Abstract
Our previous results demonstrated that B cells from a patient (pt1) with non–X-linked hyper-IgM syndrome (HIGM) possess an atypical CD23lo phenotype that is unaffected by CD40-mediated activation. To investigate the molecular mechanism underlying defective CD23 expression in pt1 B cells, we used lymphoblastoid cell lines that express LMP1 under the control of a tetracycline-inducible promoter (LCLtet). Our analysis revealed that the CD23lo phenotype in the pt1-LCLtet cells is a direct consequence of diminished CD23 transcription. We demonstrate a marked decrease in c-Rel–containing complexes that bind to the proximal CD23a/b promoters in pt1-LCLtet extracts, resulting from an overall lower expression of c-Rel in pt1-LCLtet cells. Analysis of c-Rel mRNA revealed relatively equal amounts in pt1-LCLtet and control LCLtet cells, indicating that diminished c-Rel protein expression is unrelated to decreased transcription. Finally, a critical role for c-Rel in CD23 regulation was demonstrated by effectively altering c-Rel expression that resulted in the direct modulation of CD23 surface expression. Collectively, these findings demonstrate that low levels of c-Rel are the underlying cause of aberrant CD23 expression in pt1 B cells and are likely to play a critical role in the pathophysiology of this form of HIGM.
Introduction
Differentiation of B cells in response to T-cell–dependent (TD) antigens requires cognate interactions between CD40 expressed on B cells and CD40 ligand (CD154, CD40L) expressed on activated CD4+ T cells. Antigen-activated B cells respond to this interaction by undergoing proliferation, homotypic adhesion, and class-switch recombination (CSR) to produce new clonatypic Abs (reviewed in Harnett1 ). The critical nature of the CD40:CD40L interaction for TD immune responses is illustrated by an absence of CSR and deficient humoral immunity in humans and mouse models lacking functional CD40 or CD40L.2-9 In particular, patients with X-linked hyper-IgM syndrome (XHIGM or HIGM-1) harbor mutations in the CD40L gene with impaired immunity characterized by frequent upper and lower respiratory tract infections, and other conditions including Pneumocystis carinii–mediated interstitial pneumonia, acute or chronic diarrhea associated with cryptosporidium infection, neutropenia, liver and biliary tract tumors, and autoimmunity (reviewed in Etzioni and Ochs10 ). Other forms of HIGM occur as a consequence of autosomal recessive transmission that results in defects in both CD40 signaling and CSR due to mutations in activation-induced cytidine deaminase (AID),11-13 CD40,7 or NF-κB essential modulator (NEMO).7,14-16
We have had a long-standing interest in defining the underlying mutation in a female patient (pt1) with undefined non–X-linked HIGM.17 Previously, we reported that this patient has a defect in B-cell activation that specifically targeted a subset of CD40-induced responses, including selective impairment of CD23 expression. Unlike other HIGM B cells, defective GL transcription and CSR were capable of being complemented in vitro by extended signaling through CD40.17 Also, there was a clear defect in cellular proliferation in response to mitogenic signals (K.T.L. and L.R.C., unpublished observation, August 2004). Together, these observations suggested that the pt1 mutation negatively affected the B cells' ability to respond to a normal threshold of CD40 activation signals resulting in antigen-dependent differentiation requiring a higher intensity of signaling ordinarily not achieved under physiological conditions.17
Our most recent work has used a model system to further define the pt1 mutation. Specifically, a set of Epstein-Barr virus (EBV)–transformed lymphoblastoid cell lines (LCLs) was generated from pt1 and control B cells that express the viral transforming protein latent membrane protein 1 (LMP1) under the control of a tetracycline (tet)–inducible promoter (LCLtet). Because LMP1 activation mimics signals through the CD40 pathway, LCLtet cells allow for independent comparisons of downstream functions in response to regulated LMP1 and CD40 signals. Characterization of these cells revealed a CD23lo/CD38hi surface phenotype of generated pt1-LCLtet cells that was distinctly different from the typical CD23hi/CD38lo activated phenotype of control LCLtet cells18 and conventional EBV-immortalized LCLs.19 We also observed that the pt1-LCLtet cells retained the CD23lo/CD38hi phenotype after extended periods of culture with LMP1 or CD40 signals. This was in direct contrast to the low mitogenic potential of these cells, which was restored with signals through LMP1 but not CD40. Thus, the CD23lo phenotype was maintained in pt1-LCLtet cells concurrently with the LMP1-mediated complementation of other phenotypic characteristics suggesting that the pt1 defect was closely linked to CD23 expression.
