C) Expression of informative DC Ags on monocytes cultured with GM-CSF, IL-4, and HGF, with GM-CSF and IL-4, or with HGF alone. Numbers indicate MFI of mAb staining measured in a representative experiment of 4 with similar results.
Morphology and regulatory features of HGF-differentiated monocytes. (A) GM4-DCs and HGFMo were stained with May-Grunwald-Giemsa and visualized under an AX70 optical microscope (Olympus, Tokyo, Japan) equipped with a 100 ×/1.25 NA objective lens. Image acquisition was performed with an Optronics digital camera (Olympus) and ImagePro Plus software (Media Cybernetics, Silver Spring, MD). Similar to conventional GM4-DCs, HGFMo displayed an eccentric nucleus and an irregular cytoplasmic outline with tiny projections. (B) Expression of ILT3 and CCR6 in monocytes exposed to GM-CSF and IL-4 or to HGF. Numbers indicate MFI of mAb staining measured in a representative experiment of 9 with similar results. (C) After 6 days of culture, DCs were treated with 1 μg/mL lipopolysaccharide (LPS) for 24 hours. Culture supernatants were harvested and used for ELISAs. Results are expressed as the mean ± SD recorded in 8 independent experiments performed in duplicate and depict IL-12p70, IL-10, and TGF-β release by GM4-DCs, HGFMo, or monocytes maintained in the presence of BIT serum substitute alone. *P < .001 compared with immunogenic GM4-DCs. (D) Semiquantitative RT-PCR for IL-10 in GM4-DCs, HGFMo, and monocytes maintained in the presence of BIT serum substitute alone. Th1-like and Th2-like human T cells, differentiated as previously detailed, were used as controls for IL-10 expression levels.13
Morphology and regulatory features of HGF-differentiated monocytes. (A) GM4-DCs and HGFMo were stained with May-Grunwald-Giemsa and visualized under an AX70 optical microscope (Olympus, Tokyo, Japan) equipped with a 100 ×/1.25 NA objective lens. Image acquisition was performed with an Optronics digital camera (Olympus) and ImagePro Plus software (Media Cybernetics, Silver Spring, MD). Similar to conventional GM4-DCs, HGFMo displayed an eccentric nucleus and an irregular cytoplasmic outline with tiny projections. (B) Expression of ILT3 and CCR6 in monocytes exposed to GM-CSF and IL-4 or to HGF. Numbers indicate MFI of mAb staining measured in a representative experiment of 9 with similar results. (C) After 6 days of culture, DCs were treated with 1 μg/mL lipopolysaccharide (LPS) for 24 hours. Culture supernatants were harvested and used for ELISAs. Results are expressed as the mean ± SD recorded in 8 independent experiments performed in duplicate and depict IL-12p70, IL-10, and TGF-β release by GM4-DCs, HGFMo, or monocytes maintained in the presence of BIT serum substitute alone. *P < .001 compared with immunogenic GM4-DCs. (D) Semiquantitative RT-PCR for IL-10 in GM4-DCs, HGFMo, and monocytes maintained in the presence of BIT serum substitute alone. Th1-like and Th2-like human T cells, differentiated as previously detailed, were used as controls for IL-10 expression levels.13
HGF suppresses monocytic IL-12p70 release and up-regulates IL-10 production
Immunogenic DCs are functionally defined by the ability to secrete IL-12p70.1 In sharp contrast with immunogenic GM4-DCs, HGFMo released very low levels of IL-12p70 (Figure 2C). DCs differentiated in the presence of GM-CSF, IL-4, and HGF produced intermediate levels of IL-12p70 (data not shown), suggesting that the inhibitory effect of HGF on IL-12p70 production might be maximal in the absence of exogenous IL-4.
As shown in Figure 2D, HGF treatment significantly enhanced IL-10 mRNA expression, whereas TGF-β1 mRNA expression levels were similar in HGFMo compared with fully immunogenic GM4-DCs (data not shown). Analysis of culture supernatants (Figure 2C) indicated high production of IL-10 protein by HGFMo and comparable TGF-β1 production to GM4-DCs. Thus, HGF induced the differentiation of IL-10-producing accessory cells with an impaired ability to release immunostimulatory IL-12p70.
