Abstract
B-cell lymphomas are presently diagnosed according to the WHO criteria based on morphologic, immunophenotype and cytogenetic findings. However, the precise distinction among common lymphoma types is frequently a difficult task, and there are areas of overlapping and heterogeneity between them. Here we have analyzed whether gene expression profiling (GEP) data, solely considered, could be used to validate the currently used B-cell lymphoma classification, or proposing new lymphoma types, and for identifying functional signatures or genes defining these GEP-based lymphoma classification.
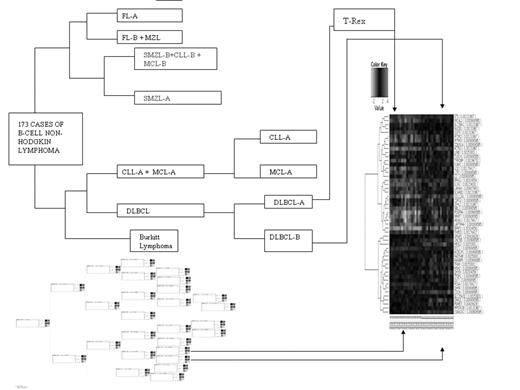
To this aim, we collected Gene Expression Profiling (GEP) for 173 cases of B-cell NHL, including BL (9), DLBCL (36), MALT (3), MCL (20), CLL (38), FL (33), MZL (6) and SMZL (29). The gene expression data for lymphoma cases was normalized against an average gene expression of reactive lymph nodes, except the SMZL which was normalized against normal spleen (3 cases). The analysis of the cases was done using Cluster Accuracy Analysis Tool (CAAT) (Cunningham P., 2005), that enabled us not only to compare gene expression between each node starting from root but also to identify new classes within existing lymphoma diagnosis defined by an internal validation method called Silhouette Width index (Julia Handl. et al, 2005). Using this approach each cluster could be represented by so called silhouette, which is based on the comparison of its tightness and separation. The average silhouette width could be applied for evaluation of clustering validity and can also be used to decide how good the number of selected clusters is.
Using this approach, we obtained the following categorization of lymphoma cases:
Using T-Rex (Herrero J & Dopazo J., 2005) to compare differential expression between the categories obtained by CAAT, and FatiScan analysis (Al-Shahrour, F., 2006), we identified genes that were differentially expressed between molecular categories of lymphoma types, assigning them to Gene Ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes)-defined pathways. The functional signatures that were identified as distinguishing between these lymphoma types were defining cell cycle, cytokine-cytokine receptor interaction, T-cell receptor, B-cell receptor, cell adhesion, NF-kB activation, and other significant interactions. Comparison between these lymphoma clusters following this definition yielded large number of genes distinguishing them, this list including already known genes and a large number of new potential markers.
Disclosure: No relevant conflicts of interest to declare.
Author notes
Corresponding author