CD23 is a type II integral membrane glycoprotein expressed on a number of cell types and is up-regulated in response to multiple activation pathways.20-22 The human CD23 protein is expressed as 2 isoforms, CD23a and CD23b, which are generated by the use of alternative transcription initiation sites followed by differential splicing.23 The CD23a promoter is constitutively active at low levels in B cells, while the CD23b promoter is activated by a number of stimuli, in particular IL-4, in multiple lineages including T cells, B cells, monocytes, and polymorphonuclear leukocytes.23 The 2 CD23 species differ at the N-terminal amino acids of the intracytoplasmic region (7 for CD23a and 6 for CD23b), which corresponds to distinct intracellular trafficking properties of the 2 proteins.23-25
CD23 was originally identified in newly immortalized LCLs as a protein that is rapidly induced upon EBV infection and expressed at high levels in immortalized B cells.22,26,27 The expression of CD23 appears to be closely associated with the immortalizing functions of EBV based on the observation that a naturally occurring EBV mutant, P3HR-1, carrying a deletion in the EBNA-2 protein, failed to induce CD23 expression and immortalize B cells in vitro.28 Analysis of EBV-transformed B cells from various differentiation stages shows a high level of surface CD2329 and expression of both CD23a/b isoforms.30
Numerous responsive elements have been identified in the proximal CD23a and CD23b promoter regions. These binding sites correspond to a range of transcriptional regulatory factors serving as targets for signaling pathways involved with CD23 gene expression. Both CD23a and CD23b promoters contain Rel/NF-κB31 and canonical STAT6 binding sites.31,32 Rel/NF-κB transcription factor family proteins are essential for promoting cell survival, proliferation, and differentiation (reviewed in Xiao and Ghosh33 ). These factors exist primarily in the cytoplasm as inactive complexes with IκB proteins. In response to cellular stimuli, including LMP1 activation, Rel/NF-κB dimers translocate to the nucleus where they bind to specific motifs in the regulatory regions of target genes. c-Rel, a lymphoid-specific Rel/NF-κB family member, has previously been implicated in CD23 expression. B cells from c-Rel–deficient mice consistently exhibited lower levels of CD23 in resting or activated states when compared with wild-type B cells.34 Other studies have revealed roles for c-Rel in both cell-mediated and humoral immunity.35 In particular, mice with inactivated or mutated c-Rel, generated by gene targeting, possess mature B cells with impaired activation and function associated with defects in isotype switching and cytokine expression.36
In this study, we attempt to further characterize the underlying defect in pt1 B cells by identifying the molecular mechanism that results in aberrant CD23 expression. We show that reduced CD23 expression is attributable to a lower level of c-Rel activity in pt1-LCLtet cells that directly affects the transcriptional activity of both the CD23a and CD23b promoters. Furthermore, with respect to CD23 expression in our system, c-Rel activity appears to be nonoverlapping with other Rel/NF-κB family members and the viral protein EBNA-2. These findings extend our previous understanding of the pt1 immunodeficiency to include a defect in c-Rel expression and activity.
Patient, materials, and methods
Cell lines
The institutional review board of Rutgers University has approved the human subjects protocol for this work (no. 02-027), and informed consent in accordance with the Declaration of Helsinki and assent were obtained from pt1 and her parents before enrollment in the study. Mini-EBV transformation (LCLtet) of primary B cells from pt1 and controls (D11 and C2 clones) was carried out with recombinant plasmid p1852 as previously described.18,37 Standard EBV-transformed LCLs and LCLtet cell lines were maintained in RPMI Complete (10% fetal bovine serum [FBS], 1% penicillin/streptomycin, 1% l-glutamine, 1% sodium pyruvate; Mediatech, Herndon, VA). LCLtet cells were also cultured with 1 μg/mL tetracycline (+tet). For –tet samples, cells were removed from tet-containing media and cultured in RPMI Complete for 4 days. All cultures were grown at 37°C, in 5% CO2.
Flow cytometry
Cells (5 × 105) were washed in fluorescence-activated cell sorter (FACS) buffer (3% FBS/0.1% NaN3/1X PBS) and incubated with equal saturating amounts of FITC-conjugated mAb against CD23 or the matched isotype control (Ancell, Bayport, MN). Samples were analyzed for cell surface expression by flow cytometry using Cell Quest software on a FACSCalibur system (Becton Dickinson, San Jose, CA). Additionally, denoted samples were further cell sorted and isolated by using a FACSVantage system with DiVa software (Becton Dickinson).
Quantitative real-time polymerase chain reaction (qPCR)
Forward primer sequences used were as follows: CD23A, 5′-TCCACTAACCAGAGCTGT-3′ and CD23B, 5′-CAGAGGCCAAATAGAACAGG-3′; the same reverse primer was used for both transcripts: 5′-GCAGCACGATCTGAGTCCCACG-3′. The primer sequences for c-Rel were forward, 5′-CGAACCCAATTTATGACAACCG-3′ and reverse, 5′-TTTTGTTTCTTTGCTTTATTGCCG-3′. Total RNA from LCLtet cells was isolated using Trizol (Invitrogen, Carlsbad, CA) and reverse transcribed with SuperScript II Reverse Transcriptase (Invitrogen). cDNA dilutions were used as templates for triplicate reactions ran on an ABI 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA) using SYBR Green PCR Mix (Applied Biosystems). The cycling conditions were 10 minutes at 95°C, followed by 35 cycles of 15 seconds at 95°C, 30 seconds at 60°C (CD23A and c-Rel) or 56°C (CD23B), and 30 seconds at 72°C. A melt curve peak analysis was performed for each amplicon. Threshold cycle (Ct) values of the target genes were compared with a standard curve and normalized to an internal control gene, metastatic lymph node 51.
Transfection of LCLtet cells
Luciferase transfections. LCLtet cells (5 × 106) were incubated with 50 μg pGL3E reporter constructs (Promega, Madison, WI) that contains nucleotides –473 to 85 of the human CD23a promoter (pGL23a) or nucleotides –338 to –23 of the human CD23b promoter (gifts from S. Lederman, Columbia University), and either 1 μg pRL-SV40 or pRL-TK control plasmids (Promega). Electroporation was performed at 960 μF and 250 mV. Cells were transferred to RPMI Complete with tet and harvested 48 hours after transfection for analysis of luciferase activity using a Dual Luciferase Assay kit (Promega) and a Lumat Luminometer (Berthold Tech, Oak Ridge, TN). Raw data were normalized to the Renilla luciferase efficiency for each experiment.
c-Rel transfections. Using the Amaxa Biosystems Transfection System (Cologne, Germany), LCLtet cells (2 × 106) were suspended in 100 μL Nucleofector V solution with 3 μg human c-Rel plasmid (pJDCMV19SV-hc-Rel),38 a gift from C. Gélinas (UMDNJ), or 7.5 nM small-interfering RNA oligos for c-Rel (siRNAc-Rel) or control sequence (siRNActrl), (Santa Cruz Biotechnology, Santa Cruz, CA) and then electroporated using program O-06 (plasmid) or U-15 (siRNA). Cells were transferred to RPMI Complete with tet and harvested 24 hours after transfection. For transfection efficiency, LCLtet cells were cotransfected with green fluorescent protein and analyzed by flow cytometry. The transfection efficiency ranges between 20% and 30% for each experiment.
