Abstract
We previously reported that mice made deficient for the transcriptional factor USF2 fail to express hepcidin 1 and hepcidin 2 genes as a consequence of targeted disruption of the Usf2 gene lying just upstream in the locus. These mice developed an iron overload phenotype with excess iron deposition in parenchymal cells and decreased reticuloendothelial iron. At that time, although the role of USF2 was still confounding, we proposed for the first time the role of hepcidin as a negative regulator of iron absorption and iron release from macrophages. Accordingly, we subsequently demonstrated that hyperexpression of hepcidin 1, but not hepcidin 2, resulted in a profound hyposideremic anemia. To analyze the consequences of hepcidin 1 deletion on iron metabolism without any disturbance due to USF2 deficiency, we disrupted the hepcidin 1 gene by targeting almost all the coding region. Confirming our prior results, Hepc1–/– mice developed early and severe multivisceral iron overload, with sparing of the spleen macrophages, and demonstrated increased serum iron and ferritin levels as compared with their controls.
Introduction
Hepcidin is a small circulating 25-amino-acid cysteine-rich peptide first identified in human blood1 and urine.2 Hepcidin is the product of the HAMP gene, which consists of 3 exons and encodes a precursor protein of 84 aa (for a review see Nicolas et al3 and Ganz4 ). The hepcidin gene is expressed in the hepatocytes, secreted in the circulation, and cleared by the kidney. Whereas humans, rats, dogs, and pigs have a single HAMP gene, there are 2 duplicated genes in mice, Hepc1 and Hepc2.5,6 In mammals, convincing evidence indicates that hepcidin constitutes the master regulator of iron homeostasis; the circulating peptide acts to limit gastrointestinal iron absorption and serum iron by inhibiting dietary intestinal iron absorption and iron recycling by the macrophages.7 As befits an iron-regulatory hormone, hepcidin synthesis is induced by iron stores and inflammation and inhibited by anemia and hypoxia.5,8-10 Most of the iron overload syndromes known to date (primary hemochromatosis and secondary iron overloads) imply a reduction of hepcidin secretion. In contrast, hypersecretion of hepcidin seems to play a determining role in anemia of inflammation (for a review see Ganz11 ). Four years after the discovery of the peptide, its mechanism of action was elucidated.12 To limit iron egress, hepcidin binds to ferroportin, the transmembrane iron transporter necessary for iron transfer out of intestinal epithelial cells and macrophages,13 thereby inducing its internalization and subsequent degradation, leading to decreased export of cellular iron.12,14,15
Much of the data concerning the involvement of hepcidin in iron metabolism were initially generated in mouse models. We previously reported that mice made deficient for the transcriptional factor USF2 fail to express either the Hepc1 or Hepc2 gene, as a consequence of targeted disruption of the Usf2 gene lying just upstream of the hepcidin genes.7 These mice presented with increased liver iron levels and developed an iron overload phenotype similar to that observed in hereditary hemochromatosis, with increased circulating iron, increased transferrin saturation, and decreased reticuloendothelial iron. We assume that the phenotype was not due to USF2 deficiency since an independent Usf2 knockout (KO) line expressed a normal amount of hepcidin mRNA and had normal iron metabolism.16 We thus proposed the role of hepcidin as a negative regulator of iron absorption and iron release from macrophages. This hypothesis was further supported by our demonstration that transgenic mice overexpressing Hepc1 were born with severe iron deficiency.16 Interestingly, we recently showed that transgenic mice overexpressing Hepc2 presented with normal iron metabolism, suggesting that only Hepc1 is able to regulate iron homeostasis in mice.6
To analyze the consequences of Hepc1 deletion on iron metabolism without any disturbance due to USF2 deficiency (only 10% of Usf2–/– mice survive), we decided to disrupt the Hepc1 gene by targeting deletion of exons 1 and 2 by classic homologous recombination. Homozygous mice presented with normal viability and developed multivisceral iron overload, with sparing of the spleen macrophages. The mutant mice also demonstrated increased serum iron and ferritin levels as compared with their controls. The Hepc1 KO mouse model will facilitate investigation into the pathogenesis of iron overload in hemochromatosis and provide opportunities to evaluate therapeutic strategies for prevention or correction of iron overload.