HGFMo induce hyporesponsiveness in allogeneic CD4+ T cells
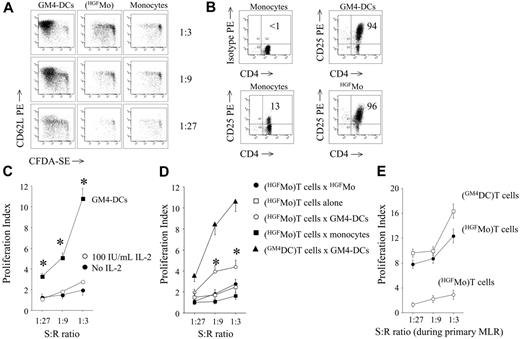
In order to test whether HGFMo might activate the proliferation of allogeneic T cells, a functional hallmark of immunogenic DCs, we established MLR cultures with highly purified naive CD4+CD25- T cells (conventional T cells). As shown in Figure 3A, HGFMo possessed a diminished ability to induce the proliferation of CD4+CD25- T cells (proliferation index equal to 2.5 ± 1.1, 1.67 ± 0.6, and 1.41 ± 0.5 at 1:3, 1:9, and 1:27 T-cell/DC ratios, respectively) compared with immunogenic GM4-DCs (proliferation index equal to 14.5 ± 4, 10.8 ± 3, and 5.6 ± 2 at 1:3, 1:9, and 1:27 T-cell/DC ratios, respectively; P < .001). CD4+CD25- T cells challenged with HGFMo up-regulated CD25 at levels that were comparable with those measured on T cells stimulated with GM4-DCs (Figure 3B). Interestingly, T-cell hyporesponsiveness to HGFMo could not be reverted by exogenous IL-2 (Figure 3C).
T-cell allostimulatory capacity of HGFMo. Allogeneic CD4+CD25-T cells were first labeled with the fluorescent dye CFSE and then cultured for 7 days at increasing stimulator-to-responder (S/R) ratios with cytokine-differentiated monocytes (primary MLR). (A) CFSE-labeled T cells activated as detailed were counterstained with PE-conjugated anti-CD62L mAb prior to flow cytometry analysis of CFSE dye dilution. Results from 1 representative experiment of 9 with similar results are shown. (B) Expression levels of CD25 on DC-challenged CD4+ T cells. Markers were set according to the proper isotypic control. The percentage of CD4+CD25+ T cells in 1 representative experiment of 9 with similar results is indicated. (C) Effect of exogenous IL-2 on T-cell hyporesponsiveness. CFSE-labeled T cells initially challenged with HGFMo were recovered from the primary MLR and treated with 100 IU/mL IL-2 for an additional 72 hours. The proliferation index (PI) was calculated as previously detailed.27 Results (mean ± SD) are representative of 3 independent experiments performed in duplicate. T-cell proliferation in response to GM4-DCs is shown as control. *P < .01 compared with T cells initially activated with HGFMo and subsequently exposed to IL-2. (D) Ag-specific restimulation of DC-activated T cells. T cells initially challenged with cytokine-differentiated DCs in the primary MLR were recovered and left untouched (□) or restimulated either with same-donor GM4-DCs (○), with HGFMo (•), or with monocytes that were cryopreserved at the start of the experiment (▪). Control cultures were established with T cells activated with GM4-DCs both in the primary and in the secondary MLR (▴). Results (mean ± SD) are representative of 4 independent experiments performed in duplicate. *P < .01 compared with T cells restimulated with HGFMo. (E) Secondary polyclonal stimulation of DC-activated T cells. T cells initially challenged with HGFMo in the primary MLR (•) were recovered and restimulated for 72 hours with 12.5 μL Dynabeads CD3/CD28 per 1 × 106cells in the presence of 10 IU/mL IL-2. Control cultures consisted of T cells initially activated with GM4-DCs (□) and then restimulated under the same experimental conditions. The proliferation of T cells in response to HGFMo in the primary MLR is also shown (○).
T-cell allostimulatory capacity of HGFMo. Allogeneic CD4+CD25-T cells were first labeled with the fluorescent dye CFSE and then cultured for 7 days at increasing stimulator-to-responder (S/R) ratios with cytokine-differentiated monocytes (primary MLR). (A) CFSE-labeled T cells activated as detailed were counterstained with PE-conjugated anti-CD62L mAb prior to flow cytometry analysis of CFSE dye dilution. Results from 1 representative experiment of 9 with similar results are shown. (B) Expression levels of CD25 on DC-challenged CD4+ T cells. Markers were set according to the proper isotypic control. The percentage of CD4+CD25+ T cells in 1 representative experiment of 9 with similar results is indicated. (C) Effect of exogenous IL-2 on T-cell hyporesponsiveness. CFSE-labeled T cells initially challenged with HGFMo were recovered from the primary MLR and treated with 100 IU/mL IL-2 for an additional 72 hours. The proliferation index (PI) was calculated as previously detailed.27 Results (mean ± SD) are representative of 3 independent experiments performed in duplicate. T-cell proliferation in response to GM4-DCs is shown as control. *P < .01 compared with T cells initially activated with HGFMo and subsequently exposed to IL-2. (D) Ag-specific restimulation of DC-activated T cells. T cells initially challenged with cytokine-differentiated DCs in the primary MLR were recovered and left untouched (□) or restimulated either with same-donor GM4-DCs (○), with HGFMo (•), or with monocytes that were cryopreserved at the start of the experiment (▪). Control cultures were established with T cells activated with GM4-DCs both in the primary and in the secondary MLR (▴). Results (mean ± SD) are representative of 4 independent experiments performed in duplicate. *P < .01 compared with T cells restimulated with HGFMo. (E) Secondary polyclonal stimulation of DC-activated T cells. T cells initially challenged with HGFMo in the primary MLR (•) were recovered and restimulated for 72 hours with 12.5 μL Dynabeads CD3/CD28 per 1 × 106cells in the presence of 10 IU/mL IL-2. Control cultures consisted of T cells initially activated with GM4-DCs (□) and then restimulated under the same experimental conditions. The proliferation of T cells in response to HGFMo in the primary MLR is also shown (○).