Nuclear extracts and electromobility shift assays (EMSAs)
Nuclear extracts and EMSAs were carried out as previously described.39 The following end-labeled probes for the CD23a and CD23b promoters were used in EMSAs: CD23b-I, 5′-CGACCCTTAGCTACTGCCTTTCACCCAGAAGA-3′; CD23b-II, 5′-CGAAGCGGGGCTCCCCAGTCCCTCTCTGGGAAAGAGGGTGA-3′; CD23b-III, 5′-CGATTTCTAAGAAAGGGACTGGTGTGAGTAAGGAGGTGAGGC-3′; and CD23a–NF-κB, 5′-AATAATAACACGGACTTCACCGGGTGTGGGGAGCA-3′. Binding reactions were prepared using 3 μg extract, 1 μg poly dI-dC in binding buffer (10 mM Tris-Cl [pH 7.5], 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 5% glycerol) in the presence or absence of competitor oligos for 15 minutes at 25°C, or the presence of 2 μg or 6 μg antibody for 90 minutes. The following rabbit polyclonal NF-κB antibodies (Santa Cruz Biotechnology) were used in supershift assays: p50, p65, p52, c-Rel, and RelB, with purified rabbit IgG as a control. Probe (3 × 104 cpm) was added to reactions and incubated for 20 minutes at 25°C. Samples were loaded on a 6% acrylamide gel and visualized by autoradiography.
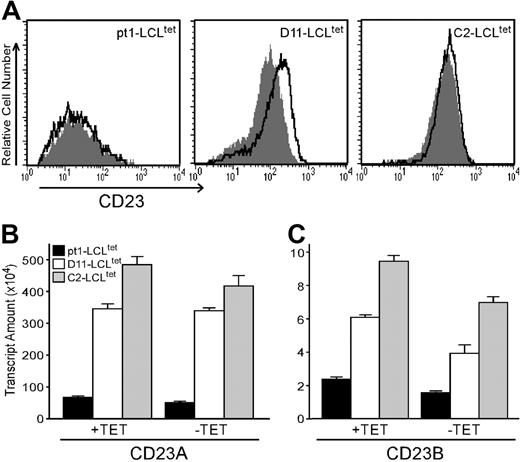
Pt1-LCLtetcells have diminished expression of CD23 protein and transcripts with LMP1 activation. (A) Cell surface expression of CD23. pt1-LCLtet and control LCLtet cells were cultured continuously in the presence of LMP1 (+tet, outlined peak) or in the absence of LMP1 (–tet, shaded peak) for 4 days. Cells were then stained with anti-CD23 and analyzed by flow cytometry. (B) Relative transcript levels of CD23a and CD23b. RNA from pt1-LCLtet cells (▪) and control LCLtet cells (D11: □; C2: ▦) was reverse-transcribed and analyzed by quantitative real-time PCR with probe sets specific for CD23a and CD23b. Note the scale difference of the y-axis. Results are expressed as relative transcript amounts and standard deviation of 3 independent experiments.
Pt1-LCLtetcells have diminished expression of CD23 protein and transcripts with LMP1 activation. (A) Cell surface expression of CD23. pt1-LCLtet and control LCLtet cells were cultured continuously in the presence of LMP1 (+tet, outlined peak) or in the absence of LMP1 (–tet, shaded peak) for 4 days. Cells were then stained with anti-CD23 and analyzed by flow cytometry. (B) Relative transcript levels of CD23a and CD23b. RNA from pt1-LCLtet cells (▪) and control LCLtet cells (D11: □; C2: ▦) was reverse-transcribed and analyzed by quantitative real-time PCR with probe sets specific for CD23a and CD23b. Note the scale difference of the y-axis. Results are expressed as relative transcript amounts and standard deviation of 3 independent experiments.
Protein immunoblots
Whole-cell lysate was prepared by lysis in buffer containing 50 mM Tris-HCl (pH 8), 1 mM EDTA, 1% NP-40, 150 mM NaCl, 1 mM Na3VO4, 50 mM NaF, and protease inhibitors. For Western analysis, equal amounts of protein were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene difluoride (PVDF) membrane (Schleicher and Schuell, Keene, NH), and probed with antibodies against c-Rel and actin (Santa Cruz Biotechnology). Horseradish peroxidase–conjugated secondary antibodies (BioRad, Hercules, CA) were used for detection by enhanced chemiluminescence (ECL; Amersham, Piscataway, NJ).
Results
Diminished CD23 expression is directly related to decreased transcriptional activity of both the CD23a and CD23b promoters
One manifestation of the pt1 B-cell defect is uncharacteristically low CD23 expression in the presence of constitutive LMP1 signals18 (Figure 1A). To determine whether low CD23 surface expression is due to reduced transcription, pt1-LCLtet and control LCLtet cells were analyzed for CD23a/b expression by qPCR. We observed that control and pt1-LCLtet cells expressed both CD23a and CD23b transcripts along with an expected NF-κB–mediated up-regulation of the CD23b transcript in all cell lines in response to LMP1 signals31,40 (Figure 1B-C). However, control LCLtet lines expressed markedly higher levels of both transcripts in the presence (+tet) or absence (–tet) of LMP1, relative to the pt1-LCLtet cells. Thus, the reduction in CD23 surface expression observed in the pt1-LCLtet cells corresponds to reduced transcriptional activity from both the CD23a and CD23b promoters.