Study design
Targeted disruption of the murine Hepc1 gene
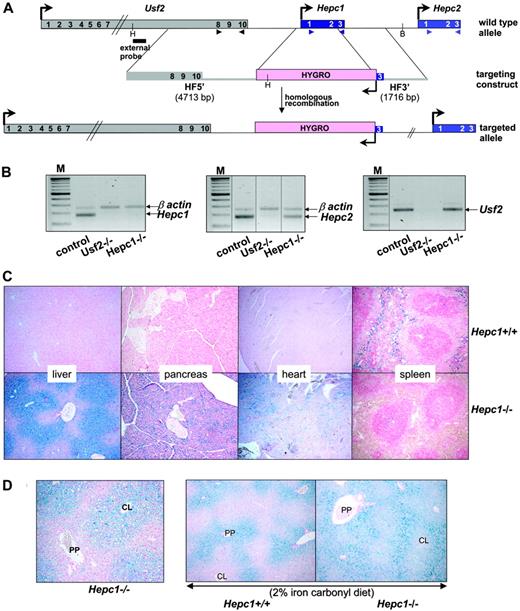
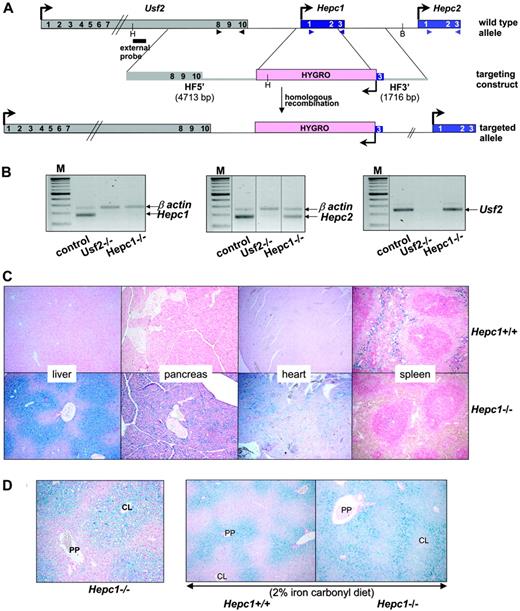
The gene-targeting vector was constructed by replacement of the 2 first exons and part of exon 3 of the Hepc1 gene with a hygromycin cassette under the control of the PGK promoter. This cassette is flanked in the 5′ direction by a 4.7-kb homologous arm and in the 3′ direction by a 1.7-kb homologous arm. Isogenic homologous DNA arms were obtained by long-distance genomic polymerase chain reaction (PCR; Expand Long Template PCR System; Roche, Mannheim, Germany) using mouse 129/Sv genomic DNA as template and subcloned. Primers for PCR-made fragments were as follows: for the 5′ homologous fragment (HF), forward 5′-CGGGGTACCTGATCAGAGTACCAGCCAGGACAGAGCC-3′ and reverse 5′-CGGGGTACCTGGCTGTCTAGGAGCCAGTCCC-3′ and for the 3′ HF, forward 5′-ATTTGCGGCCGCTGCTGTAACAATTCCCAGTG-3′ and reverse 5′-ATTTGCGGCCGCTGATCAGCTAGAAATCAAGAGGCCCTGG-3′. The 2 PCR-amplified homologous fragments were sequenced to confirm sequence fidelity. Details of vector construction are available on request. Linearized vector (25 μg) was electroporated into CK35 embryonic stem (ES) cells.17 Resistant clones were subjected to recombinant selection by PCR and correct recombination was further confirmed by Southern blot analysis. ES-cell clones with normal karyotypes were injected into C57BL/6 blastocysts and the obtained chimeric males were bred with C57BL/6 females to produce outbred F1 offsprings carrying the modified Hepc1 allele. Subsequent genotyping was carried out by PCR analysis on DNA extracted from tail samples. Detailed information for genotyping is available on request. All the studies were carried out on F1 hybrids on a mixed C57BL/6 × 129 background.
Animals
All animals were cared for in accordance with the European convention for the protection of laboratory animals. Animals were maintained in a temperature- and light-controlled environment and were given free access to tap water and food (standard laboratory mouse chow, AO3, iron content 280 mg/kg; Usine d'Alimentation Rationelle [UAR], Epinay-sur-Orsay, France). Iron overload was induced by adding 2% carbonyl iron (reduced pentacarbonyl iron) to the diet (AO3; UAR) for 14 days.