When HGFMo-primed T cells were restimulated with the original HGFMo, negligible modifications of T-cell proliferation occurred compared with the primary T-cell response to HGFMo (Figure 3D). T cells primed with HGFMo and then restimulated with GM4-DCs proliferated more vigorously than T cells re-stimulated with HGFMo (Figure 3D). As shown in Figure 3E, secondary polyclonal stimulation of HGFMo-primed T cells with anti-CD3/CD28 Abs translated into restoration of proliferation.
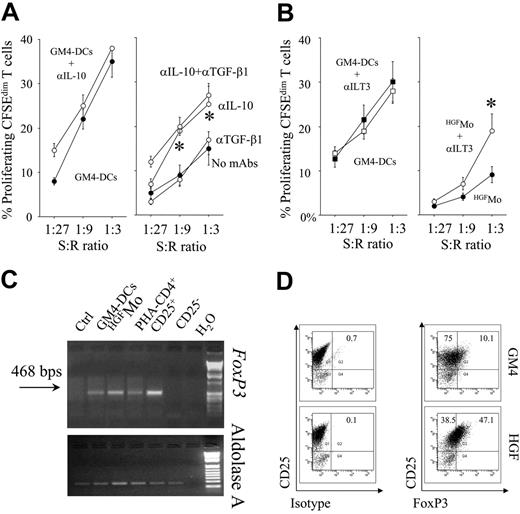
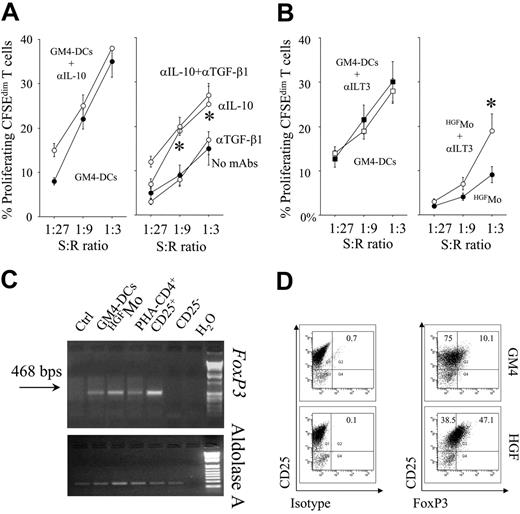
Notably, partial abrogation of T-cell hyporesponsiveness occurred in cultures containing blocking mAbs to IL-10 but not to TGF-β1, suggesting that production of IL-10 by HGFMo might be 1 mechanism responsible for the down-regulation of T-cell proliferation (Figure 4A). Moreover, blockade of ILT3 on HGFMo was also associated with partial recovery of T-cell proliferation in primary MLR at high stimulator-to-responder ratios (Figure 4B), indicating that, in addition to cytokine-mediated mechanisms, expression of ILT3 by HGFMo might be involved in their regulatory activity.
Phenotype and cytokine secretion profile of CD4+ T cells primed with HGFMo
In order to determine the expression of FoxP3 on T cells primed with the different DC preparations, T cells were subjected to semiquantitative RT-PCR studies. Both FoxP3 mRNA signals and protein levels were up-regulated in T cells challenged with HGFMo compared with T cells stimulated with GM4-DCs (Figure 4C,D). Expression of FoxP3 in CD4+CD25+ Treg cells from a healthy control subject, used as positive control, is depicted in Figure S3.