Low CD23 promoter activity is consistent with limiting amounts of a transcription factor
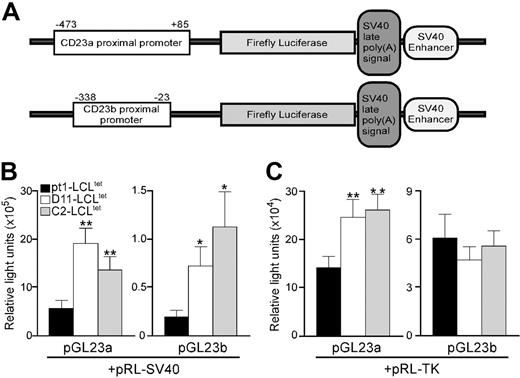
To identify sequences in the CD23a and CD23b promoters responsible for diminished endogenous CD23 transcription, transient transfections were conducted with expression constructs containing 558 bp (–473 to 85) and 315 bp (–338 to –23) of the CD23a and CD23b proximal promoters (designated pGL23a and pGL23b constructs, respectively) (Figure 2A). The promoter constructs, as well as the control pRL-SV40 plasmid expressing the Renilla luciferase gene under the control of the SV40 promoter, were introduced into the pt1-LCLtet and control LCLtet cell lines. Analysis of 3 independent experiments revealed that the CD23a promoter was between 9- to 14-fold more active than the CD23b promoter in all cell lines (Figure 2B). However, when the level of activity of the 2 promoters was compared between the pt1-LCLtet and control cell lines, both promoters were significantly more active in the controls, compared with pt1-LCLtet cells (Figure 2B). These results indicated that a trans-acting factor necessary for CD23 expression is either missing or expressed at a reduced level in the pt1-LCLtet cells.
To distinguish between 2 possibilities, a second set of transfections was carried out using the pRL-tk control plasmid, which expresses the Renilla luciferase gene under the control of the thymidine kinase promoter. We reasoned that if a specific transcription factor was absent in the pt1-LCLtet cells, cotransfecting with a different control plasmid would have no effect on the activity of the pGL23a/b constructs. Alternatively, if a transcription factor was limiting, then possible competition between the CD23 and the SV40 promoters could influence expression of the promoter constructs. Surprisingly, use of pRL-tk control plasmid eliminated any observed difference in the CD23b promoter activity between pt1 and control cells while markedly diminishing the difference in CD23a promoter activity (Figure 2C). This finding was consistent with a limiting amount of a common transcription factor necessary for CD23a, CD23b, and SV40 promoter function being expressed at a reduced level in the pt1 B cells.
Atrans-acting factor necessary for optimal CD23 transcription is limiting in pt1-LCLtetcells. (A) Schematic diagram of the pGL23a and pGL23b constructs denoting the approximate positions of the specific promoters and downstream genes encoded in the plasmid. LCLtet cells (pt1: ▪; D11: □; and C2: ▦), cultured continuously in the presence of tet, were transiently cotransfected with pGL23a/b constructs and either the pRL-SV40 (B) or the pRL-TK (C) control plasmids. Luciferase activity was assayed 48 hours after transfection. Results are shown as relative light units with SEM of at least 3 independent experiments with P < .1 (*) or P < .05 (**).
Atrans-acting factor necessary for optimal CD23 transcription is limiting in pt1-LCLtetcells. (A) Schematic diagram of the pGL23a and pGL23b constructs denoting the approximate positions of the specific promoters and downstream genes encoded in the plasmid. LCLtet cells (pt1: ▪; D11: □; and C2: ▦), cultured continuously in the presence of tet, were transiently cotransfected with pGL23a/b constructs and either the pRL-SV40 (B) or the pRL-TK (C) control plasmids. Luciferase activity was assayed 48 hours after transfection. Results are shown as relative light units with SEM of at least 3 independent experiments with P < .1 (*) or P < .05 (**).
A Rel/NF-κB–related complex possesses a unique binding pattern and affinity within regions of the CD23a and CD23b promoters in pt1-LCLtet cells
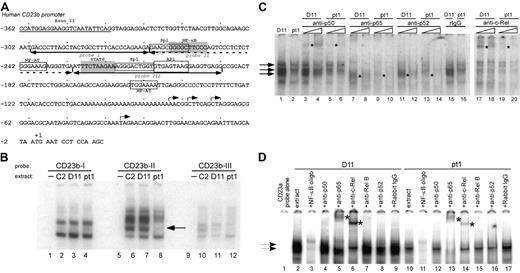
The CD23a and CD23b promoter regions have been extensively characterized and shown to contain multiple responsive binding sites that lie immediately proximal to the transcription initiation sites and upstream of the CD23a and CD23b coding sequences.31,40-44 We initially examined 3 adjacent regions of the CD23b proximal promoter since it has been previously shown to be inducible by NF-κB activation. The regions analyzed contain functional NF-κB and STAT6 sites as well as putative binding sites for Sp1, NF-AT, and AP1 (Figure 3A).31,32,40,45 The 3 adjacent probes covered 109 nucleotides of upstream sequence (–190 to –299), and there were no significant differences in factors binding to probe I (–299 to –269) (Figure 3B, lanes 1-4) or probe III (–228 to –190) (lanes 9-12). However, EMSA carried out with probe II (–268 to –229) (Figure 3B, lanes 5-8) containing a known NF-κB site and putative NF-AT and Sp1 binding sites revealed an absence of a middle complex in the pt1 extract that was clearly present in the control extracts (lanes 6-8).