Hematologic analysis of mice
Blood was obtained by retro-orbital phlebotomy before killing of mice and collected in heparinized tubes (capiject T-MLH; Laboratoires Terumo, Guyancourt, France). Blood-cell counts and erythrocyte parameters were determined using a MaxM coulter automatic analyzer (Coulter, Hialeah, FL).
Reverse transcription and reverse transcriptase (RT)–PCR
Total RNA and double-stranded cDNA were prepared as previously described.7 PCR conditions were 25 cycles of denaturation at 94°C for 30 seconds, annealing at 50°C for 30 seconds, and primer extension at 72°C for 30 seconds. Following PCR, the amplified products (171 bp for Hepc1 and Hepc2 and 250 bp for β-actin and Usf2) were separated by electrophoresis on 1.5% agarose gel. Sequences of the primers were as follows: Hepc1, 5′-CCTATCTCCATCAACAGATG-3′ (forward) and 5′-AACAGATACCACACTGGGAA-3′ (reverse); Hepc2,5′-CCTATCTCCAGCAACAGATG-3′ (forward) and 5′-AACAGATACCACAGGAGGGT-3′ (reverse); β-actin, 5′-AGCCATGTACGTAGCCATCC-3′ (forward) and 5′-TTTGATGTCACGCACGATTT-3′ (reverse). For Usf2, the PCR conditions were the same except the annealing temperature was 55°C. The primers used were as follows: 5′-TCATGCAGACAACAGCAAGACA-3′ (forward) and 5′-TCACTGCCGGGTACTCTCGC-3′ (reverse).
Iron measurements and immunohistochemistry
Serum and tissue iron concentrations were determined as previously described18 using the “IL test” (Instrumentation Laboratory, Lexington, MA). For histology, tissues were fixed in 4% formaldehyde and embedded in paraffin. Sections were immersed in Perls solution (1:1, 2% HCl and 2% potassium ferrocyanide) to visualize ferric (non-heme) iron and counterstained with nuclear fast red using standard procedures. Quantification of serum ferritin levels was performed on an Olympus AU400 automat, using the Olympus human ferritin assay kit (Olympus, Hamburg, Germany).
Results and discussion
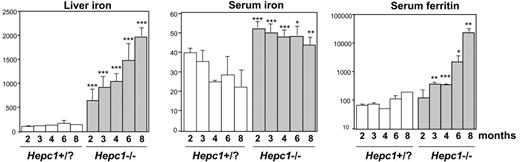
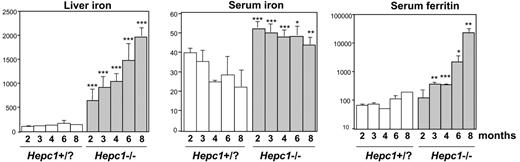
Targeted disruption of Hepc1 results in the same iron disorders as those observed in Usf2 KO mice. Targeted disruption of Hepc1 was obtained by replacing exons 1 and 2 and part of exon 3 with an hygromycin resistance cassette using homologous recombination in ES cells. Correctly targeted clones were identified by Southern blot analysis (using the indicated 5′ probe, Figure 1A) and PCR analysis (data not shown). Chimeric animals were bred with C57BL/6 females to produce inbred F1 offspring carrying the modified Hepc1 allele. Germline transmission of the targeted allele was confirmed by PCR analysis (not shown). We interbred Hepc1+/– animals to produce Hepc1–/– offspring, and obtained approximately 25% Hepc1–/– animals, indicating that there was no significant prenatal lethality. To assess the specificity of the Hepc1 targeting deletion, we checked by RT-PCR the absence of Hepc1 transcript in the liver of Hepc1–/– animals (Figure 1B). Furthermore, we showed that, in contrast to the inhibition of Hepc1 and Hepc2 gene transcription observed in the Usf2–/– mice, the expression of the neighboring genes of Hepc1, namely Hepc2 and Usf2, were still expressed in Hepc1–/– mice (Figure 1B). The mutant mice developed normally, showed no evidence of abnormalities, and both Hepc1–/– females and males appeared fertile. The only visible phenotype was a statistical loss of body weight beginning at the age of 8 months (not shown). The hematologic parameters of the mutant mice were followed through development (red blood cell, hemoglobin, mean cell volume, and hematocrit); although they were slightly increased in the first months, all the indices returned to normal value at the age of 6 months (not shown). Tissue iron accumulation was assessed by Perls staining. As shown in Figure 1C, massive iron accumulation was present in the livers and pancreata