We next examined whether T cells activated with HGFMo in the primary MLR acquired the functional characteristics of Treg cells. To this end, we measured cytokine production by T cells after challenge with cytokine-differentiated DCs. As shown in Figure 5A, IL-10 levels were higher in supernatants of MLR cultures containing HGFMo. Conversely, TGF-β1 levels were comparable in MLR cultures performed with HGFMo and with immunogenic GM4-DCs. Further stimulation of CD4+ T cells with same-donor HGFMo in a secondary MLR was associated with similar increases in the amount of released IL-10, when compared with T cells initially activated with GM4-DCs and then rechallenged with same-donor GM4-DCs (Figure 5B). The prototypic T helper type 1 (Th1) cytokines IL-2 and IFN-γ were detected at low levels in supernatants of primary MLR performed with HGF-DCs, compared with levels recorded in supernatants of MLR containing immunogenic GM4-DCs (Figure 5A). Conversely, IL-5 levels were significantly higher in supernatants of HGFMo-containing MLR cultures (Figure 5A). Both HGFMo and HGFMo-primed CD4+ T cells contributed to IL-10 production in primary MLR cultures, as indicated by IL-10 detection at the single-cell level (Figure 5C). Collectively, CD4+ T cells that developed after activation with HGFMo possessed an IL-10+TGF-β+IL-5+IL-4lowIL-2lowIFN-γlow cytokine secretion profile.
Regulatory activity of HGFMo and HGFMo-differentiated T cells. Primary MLR cultures were performed with HGFMo and purified, CFSE-labeled CD4+CD25- T cells in the presence or in the absence of either neutralizing antibodies to IL-10/TGF-β (A) (each at 10 μg/mL) or blocking antibodies to ILT3 (B) (10 μg/mL). The percentage of proliferating, CFSEdimT cells is depicted on the y-axis. *P < .01 compared with MLR cultures performed in the absence of the blocking antibodies. T-cell proliferation to GM4-DCs both in the presence and in the absence of the blocking Abs to IL-10 (A) and ILT3 (B) is also shown. Error bars indicate mean and SD. (C-D) DC-primed T cells were used for RT-PCR and flow cytometry studies of FoxP3 expression. Freshly isolated CD4+CD25+ and CD4+CD25- T cells served as positive and negative control for FoxP3 expression, respectively (Figure S3). One representative experiment of 4 with similar results is shown.
Regulatory activity of HGFMo and HGFMo-differentiated T cells. Primary MLR cultures were performed with HGFMo and purified, CFSE-labeled CD4+CD25- T cells in the presence or in the absence of either neutralizing antibodies to IL-10/TGF-β (A) (each at 10 μg/mL) or blocking antibodies to ILT3 (B) (10 μg/mL). The percentage of proliferating, CFSEdimT cells is depicted on the y-axis. *P < .01 compared with MLR cultures performed in the absence of the blocking antibodies. T-cell proliferation to GM4-DCs both in the presence and in the absence of the blocking Abs to IL-10 (A) and ILT3 (B) is also shown. Error bars indicate mean and SD. (C-D) DC-primed T cells were used for RT-PCR and flow cytometry studies of FoxP3 expression. Freshly isolated CD4+CD25+ and CD4+CD25- T cells served as positive and negative control for FoxP3 expression, respectively (Figure S3). One representative experiment of 4 with similar results is shown.
Cytokine production by T cells activated with HGFMo. (A) Supernatants of primary MLR cultures containing cytokine-differentiated DCs (indicated in the x-axis) and CD4+CD25- T cells were used for the measurement of prototypic Th1-, Th2-, and Tr1-related cytokines. Data represent mean ± SD recorded in 6 independent experiments performed in duplicate. *P < .01 compared with MLR cultures performed with GM4-DCs. (B) IL-10 levels were measured in the supernatant of secondary MLR cultures performed with T cells recovered from the primary MLR and same-donor, cytokine-differentiated DCs (indicated in the x-axis). *P < .01 compared with secondary MLR cultures performed with GM4-DCs. (C) Cells recovered from primary MLR cultures established with HGFMo (1:3 stimulator-to-responder ratio) were counterstained with PE-conjugated anti-CD4 or anti-CD86 mAbs, fixed/permeabilized as detailed in “Materials and methods,” and labeled with FITC-conjugated anti-IL-10 mAbs. The percentage of IL-10+ cells is indicated in each histogram. One representative experiment of 3 with similar results is shown.
Cytokine production by T cells activated with HGFMo. (A) Supernatants of primary MLR cultures containing cytokine-differentiated DCs (indicated in the x-axis) and CD4+CD25- T cells were used for the measurement of prototypic Th1-, Th2-, and Tr1-related cytokines. Data represent mean ± SD recorded in 6 independent experiments performed in duplicate. *P < .01 compared with MLR cultures performed with GM4-DCs. (B) IL-10 levels were measured in the supernatant of secondary MLR cultures performed with T cells recovered from the primary MLR and same-donor, cytokine-differentiated DCs (indicated in the x-axis). *P < .01 compared with secondary MLR cultures performed with GM4-DCs. (C) Cells recovered from primary MLR cultures established with HGFMo (1:3 stimulator-to-responder ratio) were counterstained with PE-conjugated anti-CD4 or anti-CD86 mAbs, fixed/permeabilized as detailed in “Materials and methods,” and labeled with FITC-conjugated anti-IL-10 mAbs. The percentage of IL-10+ cells is indicated in each histogram. One representative experiment of 3 with similar results is shown.