To determine whether NF-κB activity was limiting in the pt1-LCLtet cells, we used different competitor oligos for consensus factor binding sites and observed that all of the major complexes formed with the CD23b-II probe were competed with cold NF-κB consensus oligo (data not shown). To define whether specific Rel/NF-κB family members were limiting in the pt1-LCLtet cells, EMSA was carried out with the CD23b-II probe, extract from pt1-LCLtet and control D11-LCLtet cells, and increasing concentrations of Rel/NF-κB antibodies (Figure 3C). The most apparent difference was a virtual absence of supershifting with the c-Rel antibody in pt1-LCLtet cells compared with the D11-LCLtet cells (Figure 3C, compare lanes 17 and 18 with 19 and 20). Although there appeared to be other subtle differences in intensity with anti-p50, -p65, and -p52, the difference in c-Rel binding was the most striking and suggested a decreased level of c-Rel activity in the pt1-LCLtet cells. We further confirmed this finding by analyzing the CD23a promoter fragment containing a putative NF-κB site (–109 to –76) and showed that c-Rel binding was significantly decreased as well (Figure 3D, compare lanes 6 and 14).
A distinct NF-κB–related complex binds to the proximal CD23 promoter region in pt1-LCLtetcells. (A) Schematic representation of the proximal CD23b promoter is shown, highlighting putative transcription factor binding sites for Sp1, NF-κB, NF-AT, STAT6, and AP1. Underscoring arrows indicate the recognition sites of probes used in the following EMSAs. (B) Nuclear extracts from LCLtet cells were incubated with probes CD23b-I (lanes 1-4), CD23b-II (lanes 5-8), and CD23b-III (lanes 9-12). Lane 8, denoted by an arrow, reveals a unique complex formation binding to probe CD23b-II in pt1 extract. (C) Further analysis of the CD23b-II probe was performed with control D11 and pt1-LCLtet nuclear extracts. The following antibodies were added in increasing amounts to the indicated lanes: p50 (lanes 3-6), p65 (lanes 7-10), p52 (lanes 11-14), purified rabbit IgG (lanes 15-16), and c-Rel (lanes 17-20). (D) EMSA with a probe that spans the identified NF-κB binding site in the CD23a promoter (nucleotides –109 to –76). An unlabeled NF-κB competing oligonucleotide was added in 15-fold excess (lanes 3 and 11). The following supershifting antibodies were added to the indicated lanes: p50 (lanes 4 and 12), p65 (lanes 5 and 13), c-Rel (lanes 6 and 14), RelB (lanes 7 and 15), p52 (lanes 8 and 16), and purified rabbit IgG (lanes 9 and 17). Arrows indicate NF-κB–related complexes, and supershifted complexes are denoted by asterisks.
A distinct NF-κB–related complex binds to the proximal CD23 promoter region in pt1-LCLtetcells. (A) Schematic representation of the proximal CD23b promoter is shown, highlighting putative transcription factor binding sites for Sp1, NF-κB, NF-AT, STAT6, and AP1. Underscoring arrows indicate the recognition sites of probes used in the following EMSAs. (B) Nuclear extracts from LCLtet cells were incubated with probes CD23b-I (lanes 1-4), CD23b-II (lanes 5-8), and CD23b-III (lanes 9-12). Lane 8, denoted by an arrow, reveals a unique complex formation binding to probe CD23b-II in pt1 extract. (C) Further analysis of the CD23b-II probe was performed with control D11 and pt1-LCLtet nuclear extracts. The following antibodies were added in increasing amounts to the indicated lanes: p50 (lanes 3-6), p65 (lanes 7-10), p52 (lanes 11-14), purified rabbit IgG (lanes 15-16), and c-Rel (lanes 17-20). (D) EMSA with a probe that spans the identified NF-κB binding site in the CD23a promoter (nucleotides –109 to –76). An unlabeled NF-κB competing oligonucleotide was added in 15-fold excess (lanes 3 and 11). The following supershifting antibodies were added to the indicated lanes: p50 (lanes 4 and 12), p65 (lanes 5 and 13), c-Rel (lanes 6 and 14), RelB (lanes 7 and 15), p52 (lanes 8 and 16), and purified rabbit IgG (lanes 9 and 17). Arrows indicate NF-κB–related complexes, and supershifted complexes are denoted by asterisks.
Decreased c-Rel expression in pt1-LCLtet cells is not a function of transcriptional regulation
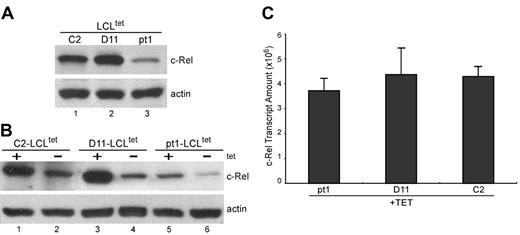
To determine whether the defect in c-Rel binding activity is related to decreased expression or aberrant nuclear translocation, pt1-LCLtet and control LCLtet cells were grown in the presence of tet, and c-Rel expression was detected in whole-cell lysate by Western analysis (Figure 4A). We observed a reduced amount of c-Rel in pt1 cells, although all other Rel/NF-κB subunits were expressed at equal levels, relative to control cells (data not shown). Examination of the nuclear fraction of cells grown in the absence of LMP1 (–tet) and then transferred to +tet media demonstrated that LMP1 signals were capable of regulating c-Rel nuclear translocalization, indicating no intrinsic defect in NF-κB activation (Figure 4B). Thus, low c-Rel activity in pt1-LCLtet cells is related to the absolute amount of c-Rel and not a deficiency in functional nuclear recruitment. To further analyze the basis of decreased c-Rel expression, we used qPCR with mRNA from cells grown in +tet media (Figure 4C). Surprisingly, the level of c-Rel mRNA in pt1 and control samples was approximately equal, suggesting that differences in c-Rel expression were mediated by translational and/or posttranslational processes, and not by specific variations in transcription or RNA stability.