of 4-month-old Hepc1–/– mice and significant iron was also detected in the hearts. In contrast, splenic iron content was markedly decreased in Hepc1–/– mice as compared with wild-type mice. Excess non-heme iron was also present in kidney, lung, muscle, and brain (data not shown). Further examination of liver iron accumulation showed that iron accumulated in liver parenchymal cells (hepatocytes), with sparing of the resident macrophages (Kupffer cells). Surprisingly, iron deposition was found more prominent in the centrolobular areas (Figure 1D, left panel). To determine whether this iron deposition was due to an intrinsic incapacity of Hepc1–/– mice to load iron in the periportal region, control and mutant animals were given a 2% iron carbonylrich diet for 14 days. It is indeed well documented that dietary carbonyl iron overload is associated with specific iron deposition in the periportal hepatocytes. This periportal staining was evidenced in the experimentally iron-loaded control mice (Figure 1D) as well as in the Hepc1–/– mice, leading to a panlobular staining of the hepatic lobule of the mutant mice. We then examined the age-dependent variation of hematologic and iron parameters in control and Hepc1–/– mice from 2 to 8 months of age. Controls consist of both Hepc1+/+ and Hepc1+/– since no significant difference was observed between these animals. As shown in Figure 2, liver iron content was already 5-fold increased in 2-month-old mutant mice and continued to increase over the duration of the study. At 8 months, Hepc1–/– mice had almost 15-fold more liver iron than control mice. At that age, mutant mice presented with a 30-fold increase of iron in the pancreas (2056 ± 85 μg iron/g of wet tissue in Hepc1–/– mice, n = 3, versus 66 ± 6 in control mice, n = 4; P < 10–7), a 5.5-fold increase in the heart (513 ± 145 μg iron/g of wet tissue in Hepc1–/– mice, n = 3, versus 92 ± 30 in control mice, n = 4; P = .002) whereas splenic iron was decreased by almost 3-fold (273 ± 30 μg iron/g of wet tissue in Hepc1–/– mice, n = 3, versus 784 ± 23 in control mice, n = 4; P < .001). Serum ferritin levels increased in parallel to the liver iron increase. In contrast, serum iron increased at 2 months and remained constant during the following months.
Generation of the Hepc1 knockout mice and phenotypic exploration. (A) Schematic representation of the targeting strategy. The structure of the Usf2/Hepc1/Hepc2 locus is shown at the top,6 the targeting construct in the middle and the resulting targeted allele at the bottom. The genes are represented by colored rectangles and the arrows represent the direction of the transcription. Restriction sites for HindIII (H) and BglII (B), used for Southern blot genotyping, and probe location are indicated. The arrowheads indicate the location of the primers designed for the RT-PCR (B). The scheme is not drawn to scale. HF indicates homologous fragments. (B) Level of specific Hepc1, Hepc2, and Usf2 transcripts in the livers of controls, Usf2–/– and Hepc1–/– mice age 6 to 8 months, as measured by RT-PCR. Following PCR, the amplified products (171 bp for Hepc1 and Hepc2 and 250 bp for β-actin and Usf2) were separated by electrophoresis on 1.5% agarose gel. M indicates 100-bp DNA ladders. The vertical dotted lines indicate assembly of noncontiguous lanes. (C) Perls staining of liver, pancreas, heart, and spleen sections. Typical liver, pancreas, and spleen sections (original magnification ×10) from Hepc1+/+ and Hepc1–/– mice age 4 months. Non-heme iron stains blue. (D) Perls staining of liver sections (original magnification ×10). Left, typical liver section of a 2-month-old Hepc1–/– animal. CL, centrolobular; PP, periportal. Right, liver sections from 2-month-old Hepc1–/– and Hepc1–/– animals submitted to an iron-rich diet for 14 days.