T cells differentiated with HGFMo acquire cell-contact-dependent suppressor activity. (A-B) CD4+ T cells activated for 7 days with HGFMo in the primary MLR were added to an independently generated MLR, consisting of autologous CD4+CD25- T cells and the same allogeneic GM4-DCs used in the primary MLR (DC/T-cell ratio equal to 1:3). The ratio of DC-activated T cells recovered from the primary MLR to autologous CD4+CD25- T cells plated in the secondary MLR is indicated. Control cultures were established in the absence of added T cells (B). CFSE fluorescence and background staining with isotype-matched fluorochrome-conjugated irrelevant mAbs in unstimulated CD4+ T cells are shown in panel A. The PI of alloreactive T cells is reported. One representative experiment of 4 with similar results is shown. (C) CD4+ T cells activated with HGFMo in the primary MLR were plated in the upper compartment of a Transwell and were restimulated with the same HGFMo used in the primary MLR (DC/T-cell ratio equal to 1:3). Autologous unstimulated CD4+CD25- T cells were plated in the lower compartment of the Transwell and were activated with third-party immunogenic GM4-DCs (DC/T-cell ratio equal to 1:3). The PI of alloreactive T cells is reported. One representative experiment of 4 with similar results is shown.
T cells differentiated with HGFMo acquire cell-contact-dependent suppressor activity. (A-B) CD4+ T cells activated for 7 days with HGFMo in the primary MLR were added to an independently generated MLR, consisting of autologous CD4+CD25- T cells and the same allogeneic GM4-DCs used in the primary MLR (DC/T-cell ratio equal to 1:3). The ratio of DC-activated T cells recovered from the primary MLR to autologous CD4+CD25- T cells plated in the secondary MLR is indicated. Control cultures were established in the absence of added T cells (B). CFSE fluorescence and background staining with isotype-matched fluorochrome-conjugated irrelevant mAbs in unstimulated CD4+ T cells are shown in panel A. The PI of alloreactive T cells is reported. One representative experiment of 4 with similar results is shown. (C) CD4+ T cells activated with HGFMo in the primary MLR were plated in the upper compartment of a Transwell and were restimulated with the same HGFMo used in the primary MLR (DC/T-cell ratio equal to 1:3). Autologous unstimulated CD4+CD25- T cells were plated in the lower compartment of the Transwell and were activated with third-party immunogenic GM4-DCs (DC/T-cell ratio equal to 1:3). The PI of alloreactive T cells is reported. One representative experiment of 4 with similar results is shown.
T cells primed with HGFMo exert cell-contact-dependent suppressor activity
In a subsequent set of experiments, we examined whether DC-primed T cells could suppress the MLR. Responder T cells proliferated in the absence of DC-primed CD4+ T cells, as evaluated by the flow cytometry analysis of CFSE dilution in the progeny of CD4+ T cells relative to the undivided, parental CD4+ T-cell population (Figure 6A). Interestingly, HGFMo-primed CD4+ T cells inhibited the proliferation of responder T cells in a concentration-dependent fashion and maximal suppression could be demonstrated when DC-primed CD4+ T cells and responder T cells were cultured at a 1:1 ratio (Figure 6A-B). Finally, coculture of putative Treg cells and responder T cells with a Transwell insert translated into the abrogation of the inhibitory activity on responder T cells (Figure 6C). Taken together, these data suggest that single-time stimulation of naive CD4+ T cells with HGFMo induced the appearance of Treg cells whose suppressor activities were largely dependent on cell-to-cell contact.
HGF induces a unique gene signature in HGFMo
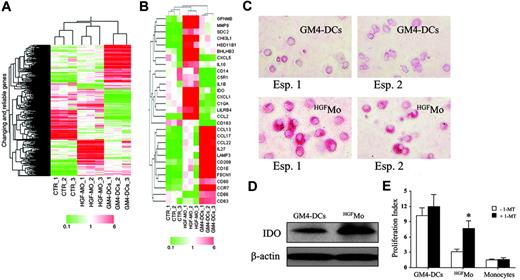
We next assessed the gene-expression profile of HGFMo and GM4-DCs as well as untreated monocytes using the Affymetrix HG-U133A GeneChip array. In order to stringently identify genes specifically associated with treatment, an ANOVA test was applied to the probelist coming from the first low-level filter described in “Materials and methods.” Specifically, a Welch t test at a confidence level of .05 with the Benjamini and Hochberg correction of the familywise error rate was applied along with the Tukey post-hoc test. The ANOVA analysis led to the identification of 672 significant probesets (Figure 7A, Table S1). Among them, 159 probesets showing a signal increase greater than 2-fold in HGF-treated samples were selected as up-regulated by HGF (Table S2). The Tukey post-hoc test led us to determine which pairs among the groups under study have expression means that are statistically significant. Probesets showing statistically significant differences identified by post-hoc test are shown in Figure 7B.