Detection of reduced c-Rel protein expression that is not related to transcript levels. (A) Protein expression of c-Rel. Whole-cell lysate from LCLtet cells, cultured in the presence of tet, was analyzed by Western blot and probed with antibodies against c-Rel and actin, which served as an internal loading control. (B) Nuclear translocation of c-Rel with LMP1 stimulation. Nuclear extract from LCLtet cells, cultured in the presence (+tet) or absence (–tet) of LMP1 was also analyzed for c-Rel expression by immunoblot. (C) Relative transcript levels of c-Rel. RNA from pt1 and control LCLtet cells was reverse-transcribed and analyzed by qPCR with probes specific for c-Rel. Results are expressed as relative transcript amounts and standard deviation of 2 independent experiments.
Detection of reduced c-Rel protein expression that is not related to transcript levels. (A) Protein expression of c-Rel. Whole-cell lysate from LCLtet cells, cultured in the presence of tet, was analyzed by Western blot and probed with antibodies against c-Rel and actin, which served as an internal loading control. (B) Nuclear translocation of c-Rel with LMP1 stimulation. Nuclear extract from LCLtet cells, cultured in the presence (+tet) or absence (–tet) of LMP1 was also analyzed for c-Rel expression by immunoblot. (C) Relative transcript levels of c-Rel. RNA from pt1 and control LCLtet cells was reverse-transcribed and analyzed by qPCR with probes specific for c-Rel. Results are expressed as relative transcript amounts and standard deviation of 2 independent experiments.
Increased levels of c-Rel in pt1-LCLs expressing high levels of CD23
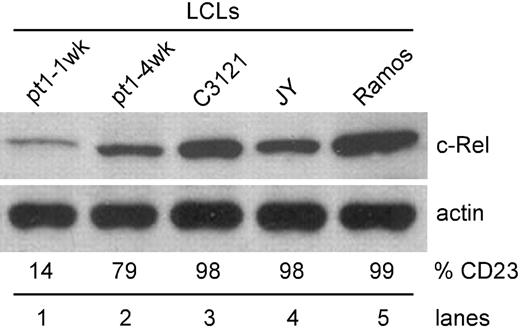
To ensure that the low c-Rel phenotype was not an artifact of the pt1-LCLtet line, but an intrinsic feature of the pt1 B cell, expression of c-Rel was analyzed in conventional EBV-transformed LCLs, immortalized from an independent sample of pt1 blood. By using these cells, we were able to assess c-Rel expression in cells that were newly immortalized (CD23lo/CD38hi) and in culture for 1 week (pt1-1wk) and cells that had acquired a lymphoblastic (CD23hi/CD38lo) phenotype upon extended growth in culture (pt1-4wk).18 As shown in Figure 5, newly immortalized pt1-LCLs (lane 1) express significantly less c-Rel than other CD23hi LCLs (lanes 3-4) or transformed B cell lines (lane 5). Also, as the pt1-LCLs were grown in culture and acquired a lymphoblastic phenotype, a population emerged in which CD23 was up-regulated, concurrent with an increase in c-Rel expression to levels significantly greater than newly immortalized LCLs (Figure 5, lane 2). Therefore, a defect in c-Rel expression appears to play a key role in maintaining the CD23lo phenotype in both the newly immortalized pt1-LCL and pt1-LCLtet cells.
Up-regulation of CD23 enhances c-Rel expression. Using established LCLs, whole-cell lysate was analyzed for c-Rel expression by Western blot. Pt1-1wk LCLs were cultured for 1 week and pt1-4wk LCLs, with a lymphoblastic phenotype, were cultured for 4 weeks prior to analysis. C3121 and JY are EBV positive, and Ramos is an EBV-negative B-cell line. Cell surface expression of CD23 (%CD23) was also examined by flow cytometry. Values represent percent positive cells.
Up-regulation of CD23 enhances c-Rel expression. Using established LCLs, whole-cell lysate was analyzed for c-Rel expression by Western blot. Pt1-1wk LCLs were cultured for 1 week and pt1-4wk LCLs, with a lymphoblastic phenotype, were cultured for 4 weeks prior to analysis. C3121 and JY are EBV positive, and Ramos is an EBV-negative B-cell line. Cell surface expression of CD23 (%CD23) was also examined by flow cytometry. Values represent percent positive cells.
Effects of c-Rel regulation on CD23 cell surface expression in LCLtet cells
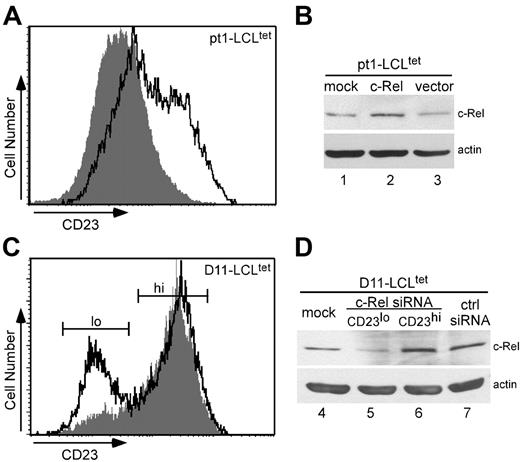
A series of studies was conducted to investigate the direct role of c-Rel in CD23 expression. Pt1-LCLtet cells were grown in the presence of tet, transiently transfected with a human c-Rel expression plasmid, and subsequently analyzed for surface CD23 expression. Overexpression of c-Rel in pt1-LCLtet cells (Figure 6A, outlined peak) showed an approximate 30% increase in surface CD23 expression over cells transfected with the empty vector (shaded peak). These results were further confirmed by Western analysis, which revealed an increase in c-Rel protein levels in transfected pt1-LCLtet cells (Figure 6B, lane 2) in comparison with mock (lane 1) or empty vector (lane 3) transfectants.