Generation of the Hepc1 knockout mice and phenotypic exploration. (A) Schematic representation of the targeting strategy. The structure of the Usf2/Hepc1/Hepc2 locus is shown at the top,6 the targeting construct in the middle and the resulting targeted allele at the bottom. The genes are represented by colored rectangles and the arrows represent the direction of the transcription. Restriction sites for HindIII (H) and BglII (B), used for Southern blot genotyping, and probe location are indicated. The arrowheads indicate the location of the primers designed for the RT-PCR (B). The scheme is not drawn to scale. HF indicates homologous fragments. (B) Level of specific Hepc1, Hepc2, and Usf2 transcripts in the livers of controls, Usf2–/– and Hepc1–/– mice age 6 to 8 months, as measured by RT-PCR. Following PCR, the amplified products (171 bp for Hepc1 and Hepc2 and 250 bp for β-actin and Usf2) were separated by electrophoresis on 1.5% agarose gel. M indicates 100-bp DNA ladders. The vertical dotted lines indicate assembly of noncontiguous lanes. (C) Perls staining of liver, pancreas, heart, and spleen sections. Typical liver, pancreas, and spleen sections (original magnification ×10) from Hepc1+/+ and Hepc1–/– mice age 4 months. Non-heme iron stains blue. (D) Perls staining of liver sections (original magnification ×10). Left, typical liver section of a 2-month-old Hepc1–/– animal. CL, centrolobular; PP, periportal. Right, liver sections from 2-month-old Hepc1–/– and Hepc1–/– animals submitted to an iron-rich diet for 14 days.
Age-dependent examination of hematologic and iron parameters. Liver iron (μg iron/g wet tissue), serum iron (μM), and ferritin (ng/mL) were determined in control (Hepc1+/+ and Hepc1+/– mice) and Hepc1–/– mice between 2 and 8 months of age. Statistical analysis was performed using Student t test (unpaired, 2-tailed). *P < .05; **P < .01; ***P < .001 as compared with sex- and age-matched animal; there were at least 5 animals in each group.
Age-dependent examination of hematologic and iron parameters. Liver iron (μg iron/g wet tissue), serum iron (μM), and ferritin (ng/mL) were determined in control (Hepc1+/+ and Hepc1+/– mice) and Hepc1–/– mice between 2 and 8 months of age. Statistical analysis was performed using Student t test (unpaired, 2-tailed). *P < .05; **P < .01; ***P < .001 as compared with sex- and age-matched animal; there were at least 5 animals in each group.
Collectively, these features of iron disorders observed in Hepc1–/– mice appeared very similar to those described in the Usf2–/–mice. They confirm our first hypothesis that iron deregulation of the Usf2–/–mice was due to the suppression of Hepc1 gene expression, and highlight the nonredundant role of Hepc1 and Hepc2 in mice.
The phenotype reported here for the Hepc1–/–mice is very similar to the one recently reported for Hjv–/–mice that presented with a complete deficit in hepcidin production.19,20 These 2 mouse models are characteristic of the severe, early onset of human juvenile hemochromatosis caused by mutations in the hemojuvelin gene and, more rarely, in the hepcidin gene (for a review see Roetto and Camaschella21 ). The gradual accumulation of liver iron in the Hepc1–/–mice suggests that hepcidin is the final setpoint regulator of iron homeostasis and that, in its absence, there is no effective regulatory mechanism to decrease iron uptake. This result differs from that observed in the Hfe–/–mice, which do not completely lack hepcidin and still retain their ability to regulate intestinal iron absorption.22 The most intriguing result reported here, which seems in fact a unique feature of the Hepc–/–mice, is the centrolobular accumulation of liver iron. Indeed, when documented, the iron accumulation of the other forms of hemochromatosis, primary or secondary, appeared to be periportal. The specific zonation seen in Hepc1–/– mice is interesting but difficult to explain. In this respect, it will be interesting to analyze the specific zonation of the iron-related proteins in the liver, a still poorly investigated domain.
Prepublished online as Blood First Edition Paper, March 30, 2006; DOI 10.1182/blood-2006-02-003376.
J.-C.L.-B. and L.V. contributed equally to this work.
J.-C.L.-B. and L.V. designed and performed research, analyzed data, and wrote the paper; M.B., D.-Q.L. and G.R. performed experiments; C.H. and G.H. provided technical support for ES cells and homologous recombination; A.K. analyzed data; and S.V. designed research, analyzed data, and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.