Microarray analysis of gene expression inHGFMo. Monocytes were isolated from the peripheral blood of 3 different healthy donors as detailed in “Materials and methods” and were used in 3 independent experiments, each performed in triplicate. (A) Eisen tree map of the 672 genes that showed significant differences in their expression levels was performed using a supervised approach (ANOVA), as detailed in “Materials and methods.” A combination of 2 hierarchic clustering analyses, the “gene tree” on the left and the “condition tree” on top, is shown. Gene coloring was based on normalized signals, as indicated at the bottom of the panel. (B) Eisen tree map computed using the Pearson correlation equation on the 29 modulated probe sets passing the Tukey post-hoc test. Gene coloring was based on normalized signals, as indicated at the bottom of the panel. Detection of IDO protein expression in GM4-DCs and HGFMo by immunocytochemistry (C) and Western blot (D). One representative experiment out of 3 with similar results is shown. Cells were visualized using an AX70 digital microscope (Olympus) equipped with a 40 ×/0.65 NA objective lens. Image acquisition was performed as described for Figure 2A. (E) Effect of IDO inhibition on the proliferation of CD4+ T cells in primary MLR cultures. 1-methyl-D-tryptophan (1-MT; Sigma Chemical) was added at the beginning of the MLR at 200 μM (final concentration).64 CFSE-labeled CD4+ T cells were cultured in the presence of either GM4-DCs, HGFMo, or cytokine-untreated monocytes that were cryopreserved at the start of the experiment (stimulator-to-responder cell ratio equal to 1:3). T-cell proliferation was expressed in terms of PI, as previously detailed. One representative experiment of 3 with similar results is shown. Error bars indicate mean and SD. *P < .01 compared with MLR cultures performed in the absence of 1-MT.
Microarray analysis of gene expression inHGFMo. Monocytes were isolated from the peripheral blood of 3 different healthy donors as detailed in “Materials and methods” and were used in 3 independent experiments, each performed in triplicate. (A) Eisen tree map of the 672 genes that showed significant differences in their expression levels was performed using a supervised approach (ANOVA), as detailed in “Materials and methods.” A combination of 2 hierarchic clustering analyses, the “gene tree” on the left and the “condition tree” on top, is shown. Gene coloring was based on normalized signals, as indicated at the bottom of the panel. (B) Eisen tree map computed using the Pearson correlation equation on the 29 modulated probe sets passing the Tukey post-hoc test. Gene coloring was based on normalized signals, as indicated at the bottom of the panel. Detection of IDO protein expression in GM4-DCs and HGFMo by immunocytochemistry (C) and Western blot (D). One representative experiment out of 3 with similar results is shown. Cells were visualized using an AX70 digital microscope (Olympus) equipped with a 40 ×/0.65 NA objective lens. Image acquisition was performed as described for Figure 2A. (E) Effect of IDO inhibition on the proliferation of CD4+ T cells in primary MLR cultures. 1-methyl-D-tryptophan (1-MT; Sigma Chemical) was added at the beginning of the MLR at 200 μM (final concentration).64 CFSE-labeled CD4+ T cells were cultured in the presence of either GM4-DCs, HGFMo, or cytokine-untreated monocytes that were cryopreserved at the start of the experiment (stimulator-to-responder cell ratio equal to 1:3). T-cell proliferation was expressed in terms of PI, as previously detailed. One representative experiment of 3 with similar results is shown. Error bars indicate mean and SD. *P < .01 compared with MLR cultures performed in the absence of 1-MT.