To further assess the dependency of CD23 on c-Rel expression, control D11-LCLtet cells, which express high levels of CD23 (Figure 1A), were cultured in the presence of tet and transfected with siRNAs to c-Rel (siRNAc-Rel) or a scrambled control sequence (siRNActrl). After 24 hours, cells were evaluated for CD23 surface expression. Flow cytometry results illustrate a bimodal pattern for siRNAc-Rel-transfected D11-LCLtet cells (Figure 6C, outlined peak). Scatter plot (forward versus side cell scatter) analysis shows a heterogeneous group composed of 2 distinct viable populations corresponding to levels of CD23 expression, which were further cell sorted by flow cytometry (CD23lo and CD23hi, Figure 6C). Both populations were selected for histogram analysis. Unlike siRNAc-Rel-transfected cells, cells transfected with siRNActrl revealed a single CD23hi population (shaded peak). The predominant CD23hi peak of siRNAc-Rel-transfected cells resembles siRNActrl, whereas the smaller CD23lo peak is shifted leftward of the control. We confirmed successful siRNAc-Rel transfection by Western analysis (Figure 6D). Whole-cell lysate from mock-, siRNActrl-, and CD23hi siRNAc-Rel–transfected samples (Figure 6D, lanes 4, 7, and 6, respectively) showed no difference in expression, while CD23lo siRNAc-Rel transfectants (lane 5) expressed a lower level of c-Rel protein. Together these results reveal that modulation of c-Rel expression by overexpression and siRNA knockdown experiments directly regulate CD23 cell surface expression. The fact that we observed these results, even in the context of normal levels of other Rel/NF-κB subunits, greatly extends our understanding of the activation defects in pt1 B cells.
Discussion
Our previous work revealed a B-cell defect in a patient with undefined HIGM syndrome that is tightly linked to signaling through CD40 and characterized by aberrant CD23 expression.17
Further work revealed that pt1-LCLs initially expressed a CD23lo/CD38hi phenotype, yet over time, a predominant population of CD23hi/CD38lo cells with increased mitogenic activity emerged from the initial CD23lo population. However, using pt1-LCLtet cells expressing LMP1 under the control of a tet-inducible promoter revealed that CD23 surface expression did not increase throughout culturing even though proliferation was driven efficiently by LMP1 signals (Lu et al18 and K.T.L. and L.R.C., unpublished data, August 2004). The “unlinking” of these 2 processes in the context of LMP1 expression allowed us to study defective CD23 expression in cells that were competent for proliferation. Our results reveal that the expression of c-Rel is compromised in pt1-LCLtet cells and that this lowered expression is directly responsible for decreased CD23 transcription and surface expression. Furthermore, we have demonstrated that lowered c-Rel expression is not a consequence of reduced transcription, but likely results from a translational or posttranslational process. Thus, CD40-related B-cell dysfunction can be accounted for, in part, by a level of c-Rel expression that is below a threshold necessary for efficient cellular proliferation and activation.
c-Rel modulates CD23 expression. Transient transfection of pt1-LCLtet cells with a c-Rel expression plasmid (outlined peak) and an empty vector (shaded peak) as the negative control (A) or siRNAs for c-Rel (outlined peak) and a scrambled control sequence (shaded peak) (C). Cell surface expression of CD23 was analyzed by flow cytometry. Total cell lysate from the corresponding overexpression (B) or siRNA transfections, with the CD23lo and CD23hi siRNARel subpopulations cell sorted (D), were further examined for c-Rel expression by Western blot analysis, with actin as a loading control. Results are representative of 3 independent experiments.
c-Rel modulates CD23 expression. Transient transfection of pt1-LCLtet cells with a c-Rel expression plasmid (outlined peak) and an empty vector (shaded peak) as the negative control (A) or siRNAs for c-Rel (outlined peak) and a scrambled control sequence (shaded peak) (C). Cell surface expression of CD23 was analyzed by flow cytometry. Total cell lysate from the corresponding overexpression (B) or siRNA transfections, with the CD23lo and CD23hi siRNARel subpopulations cell sorted (D), were further examined for c-Rel expression by Western blot analysis, with actin as a loading control. Results are representative of 3 independent experiments.
The observed phenotypes of both the pt1 primary B cells and B-cell lines are consistent with what is known about the role of c-Rel in B-cell development, growth, and survival. The c-Rel protein is expressed throughout all stages of B-cell development, but its expression is highest in mature B cells (reviewed in Gilmore et al46 ). Homozygous deletion of the c-Rel gene leads to primary defects in mitogen- and antigen-induced proliferation and differentiation of B cells into antibody-expressing cells.34,47 The proliferation defect is due to arrested cell cycle progression and can be complemented by overexpression of both cyclin E and Bcl-xL.48 Cyclin E expression is induced by c-Rel–mediated activation of the cell cycle transcription factor E2F3a,49 and Bcl-xL is a direct target of activated c-Rel. Also, c-Rel–/– B cells are sensitized to several apoptotic stimuli.50-53 Accordingly, pt1-LCLtet cells have a higher spontaneous rate of apoptosis, in comparison with control cells, and express significantly lower levels of several NF-κB–related prosurvival genes including A1/Bfl-1, Mcl-1, and Bcl-xL (K.T.L. and L.R.C., unpublished data, March 2006). However, the pt1-LCLtet cells that are not undergoing apoptosis proceed normally through the cell cycle, supporting our previous observation of the reversal of cell cycle arrest by signals from LMP1 that do not depend on a threshold of c-Rel expression. Another plausible explanation includes the existence of a cellular or viral protein that is regulated by LMP1 and may bypass c-Rel activation and directly restore the ability of cells to proliferate. In support of this hypothesis are studies showing that IL-6 and IL-10 can partially alleviate the proliferation defect seen in c-Rel–deficient mice.54
The atypically low expression of c-Rel in pt1 B cells is unlinked to differential transcription and is likely related to a translational or posttranslational mechanism. This unexpected finding suggests a possible modification of components or processes in pt1-LCLtet cells associated with c-Rel regulation and turnover. The turnover of c-Rel has been shown to be regulated by the ubiquitin-proteasome (Ub-Pr)–mediated pathway, with the C-terminal sequence of c-Rel serving an important role as a target for ubiquitin conjugation and subsequent c-Rel degradation.55 In addition, c-Rel has recently been identified as a substrate for caspase-3, with cleavage sites also located in the C-terminal region.56 Phosphorylation of c-Rel increases sensitivity to both the Ub-Pr pathway and caspase-mediated cleavage. c-Rel is phosphorylated primarily on serine and threonine residues, and to a lesser extent on tyrosine residues.57-59 PEST sequences, regions enriched with serine/threonine phosphorylation sites that are recognized both by the proteasome and specific kinases, have been identified in the C-terminal region of c-Rel.60 Furthermore, mutagenizing a subset of serines revealed a critical role for these residues in the transactivating function of c-Rel.61,62 One possible scenario for the decreased level of c-Rel in pt1-LCLtet cells is that hypophosphorylation of c-Rel leads to degradation via the Ub-Pr or apoptotic pathways. Possible up-stream targets that may be affected in the pt1-LCLtet cells include (1) protein kinase C ζ (PKC ζ), recently identified as an activator of c-Rel in response to TNFα62 ; (2) NF-κB–inducing kinase (NIK), shown to phosphorylate c-Rel upon T-cell activation through CD3 and CD2863 ; and (3) the constitutively active serine/threonine protein phosphatase X (PPX; also called protein phosphatase 4 [PP4]), which associates with c-Rel to activate NF-κB–mediated transcription.64 We are currently analyzing these targets for a possible involvement in the pt1 phenotype.
Of importance, we observe no defect in c-Rel translocation into the nucleus upon LMP1 signaling, and therefore the signals involved with activation of c-Rel, in particular those mediated by PI3 kinase and Akt that are required for nuclear translocation, appear to be intact (reviewed in Gilmore et al46 ). Also, EBNA-2 can specifically up-regulate CD23 expression by binding to the transcription factor, CBF142,65 ; however, it seems clear from our results that this pathway is also contingent on c-Rel activity since in the presence of EBNA-2 we still observe decreased expression of CD23. In pt1-LCLtet cells, EBNA-2 is robustly expressed and is therefore not a factor in the observed low level of CD23 expression. It is noteworthy that the pt1-LCLtet cells show a strong dependency on c-Rel with regard to CD23 expression even though all other Rel/NF-κB family members are expressed at control levels. This observation supports others' findings that specific motifs are preferentially bound by particular Rel/NF-κB family members.66,67 The preferred binding sites for c-Rel and p65 homodimers are the same, yet c-Rel homodimers are capable of binding a broader range of recognition sequences relative to p65 homodimers.68 Also, it has been shown that direct interactions with specific coactivators regulate distinct target genes.69 Because c-Rel is differentially phosphorylated, interactions between unique coactivators and c-Rel–dependent target genes may be further mediated by posttranslational modifications.
Finally, since the major phenotype of pt1 is an absence of switched isotypes, it is noteworthy that c-Rel signals have been implicated in mediating CSR. NF-κB1/c-Rel knock-out mice show normal lymphopoiesis but severe impairment in humoral immunity characterized by low or absent serum Igs and the inability to initiate antigen-specific antibody responses.70,71 Moreover, B cells from patients having X-linked hyper-IgM syndrome with ectodermal dysplasia (XHM-ED) were found to express normal levels of c-Rel but were impaired in CD40-mediated c-Rel activation. In vitro–stimulated B cells produced normal levels of AID and germ-line transcripts but failed to undergo CSR in response to CD40L and IL-4.72 These studies support the premise that c-Rel signals are essential for CSR. Thus, there is strong precedent that HIGM can be caused by a lack of c-Rel activity. Since the defect in pt1 CSR appears to be a threshold defect that can be overcome with extended signaling through CD40, it will be of interest to determine if sustained signaling through CD40 results in the up-regulation of c-Rel in pt1 B cells.
In summary, we show that modulation of c-Rel levels is sufficient to regulate CD23 surface expression. Accordingly, the pt1 defect results in a reduced level of c-Rel that directly affects CD23 activation in pt1 B cells. We hypothesize that the c-Rel defect may be a secondary effect related to a mutation in an upstream factor that regulates c-Rel expression. Ultimately, the identification of c-Rel–related transcriptional coactivators essential for B-cell activation should offer insights into the molecular mechanisms by which the c-Rel deficiency underlies this unique form of HIGM.
Authorship
K.T.L. designed and performed research, analyzed data, and cowrote the paper; F.L.S. designed and performed research and analyzed data; R.L.D. performed research; C.S. is the supervising clinician involved in all work directly involving pt1; and L.R.C. designed research and cowrote the paper.
The authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, August 8, 2006; DOI 10.1182/blood-2006-03-008839.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Dr Céline Gélinas (Center for Advanced Biotechnology and Medicine, New Jersey) for her generosity in providing the pJDCMV19SV-hc-Rel plasmid and for critically evaluating the paper. Also, we acknowledge and thank Dr Seth Lederman (Columbia University) for the CD23a/b promoter constructs and Christina DeCoste (Princeton University) for assistance with the cell sorting. We are grateful to pt1 and her family for their continuing participation in this research. Finally, we acknowledge past and present members of the Covey laboratory for insightful criticism and continuous support.
This work was supported by a National Institutes of Health (NIH) grant (AI37081) and a Busch Biomedical Research Grant from Rutgers University to L.R.C. K.T.L. and R.L.D. were supported by fellowships from the New Jersey State Commission on Cancer Research and an NIH training fellowship (T32AI07403; principal investigator, Dr Sidney Pestka, Department of Molecular Genetics, Microbiology, and Immunology at the Robert Wood Johnson Medical School, University of Medicine and Dentistry of New Jersey), respectively.