The functional analysis of transcripts passing the Tukey posthoc test revealed that HGF treatment induced genes involved in tolerance and tumor progression. Among genes implicated in tolerance, we documented overexpression of C5R1,33 HSD11B1,34 CCL2 (Figure S4 and Table S3),35 C1QA,36 IDO (Figure S4),37 ILT3 (LILRB4; see also flow cytometry data in Figure 2B),38 and IL10 (see also flow cytometry and RT-PCR data in Figure 2C-D). Among genes potentially involved in the promotion of tumor progression, HGFMo displayed an up-regulation of CXCL1 (Figure S4),39 IL1B,40 GPNMB,41 CHI3L1,42 CXCL5,43 MMP9,44 and SDC2.45 Conversely, GM4-DCs expressed fascin (FSCN1), which is involved in the Ag-presentation activity of mature DCs,46 LAMP3, and CD209 (see also flow cytometry data in Figure 1A).47 Finally, GM4-DCs up-regulated CD80, CD86, and CD83 (see also flow cytometry data in Figure 1A) as well as mRNA signals for molecules involved in T-cell chemotaxis and migration (eg, CCL17, CCL13, CCL22, CCR7, and IL27).48,49
In light of the functional relevance of IDO overexpression by HGFMo for the induction of T-cell hyporesponsiveness,37 we explored the potential role of IDO in the suppression of T-cell proliferation. To this end, MLR cultures containing HGFMo and naive allogeneic CD4+ T cells received the IDO inhibitor 1-MT.50 As shown in Figure 7E, provision of 1-MT translated into a significant increase of CD4+ T-cell proliferation in response to HGFMo during primary allogeneic challenge. Collectively, gene expression studies suggested that HGF modulates specific signaling pathways and induces a unique gene signature in monocytes.
Discussion
The effects of HGF on cells of the immune system are largely unknown. In murine allogeneic bone marrow transplantation, HGF ameliorated acute graft-versus-host disease (aGVHD) through reduction of IL-12 serum levels and suppression of target organ IFN-α and TNF-α mRNA.51,52 Furthermore, HGF prolongs the survival of cardiac allografts by immunomodulative potencies, enhancing IL-10 and TGF-β mRNA expression during acute rejection.53 Finally, HGF improves experimental autoimmune myocarditis by inhibiting myosin-specific T-cell responses and enhancing T-cell production of IL-10.54
The studies showed herein demonstrate that HGF affects the differentiation of human monocytes toward accessory cells with DC features, endowed with regulatory activity. Such monocytes differentiated in the presence of exogenous HGF (HGFMo) manifested endocytic activity as well as unique phenotypic features, including low or undetectable expression of costimulatory molecules and CD209, and maintenance of CD14. This observation accords with previous reports on CD14+ DCs differentiated with GM-CSF/IL-15 or IL-10/IFN-α55,56 and after in vivo exposure to G-CSF.14 Importantly, addition of IL-4 to the DC culture reversed the regulatory activity of HGF on monocyte differentiation.
Human plasmacytoid DCs have been implicated in the induction of both Th1 and Treg responses depending on state of activation and maturation stimulus.8,57 Ags assigned to pDCs, namely CD123 and BDCA-2, were expressed at comparable levels on HGFMo and GM4-DCs. Collectively, monocytes cultured with HGF acquired a peculiar costimulationlowILT3+ cell-surface phenotype, distinct from that so far described in conventional DCs32 as well as in pDCs.8
HGFMo acquired an IL-10++IL-12p70- cytokine secretion profile and were poor activators of allogeneic CD4+ T-cell proliferation, as shown by CFSE dilution experiments. Production of IL-10 is a well-known mechanism responsible for immature DC-mediated inhibition of T-cell function.58 However, it has been demonstrated that in addition to autocrine IL-10 production by immature DCs, other mechanisms of T-cell suppression by DCs might be operational, including surface expression of ILT3, ILT4, or specific isoforms of CD45.5,38,59 Interestingly, HGFMo expressed high levels of ILT3, in sharp contrast with GM4-DCs (Figure 2B) and with freshly isolated, cytokine-untreated monocytes (data not shown). Blockade of surface-membrane ILT3 on HGFMo translated into partial restoration of T-cell proliferation in primary MLRs. Mechanistically, IL-10 released in MLR cultures performed with HGFMo was at least in part responsible for the suppression of T-cell proliferation, as shown by experiments with neutralizing mAbs to IL-10. Importantly, both HGFMo and HGFMo-primed CD4+ T cells contributed to IL-10 production and release in supernatants of MLR cultures, as suggested by flow cytometry analyses of intracellular cytokine expression. These findings indicate that multiple mechanisms might be involved in the induction of T-cell hyporesponsiveness by HGFMo, including secretion of immunomodulatory cytokines and expression of inhibitory surface Ags. Finally, T-cell hyporesponsiveness was not reverted by exogenous IL-2, as already reported for the anergic state of IL-10-treated T cells but at variance with anergy induced by costimulation blockade.14,59
Evidence has been provided in favor of an in vitro generation of human Treg cells from naive T cells.60 However, in vitro-differentiated Treg cells often resemble Tr1 cells rather than classic CD4+CD25+ Treg cells, as they produce high amounts of IL-10 and suppress bystander T cells in a cytokine-dependent manner. Notably, human DC subsets can induce the in vitro generation of CD4+CD25+FoxP3+ Treg cells that produce IL-10 and TGF-β and suppress T cells in a Ag-nonspecific manner.57 CD4+CD25+ Treg cells can be also expanded in vitro by Ag-processing DCs differentiated with GM-CSF.61
We first measured the expression levels of the transcription repressor FoxP3, which is preferentially detected in CD4+CD25+ Treg cells of thymic and peripheral origin and is currently considered as a master control gene for Treg-cell development.62 We used T cells that were initially depleted of the CD25+ fraction. Under these conditions, CD25 expression on developing T cells can be attributed to differentiation from the starting CD4+CD25- cell population rather than expansion of pre-existing CD4+CD25+ T cells by HGFMo. Notably, CD4+ T cells confronted with HGFMo displayed a CD25+ phenotype and expressed FoxP3 mRNA and protein. Furthermore, HGFMo-primed CD4+ T cells released high quantities of IL-10, TGF-β, and IL-5, but low levels of IL-2, IL-4 and IFN-γ. These findings suggested that CD4+ T cells activated with HGFMo acquired regulatory activity against conventional, nonregulatory T cells. Coculture experiments indicated that the addition of HGFMo-primed CD4+ T cells to an allogeneic MLR containing freshly isolated responder CD4+CD25- T cells and GM4-DCs translated into measurable suppression of responder T-cell proliferation in the presence of cell-to-cell contact.
In this respect, the relationship between IL-10-secreting Treg cells and naturally occurring CD4+CD25+ is incompletely defined. Whereas naturally occurring CD4+CD25+ Treg cells exert in vitro suppression through cell-contact-dependent mechanisms, IL-10 and/or TGF-β1 might be required for in vivo suppression, depending on specific disease models. Previously, it has been shown that IL-10-producing Treg cells might inhibit in vitro T-cell proliferation in IL-10-independent, but cellcontact-dependent, manner.63 Accordingly, CD4+CD25+ Treg cells differentiated in vitro upon challenge with HGFMo released copious amounts of IL-10 but inhibited T-cell proliferation in an IL-10-independent manner.
Gene-expression profiling led us to identify signaling pathways activated by HGF treatment. Interestingly, HGF-DCs overexpressed a set of genes implicated in immune tolerance and tumor invasion, whereas GM4-DCs up-regulated markers characteristic of mature DCs. Among genes involved in tumor progression, HGF up-regulated angiogenetic proteins, such as CXCL1, CXCL5, and IL-1β. It has been demonstrated that IL-1β promotes tumor invasiveness and angiogenesis, and also induces immune suppression in the host.40 In addition, HGFMo expressed high levels of genes implicated in matrix degradation and cell migration, such as MMP9, SDC2, and GPNMB, or osteoactivin. It is worth mentioning that osteoactivin, known as DC-associated heparan sulfate proteoglycan-integrin ligand (DC-HIL), increases the ability of tumor cells to degrade components of the extracellular matrix and increases motility, a critical aspect of invasive cancers.41
Interestingly, genes potentially involved in the induction of immune tolerance such as C5R1,33 HSD11B1,34 CCL2,35 C1QA,36 IDO,37 ILT3 (LILBR4),55 complement component C1q,36 and IL10 were up-regulated by HGF. Expression of IDO in mouse DCs inhibits T-cell clonal expansion.37,64 Interestingly, provision of the IDO inhibitor 1-MT to MLR cultures containing HGFMo translated into a significant increase of T-cell proliferation, suggesting that modulation of tryptophan catabolism in T cells might represent one additional mechanism responsible for the HGF-mediated regulation of T-cell function.
An important implication of our findings pertains to the tumor-cell-driven escape from immunosurveillance. HGF is reportedly increased in the serum of patients affected by Hodgkin disease,20 and increased serum HGF levels confer unfavorable prognosis to patients with multiple myeloma.65 Conceivably, HGF might impair DC function in vivo through induction of IL-10 release by maturing DCs,66 and DC dysfunction induced by soluble factors (ie, HGF or IL-616 ) is expected to negatively affect the efficacy of antitumor vaccination strategies. Finally, IDO may suppress antitumor immunity in mouse models of neoplastic diseases67 and transfection of IDO renders tumor cell lines immunosuppressive in vitro. From a therapeutic standpoint, HGF might represent an attractive target for cancer immunotherapy and HGF antagonists such as NK468 warrant further investigations, as they might limit tumor evasion of the host immune system.
Prepublished online as Blood First Edition Paper, March 9, 2006; DOI 10.1182/blood-2005-08-3141.
Supported by “Stem Cell Project,” Fondazione Cassa di Risparmio di Roma, Rome, Italy; Centro Nazionale delle Ricerche-Ministero dell'Istruzione, dell'Università, e della Ricerca (CNR-MIUR), Rome, Italy; Alma Med Foundation, University of Bologna, Bologna, Italy; and the Italian Association against Leukemia, Section of Bologna (BOLOGNAIL), Italy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.